Abstract
Aneuploidy, referring here to genome contents characterized by abnormal numbers of chromosomes, has been associated with developmental defects, cancer, and adaptive evolution in experimental organisms1–9. However, it remains unresolved how aneuploidy impacts gene expression and whether aneuploidy could directly bring phenotypic variation and improved fitness over that of euploid counterparts. In this work, we designed a novel scheme to generate, through random meiotic segregation, 38 stable and fully isogenic aneuploid yeast strains with distinct karyotypes and genome contents between 1N and 3N without involving any genetic selection. Through phenotypic profiling under various growth conditions or in the presence of a panel of chemotherapeutic or antifungal drugs, we found that aneuploid strains exhibited diverse growth phenotypes, and some aneuploid strains grew better than euploid control strains under conditions suboptimal for the latter. Using quantitative mass spectrometry-based proteomics, we show that the levels of protein expression largely scale with chromosome copy numbers, following the same trend observed for the transcriptome. These results provide strong evidence that aneuploidy directly impacts gene expression at both the transcriptome and proteome levels and can generate significant phenotypic variation that could bring about fitness gains under diverse conditions. Our findings suggest that the fitness ranking between euploid and aneuploid cells is context- and karyotype-dependent, providing the basis for the notion that aneuploidy can directly underlie phenotypic evolution and cellular adaptation.
Whole-chromosome or segmental aneuploidy has been observed in a wide range of organisms and conditions, from pathogenic and experimental fungal species adapting to growth inhibition, to human diseases such as cancer and Down Syndrome2–10, but how aneuploidy affects gene expression and cellular physiology remains unclear1,11–13. The budding yeast Saccharomyces cerevisiae, with its 16-chromosome complement has been a useful experimental model for addressing this question at a fundamental level. Experimental evolution in yeast suggested a correlation between the emergence of aneuploidy and adaptive phenotypes in response to various perturbations3–7,10. Transcriptome profiling demonstrated that aneuploidy causes changes in mRNA levels mostly scaling with chromosome copy numbers and well beyond for some genes7,10,11. On the other hand, two recent studies concluded that aneuploidy reduces cellular fitness irrespective of the specific karyotype11,12 and suggested that dosage compensation for proteins encoded on aneuploid chromosomes correlates with a common stress response, dubbed “proteotoxic stress”11. This raises the following conundrum: If net protein expression levels are insensitive to chromosome stoichiometry and aneuploidy inevitably impairs fitness, how might aneuploidy provide phenotypic variation and possibly fitness advantages under selective conditions?13 We aimed to resolve this conundrum using a panel of aneuploid yeast strains with a wide range of karyotypes grown under diverse conditions. In particular, our experiments were designed to answer these questions: 1) Can aneuploidy directly confer phenotypic variation and possibly improved fitness? And 2) is the proteome proportionally affected by chromosome copy number variation due to aneuploidy?
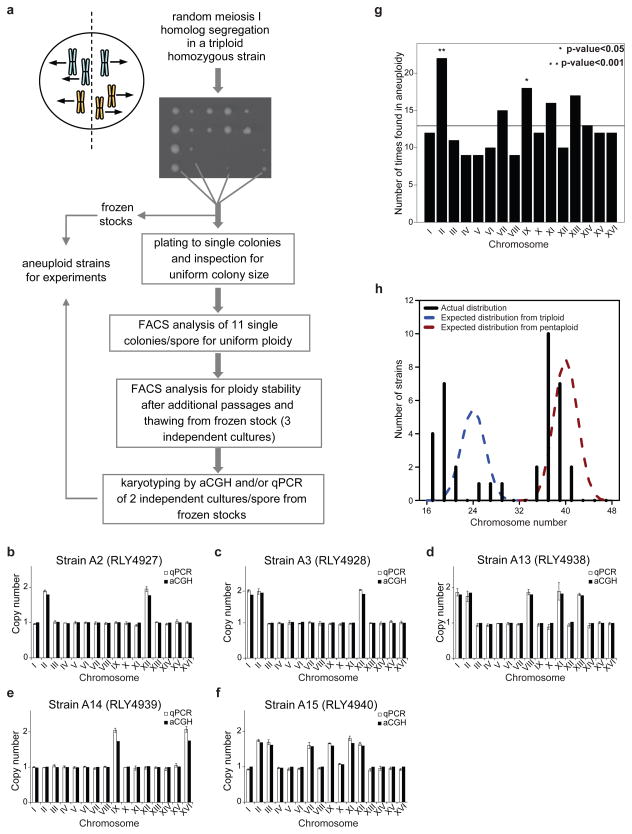
