Abstract
Mycobacterium avium subsp hominissuis, hereafter referred to as M. avium, forms biofilm, a property that, in mice, is associated with lung infection via aerosol. As M. avium might co-inhabit the respiratory tract with other pathogens, treatment of the co-pathogen-associated infections, such as in bronchiectasis, would expose M. avium to therapeutic compounds which may have their origin in other organisms sharing the natural environments. Incubation of M. avium with two compounds produced by environmental organisms, streptomycin and tetracycline in vitro at sub-inhibitory concentrations increased biofilm formation in a number of M. avium strains, although exposure to ampicillin, moxifloxacin, rifampin, and TMP/SMX had no effect on biofilm. No selection of genotypically resistant clones was observed. While bacteria incubation in presence of streptomycin upregulates the expression of biofilm-associated genes, the response to the antibiotics had no association with a regulation of a regulator (LysR) linked to the formation of biofilm in M. avium.
Biofilms are made of planktonic and sessile bacteria. While planktonic M. avium is susceptible to clarithromycin and ethambutol (clinically used antimicrobials), sessile bacteria are at least 3- to 4-fold more resistant to antibiotics. The sessile phenotype, though, is reversible, and no selection of resistant clones was observed. Mice infected through the airway with both phenotypes were infected with a similar number of bacteria, demonstrating no phenotype advantage.
M. avium biofilm formation is enhanced by commonly used compounds and, in the sessile bacterial phenotype, is resistant to clarithromycin and ethambutol, in a reversible manner.
Keywords: M. avium, biofilm, gene expression, antibiotics
INTRODUCTION
Biofilm formation is an approach used by a number of organisms to survive under conditions of nutrient depletion [1]. Mycobacterium avium subsp hominissuis, hereafter referred to as M. avium, is an environmental bacterium which commonly causes infection in individuals with chronic lung infection and Acquired Immunodeficiency Syndrome (AIDS) [2–4]. One of the characteristics of M. avium is the ability to form biofilm in the environment [5]. Several genes have been linked with the ability to develop biofilm on both polyvinyl carbonate (PVC), as well as plastic surfaces. M. avium mutants deficient in those genes are impaired in biofilm formation in vitro [6]. More recently, it has been shown that M. avium lung infection in mice appears to be greatly dependent on the ability to colonize and establish biofilm on the surface of bronchial cells [6]. This finding offers, at least partially, a possible explanation for the challenge of treating M. avium lung infection [7, 8], which is frequently unsuccessful [9].
Although the presence of biofilm can be the rationale for many of the clinical aspects of the M. avium complex lung infection, other factors may participate in and influence the formation, maintenance of biofilm and the response to therapy. M. avium is an environmental organism that exists in the presence of other bacteria, both in the environment outside of the host, as well as in the host. For example, both in the soil and water, M. avium shares the habitat with many other organisms, potentially forming complex colonies. In individuals with chronic lung disease, many pathogens, such as Pseudomonas aeruginosa, Burkholderia cepacia, Staphylococcus aureus, and others, can be encountered co-habiting the airways. It is common that those patients have many episodes of infection for which antibiotics are often prescribed. There is a possibility that some of the environmental microbes that produce natural antibacterial molecules used as human therapy share the soil with M. avium. Assuming that M. avium would have been in contact with some of the same antibacterial molecule before, it is likely that the bacterium has developed defense mechanisms, which can certainly be triggered by the presence of the same or similar antimicrobial molecules found in the environment.
The biofilm structure is composed of at least two populations of microorganisms, the planktonic and sessile. Sessile bacteria compose the biofilm, and planktonic organisms occasionally detach (both pre- and post-biofilm phase). A number of studies looking at a large number of pathogens, including Mycobacterium abscessus, have established that planktonic and sessile subpopulations have different susceptibility to antibiotics [10]. Biofilm resistance to antibiotics has been attributed to at least two main reasons. One would be that biofilm creates a barrier against the penetration of antibiotics; while the other would be that bacteria in the biofilm are not in the replicating state and, therefore, are resistant to the majority of antibiotics.
In this study, we first examined whether several antibiotics, commonly used compounds to treat infections in patients with cystic fibrosis and other chronic lung diseases, have any effect on the ability of M. avium to form biofilm. In addition, we evaluated the susceptibility of planktonic and sessile subpopulations to antibiotics frequently used to treat lung M. avium infection.
MATERIALS AND METHODS
Bacteria
M. avium strains 101, 104 and A5 (kindly provided by Kathleen Eisenach, University of Arkansas, Little Rock) were used in the reported experiments. They were isolated from the blood of an AIDS patients. Strains 3362-33 and 3362-34 are M. avium strains isolated from the lung of individuals with pulmonary pathology. They were kindly provided by Dr. Richard Wallace (Tyler TX). Bacteria were grown on Middlebrook 7H10 agar supplemented with oleic acid, albumin, dextrose and catalase (OADC), for 10 days at 37°C, harvested and used to establish a suspension of 5 × 108 cfu/mL. The suspension was then used to seed a PVC 96-well plate, as described before [5, 11, 12]. The plate was kept at room temperature for 14 days. After this period, biofilm had developed on the surface of the walls. The biofilm impaired strains 5G4 (in which MAV_3209 hypothetical membrane protein has been inactivated) and 6H9 (inactivation of sucA) were cultured in agar plates in presence of 200 mg/L of kanamycin. The strains have been previously described [6, 12] and are transposon mutants of the strain A5. M. avium clone 8G12 was obtained by screening a transposon library for clones associated with increased ability of biofilm formation on PVC. The clone 8G12 was selected because it has the ability to form biofilm 8-fold greater (relative increase compared to the parent strain A5) than the wild-type strain A5. Sequencing of the inactivated gene determined that it is MAV_2151, a transcription regulator of the LysR family (data not shown).
Antibiotics
Streptomycin and tetracycline are molecules initially isolated from environmental fungi and bacteria. Ampicillin, rifampin, moxifloxacin and trimetropin/sulfametoxazol were also used. All the compounds, with one exception, were purchased from Sigma Chemicals (St. Louis MO). Moxifloxacin was obtained from Bayer, CT. Different concentrations were obtained by dilution in Hank’s balanced salt solution (HBSS). Clarithromycin was provided by Abbott Laboratories (Chicago IL) and ethambutol was purchased from Sigma.
Antibiotic susceptibility was performed with both planktonic and sessile phenotypes, as previously described using broth microdilution method [11]. Briefly, 1 × 105 bacteria were seeded in 0.3 mL of 7H9 Middlebrook broth with OADC in a 96-well microtiter plate (Falcon). The number of bacteria for the inoculum was established by using turbidity method and confirmed by plating onto 7H10 agar. Concentrations of clarithromycin and ethambutol were used from 0.125 mg/L to 128 mg/L.
Biofilm assays and phenotypes
For the assays, we used three strains of M. avium isolated from blood of AIDS patients (MAC 101, 104, A5) and two strains isolated from lung infection (3362-33 and 3362-34). Bacteria were grown in Middlebrook 7H10 agar plates, and then 1 × 107 bacteria in 0.3 mL of HBSS were seeded in wells of 96-well PVC plates. Two hours after seeding, bacteria were exposed to either tetracycline or streptomycin at 0.5, 1 or 2 mg/L (MIC is > 128 mg/L and 8 mg/L, respectively), ampicillin 10 mg/L, moxifloxacin 1 mg/L, TMP/SMX 3 mg/L or rifampin 1 mg/L (subinhibitory concentrations). Antibiotics were added again at day 7, and PVC plates were followed for 14 days. Biofilm formation was then quantified as previously reported [5, 12], measuring the amount of biofilm by staining it with crystal violet and using the intensity of the staining to determine the exact biofilm mass, using spectrophotometry.
To determine whether the phenotype was temporary or permanent, sessile bacteria were passed on 7H10 agar plate twice and the minimum inhibitory concentration (MIC) of both clarithromycin and ethambutol determined.