To generate fully isogenic and stable aneuploid strains, containing a wide range of chromosome stoichiometries and without the need of any genetic selection, we induced meiosis in yeast strains with an odd ploidy (3N or 5N), which produces aneuploid progenies at high frequencies14,15 (Fig. 1a). To minimize other genetic variation, we generated the starting triploid and pentaploid progenitor strains by cycles of mating type switching and mating, from a single haploid S288c strain (Supplementary Fig. 1a). Absence of segmental chromosome abnormalities in the resulting polyploid progenitors was verified by array-based comparative genomic hybridization (aCGH), although the pentaploid strain had quickly lost one copy of chromosomes III and V (Supplementary Fig. 1b). To isolate aneuploid strains with stable karyotypes, we used a multi-step approach (detailed in Supplementary Methods). Briefly, each spore was first spread on a YEPD plate to form single colonies (Fig. 1a). From each resulting plate displaying uniform colony sizes eleven colonies were randomly picked and analyzed by fluorescence activated cell sorting (FACS) to identify those original spores producing colonies with uniform ploidy (Supplementary Fig. 2a,b). Karyotype stability was further verified after freezing and revival (Supplementary Fig. 2c). The karyotypes of the final aneuploid strains were determined by a novel quantitative polymerase chain reaction (qPCR)-based assay that allows accurate karyotyping in high-throughput formats (Supplementary Methods; Fig. 1b–f; Supplementary Fig. 2d). A subset of the karyotypes was also confirmed by aCGH (Fig. 1b–f; Supplementary Fig. 3). To minimize accumulation of single nucleotide mutations, all aneuploid strains were passaged no more than three times between initial derivation and experimental usage. Indeed, whole genome re-sequencing of the five aneuploid strains used for the transcriptome and proteome analyses (see below) revealed absence of mutations in coding regions that were not already present in the parental strains (Supplementary Information). The above procedure yielded 38 isogenic stable aneuploid strains (12.5% of spores analyzed) with ploidy between 1N and 3N, harboring 35 distinct karyotypes, mostly with multiple chromosomes in aneuploidy (Supplementary Fig. 4). Except for chromosomes II and IX, all 16 yeast chromosomes were equally represented as aneuploid chromosomes in this collection (Fig. 1g). The chromosome number distributions were skewed toward the left (lower number) from those expected from random meiosis I segregation (Fig. 1h), suggesting that strains with larger numbers of aneuploid chromosomes are either less viable or karyotypically unstable.
Figure 1. Generation of aneuploid yeast strains.
(a) Sporulation of a homozygous triploid strain followed by karyotype stability tests of the meiotic progenies. (b–f) Karyotypes of the five aneuploid strains used in Fig. 3, determined by qPCR (white bars; mean ± s.d.) and aCGH (black bars). (g) Distribution of aneuploid chromosomes; p-values were calculated from a binomial distribution; the horizontal line represents the expectation number assuming uniform representation (h) Karyotype (total chromosome number) distribution across the aneuploid strain collection. Black lines: observed distribution (binned every two chromosomes); blue and red dashed lines: expected binomial distributions from random homolog segregation during triploid and pentaploid sporulation, respectively.
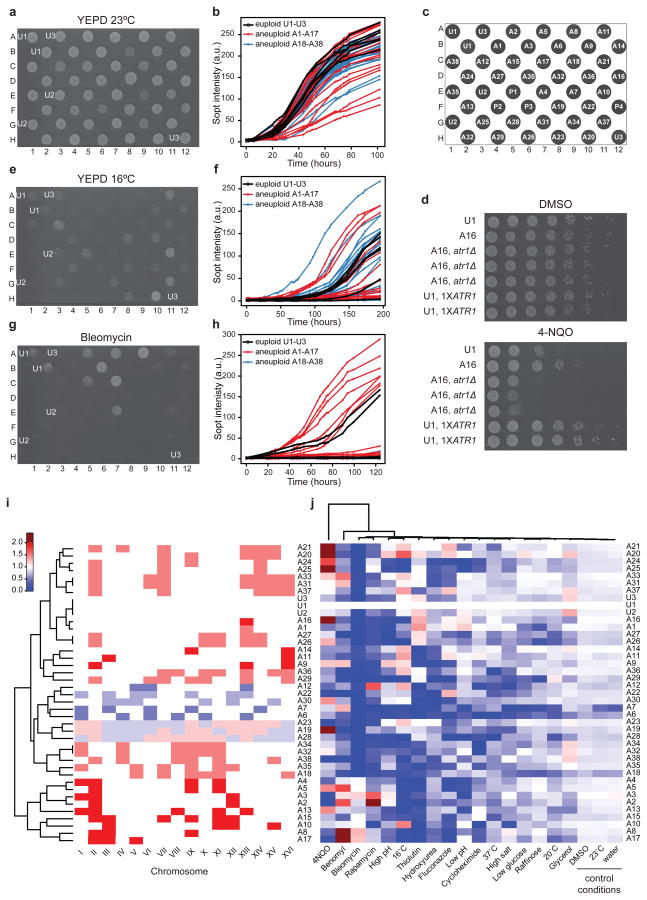
Growth comparison of the aneuploid strains with isogenic 1N, 2N and 3N euploid strains in rich media at 23 °C confirmed the previous observation that most aneuploid strains grew poorly compared with euploid controls11, although a few aneuploid strains grew similarly to the euploids (Fig. 2a–c). Next we compared the growth under conditions divergent from that optimal for euploid yeast cells, including environmental perturbations, such as extreme temperature or pH or nutrient shortage, and the presence of chemotherapeutic or antifungal drugs. Strikingly, while under every condition, most aneuploid strains grew slower than or as poorly as the euploids, under several conditions, especially those severely retarding euploid growth, some aneuploid strains showed improved growth in respect to euploids (Fig. 2e–h; Supplementary Fig. 5). Karyotyping at the end of the growth assays confirmed persistent karyotype stability in most cases (Supplementary Information). For example, several aneuploid strains grew significantly better than euploid strains in rich media at 16 °C (Fig. 2e, f), or in the presence of drugs such as rapamycin (an immunosuppressant and proposed anticancer agent), bleomycin (a chemotherapeutic compound), thiolutin (an antibiotic) or fluconazole (an antifungal drug) at concentrations inhibitory to euploid growth (Fig. 2g, h; Supplementary Fig. 5; Supplementary Table 3). The observed phenotypic diversity was unlikely to be due to differences in mating type, as euploid strains with different mating types grew similarly under the tested conditions (Supplementary Fig. 6).
Figure 2. Phenotypic profiling of aneuploid strains.
(a–b, e–h) Representative images (left) and growth curves (right) under indicated conditions. U1–U3: haploid, diploid and triploid euploid control strains, respectively. (c) Strain positions. A1–A38: aneuploid strains (see Supplementary Figure 4 for their karyotype). P1–4: four petite strains not further studied. (d) One-copy number increase of ATR1 is required and sufficient to confer resistance to 0.4μg/ml 4-NQO. (i) Clustering of strains based on karyotypic similarity. White: euploid chromosome number; red: gain over euploid number; blue: chromosome loss. (j) Clustering of conditions used in phenotypic profiling based on the fitness relative to U1. White: growth similar to U1; red: fitness gain over U1; blue: fitness loss. The strains were ordered as in (i). Scale bar applies to both (i) and (j). Analysis details in Supplementary Methods.
To understand how phenotypes relate to karyotypes, we clustered the aneuploid strains based on karyotypic similarity (Fig. 2i) and clustered the conditions used in the phenotypic profiling based on their effects on growth (Fig. 2j; Supplementary Methods). This analysis revealed that several pairs of aneuploid strains with identical (e.g. A26 and A27; A31 and A33) or similar karyotypes (e.g. A8 and A17; A1 and A16; A20 and A21) exhibited similar growth patterns across the different conditions. Second, along each growth condition (column in Fig. 2j), divergent karyotypes can be observed that exhibited improved growth compared to the haploid control under certain conditions. To pinpoint a specific mechanism linking a specific aneuploid karyotype to a specific fitness improvement, we noticed that strain A16, resistant to 4-nitroquinoline-N-oxide (4-NQO, a tumorigenic compound), had only chromosome XIII in aneuploidy. Chromosome XIII harbors the gene ATR1, encoding a transporter protein known to confer 4-NQO hyper-resistance when overexpressed16. We confirmed that an extra copy of chromosome XIII in strain A16 led to a proportional increase in mRNA expression of ATR1 both in presence and absence of 4-NQO (Supplementary Fig. 7a). Deletion of the extra copy of ATR1 from the 4-NQO resistant aneuploid strains restored expression of ATR1 to levels comparable to euploid (Supplementary Fig. 7a) and abolished their resistance to 4-NQO (Fig. 2d; Supplementary Fig. 7b–c), demonstrating that the increased ATR1 copy number was required for 4-NQO resistance. Furthermore, introducing one extra copy of the ATR1 gene expressed under its own promoter into haploid or diploid euploid strains was sufficient to confer resistance to 4-NQO (Fig. 2d; Supplementary Fig. 7b–c).
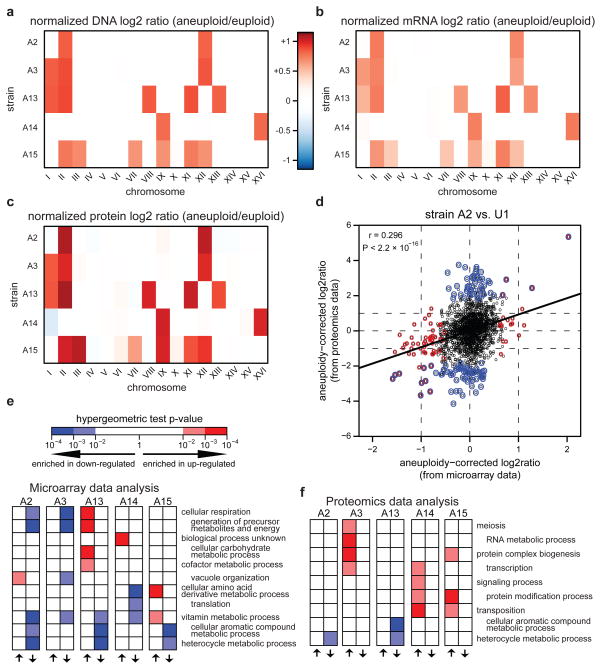
Immunoblot analysis of disomic yeast strains suggested that most proteins encoded on aneuploid chromosomes may be dosage compensated11, raising a question as to how aneuploidy might confer large phenotypic variation, as observed above. To investigate this further, we performed parallel RNA microarray and quantitative proteomics analysis on five aneuploid strains of the same mating type and exhibiting different growth rates and chromosome stoichiometries. Multidimensional Protein Identification Technology (MudPIT) analysis17 on the soluble fraction of whole-cell extracts identified ~2,000 different gene products per strain, representing ~33% of the yeast proteome with highly reproducible quantification of protein abundances across the biological replicates and broad coverage of all cellular components (Supplementary Fig. 8 and Supplementary Table 4). This analysis revealed that strains with similar karyotypes tend to have similar global proteomic changes (Supplementary Fig. 8a) and that changes in chromosome copy numbers due to aneuploidy lead to proportional changes in the chromosomal average protein expression following the same trend as the transcriptome (Fig. 3a–c; Supplementary Figs 9 and 10a), indicating a direct gene dosage effect on the proteome. Dosage compensation was also minimal and in most cases insignificant for core complex proteins18 encoded on aneuploid chromosomes (Supplementary Fig. 10b).
Figure 3. Effects of aneuploidy on the proteome.
(a–c) Heat maps of chromosome stoichiometry (a, aCGH data, Fig. 1b–f), average mRNA level (b, microarray data) and average protein level (c, proteomics data; see Supplementary Fig. 9) per chromosome of the five aneuploid strains compared to U1. (d) A correlation between protein expression and gene expression changes relative to haploid euploid strain U1 (see Supplementary Fig. 12). Outlier mRNAs and proteins (defined as in Supplementary Information) are highlighted in red and blue, respectively. (e–f) Subset of GO-Slim analysis applied to outlier genes from microarray (e) and proteomics (f) datasets (see Supplementary Methods for details). Complete results in Supplementary Fig. 12a–b. P-values were calculated from hypergeometric tests.
The mRNA and protein levels of individual genes were modestly though significantly correlated even after correcting for chromosomal copy number effect (Fig. 3d; Supplementary Fig. 11), consistent with a recent proteomic analysis of haploid versus diploid yeast19. However, among the genes with expression changes two standard deviations away from their chromosomal average (referred to as ‘outlier’ genes), only a fraction (~3–14%) were common between the microarray and the proteomics datasets (Fig. 3d; Supplementary Fig. 11). As a result, mRNA outliers and protein outliers were enriched for distinct classes of biological processes, and no specific class of genes was consistently found to be significantly enriched across all aneuploid strains (Fig. 3e–f; Supplementary Fig. 12a–b), suggesting a lack of a common gene expression response to aneuploidy. Because genes of the “response to stress” category was enriched in neither the transcriptome nor the proteome in any of our aneuploid strains, in contrast to the conclusion of the previous study11, we further performed a more stringent analysis by only considering outlier genes expressed more than three standard deviations from the chromosomal average in our microarray data. Enrichment for either “response to stress” (Gene Ontology) or “Environmental Stress Response” genes20 was found in three of the five aneuploid strains, but interestingly this enrichment correlated with neither growth rates nor the number of aneuploid chromosomes (Supplementary Fig. 12c). Taken together, the above results indicate that aneuploidy has global and complex effects at both the transcriptome and the proteome levels, and that an increase in stress gene expression is not a obligate property of aneuploid strains.
Taken together, our analysis of a large set of isogenic and stable aneuploid yeast strains with broad chromosome stoichiometries demonstrated that aneuploidy directly confers phenotypic variation and sometimes growth advantage under conditions sub-optimal for euploid cells. These observations suggest that aneuploidy does not inevitably result in growth impairment, but rather that the impact of aneuploidy on cellular fitness is both karyotype- and condition-dependent. The difference between our findings and the previous observation of a common stress signature and proliferative disadvantage across disomic yeast strains11 may be that in our study the naturally stable, multiple-chromosome aneuploidy resulted in less protein expression imbalances than single-chromosome aneuploidy maintained through continuous drug and nutrient marker selection. Furthermore, our proteomic analysis, performed in quadruplicates and quantifying thousands of proteins encoded on aneuploid chromosomes, in contrast to just 16 proteins analyzed by immuno-blotting in the previous work, revealed a whole-sale chromosome dosage effect on the proteome, consistent with a recent report of gene copy number effects on protein levels21. These findings suggest that aneuploidy is a large-effect mutation profoundly altering gene expression at the functional level. Under conditions to which euploid cells are well adapted, the large phenotypic effects caused by aneuploidy are likely to cause a reduction in fitness that could lead to rapid clearance of most aneuploid cells from the population. However, under a strong selective pressure due to adverse environmental changes or clinical drug treatments, the rise of aneuploidy, readily achieved through erroneous mitosis, can be a highly effective mechanism to generate phenotypic variation and rapid adaptation.
Methods summary
Generation of a collection of isogenic aneuploid yeast strains
Homozygous triploid and pentaploid strains were generated as described in Supplementary Fig. 1a. Aneuploid strains were generated by sporulation of the above polyploid strains, followed by karyotype stability tests and determination as described in Fig. 1a and Supplementary Fig. 2. Strains and plasmid are listed in Supplementary Information.
Karyotyping
aCGH, performed as previously described7, and qPCR were used for karyotyping. qPCR assays were designed with primers in non-coding regions on each chromosome arm (Supplementary Table 1 lists primer sequences). DNA samples were prepared by alkaline lysis, and qPCR reactions were performed in 384-well plates using a BioMek FX (Beckman Coulter) to assemble 10 μl reactions and an ABI 7900HT (Applied Biosystems) for cycling. Chromosome copy numbers were determined using a modified ΔΔCt method (Supplementary Methods).
Phenotypic profiling
Equal amounts (OD600) of aneuploid and euploid control cultures were spotted, using the Biomek FX robot, onto omnitrays containing various solid media and grown under conditions listed in Supplementary Table 2. Omnitrays representing three biological replicates of each tested condition were scanned on an HP ScanJet 4070 desktop scanner. Growth data was obtained by automated spot detection and intensity measurements.
Quantitative whole-genome proteomics
Whole-cell lysates were prepared from 50 ml cycling yeast cultures by bead-beating. High-speed supernatants were collected and precipitated. Chromatography and mass spectrometry analysis were performed as previously described22. The MS/MS datasets were searched using SEQUEST23 against a database of 11,986 sequences, consisting of 5,816 S. cerevisiae non-redundant proteins (NCBI), 177 contaminants and 5,993 decoy sequences. Relative protein levels were determined by calculating distributed Normalized Spectral Abundance Factors (dNSAFs)24.
Statistical analysis
All statistical analyses were performed in the R environment25 using standard packages and custom scripts.
Full Methods section is included within the Supplementary Information.
Supplementary Material
Acknowledgments
Authors thank C. W. Seidel for assistance with microarray data analysis, B. Fleharty and A. Peak for technical assistance with microarray hybridization, A. Perera and K. Walton for assistance in genome resequencing, W. McDowell for technical assistance with qPCR, J. Haug for technical support with flow cytometry experiments, G. Chen for technical suggestions and A. Paulson for assistance with submission of microarray and sequencing data to public repositories. This work was performed to fulfill, in part, requirements for J. Zhu’s PhD thesis research as a student registered with the Open University. This work was supported by NIH RO1GM059964 to R.L.
Footnotes
Author Contribution: N.P., G.R. and R.L. designed the study. N.P., G.R. and J. Z. performed all experiments. N.P. developed all custom R scripts. N.P., G.R., J.Z., W.D.B and B.W.S. set up the high-throughput qPCR method. W.D.B. performed all qPCR karyotyping assays. A.S. and L.F. performed mass spectrometry experiments. N.P., G.R., A.S. and L.F. analyzed proteomics data. N.P., G.R. and G.L.H. analyzed sequencing data. R.L. coordinated and supervised the project. N.P., G.R. and R.L. prepared figures and wrote the manuscript. All authors read and agreed with the paper content.
Author Information: Microarray data are deposited in ArrayExpress under accession numbers E-MTAB-318 and E-MTAB-325. Sequencing data are deposited in NCBI’s SRA database under accession number SRP003582. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to RL (rli@stowers.org).
Supplementary Information is linked to the online version of this paper at www.nature.com/nature
References
- 1.Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Selmecki A, Forche A, Berman J. Aneuploidy and Isochromosome Formation in Drug-Resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polakova S, et al. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc Natl Acad Sci U S A. 2009;106:2688–2693. doi: 10.1073/pnas.0809793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunham MJ, et al. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gresham D, et al. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rancati G, et al. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135:879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selmecki A, Gerami-Nejad M, Paulson C, Forche A, Berman J. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol Microbiol. 2008;68:624–641. doi: 10.1111/j.1365-2958.2008.06176.x. [DOI] [PubMed] [Google Scholar]
- 9.Selmecki AM, Dulmage K, Cowen LE, Anderson JB, Berman J. Acquisition of Aneuploidy Provides Increased Fitness during the Evolution of Antifungal Drug Resistance. PLoS Genet. 2009;5:e1000705. doi: 10.1371/journal.pgen.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes TR, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 11.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 12.Williams BR, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. 322/5902/703 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pavelka N, Rancati G, Li R. Dr Jekyll and Mr Hyde: role of aneuploidy in cellular adaptation and cancer. Curr Opin Cell Biol. 2010 doi: 10.1016/j.ceb.2010.06.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parry EM, Cox BS. The tolerance of aneuploidy in yeast. Genet Res. 1970;16:333–340. doi: 10.1017/s0016672300002597. [DOI] [PubMed] [Google Scholar]
- 15.St Charles J, Hamilton ML, Petes TD. Meiotic Chromosome Segregation in Triploid Strains of Saccharomyces cerevisiae. Genetics. 2010 doi: 10.1534/genetics.110.121533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mack M, et al. Genetic characterization of hyperresistance to formaldehyde and 4-nitroquinoline-N-oxide in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1988;211:260–265. doi: 10.1007/BF00330602. [DOI] [PubMed] [Google Scholar]
- 17.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 18.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 19.de Godoy LM, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 20.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Springer M, Weissman JS, Kirschner MW. A general lack of compensation for gene dosage in yeast. Mol Syst Biol. 2010;6:368. doi: 10.1038/msb.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Florens L, Washburn MP. Proteomic analysis by multidimensional protein identification technology. Methods Mol Biol. 2006;328:159–175. doi: 10.1385/1-59745-026-X:159. [DOI] [PubMed] [Google Scholar]
- 23.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal Chem. 2010;82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- 25.Ihaka R, Gentleman R. R: A Language for Data Analysis and Graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.