To obtain both subpopulations, PVC plates with M. avium biofilm were allowed to remain at rest for 14 days. Then, the supernatant was removed and centrifuged at 4,000 × g for 20 min at 4°C to collect the bacterial pellet. The pellet, made of planktonic bacteria, was washed at 4°C with Hanks’ balanced salt solution (HBSS), passed through a 24-gauge needle ten times and resuspended in fresh HBSS. The concentration was adjusted to 5 × 107 cfu/mL. The biofilm was made of sessile population of bacteria, then removed with a cotton swab and resuspended in HBSS at 4°C. The bacteria were spun down as described above, then passed through a 24-gauge needle ten times to disperse the bacteria in the suspension, and the concentration was adjusted to 5× 107 cfu/mL. Both subpopulations were maintained at 4°C for up to 3 days before the assays, in order to conserve the phenotype. Bacteria were then placed in 7H9 broth suspension with different concentrations of antibiotics and plated for quantification after 48 h. The bacterial concentration in the inoculum was determined by plating the suspensions onto 7H10 agar.
To determine whether clones of planktonic and sessile bacteria were resistant to 20 mg/L of clarithromycin (CLA) or ethambutol (ETM), bacterial suspension of 108 cfu/mL were plated onto 7H10 agar containing either clarithromycin or ethambutol. The same inoculum was plated onto 7H10 agar without antibiotics. Colonies obtained from plates with antibiotics were additionally tested individually for susceptibility, as described above.
Mouse experiments
Female C57BL/6 mice, with 20 grams of weight, were purchased from Jackson Laboratory (West Grove PA). After 1 week of acclimation, mice were infected with 108 bacteria delivered to the nostrils in 0.2 mL of HBSS. Twelve mice were used per time point. Mice were harvested at day seven and at five weeks following infection. Lungs were removed, homogenized, diluted and plated onto 7H11 agar containing trimetropim/sulfamethoxazole, polymyxin B, carbenicillin and amphotericin B, as reported [6]. Quantification of colony forming units (cfu) was performed after 14 days.
Biofilm-related genes
Since exposure to streptomycin and tetracycline resulted in increased biofilm formation, it was important to determine whether the antibiotics triggered the expression of biofilm-related genes. Both guaB2 and gtf were selected [12]. Those genes are involved in glycopeptolipid (GPL) synthesis and synthesis of lipoarabinomannan (LAM), respectively. MAC 104 was exposed to 1 mg/L streptomycin for 2 h, or 1 mg/L of tetracycline, and RNA purification and cDNA synthesis were performed, as previously described [13]. The cDNA was amplified using the following primers:
-
GuaB2: F:5′-TCA CCT GCC GCC CCG ACA ACA CGC TGC CCC-3′
R:5′-GGC ACC CGG CCC TCG ATG CCC TCG GGC ACC-3′
-
Gtf: F:5′-ATG GAG GGC GCC GAC GTG CCC-3′
R:5′-AGG ATC GCG GTG ATG CTG CCC-3′
Real-Time PCR was carried out as previously described [12]. Briefly, quantitative fluorogenic amplification of cDNA was performed using the iCycler (Bio-Rad) and SYBR green technology (Bio-Rad), according to the method previously described [12].
Statistical analysis
Experiments were repeated at least three times and the results are expressed as a mean ± standard deviation. The comparisons among experimental groups were analyzed using ANOVA and Student’s t-test when appropriate. A P value of < 0.05 was considered statistically significant.
RESULTS
Antibiotics and biofilm
It was observed that exposure to streptomycin at subinhibitory concentration resulted in increased M. avium biofilm formation. In fact, incubation in presence of antimicrobial molecules produced by environmental microorganisms may impact the formation of biofilm by other environmental bacteria. AIDS-derived M. avium strains placed in contact with tetracycline were induced to produce significantly more biofilm, although the significance of the increase in biofilm formation related to tetracycline exposure may be understated in relation to the MIC of tetracycline and the concentration of the antibiotic that M. avium was exposed to. Incubation with streptomycin at 0.5 mg/L also induced AIDS-derived M. avium to produce increased amounts of biofilm. When pulmonary strains of M. avium were exposed to either tetracycline or streptomycin at all tested concentrations, both strains showed significant increase in biofilm production (Tables 1 and 2). Exposure to ampicillin, moxifloxacin, or TMP/SMX did not result in any change in biofilm formation (Table 3). The use of rifampin was associated with a decreased the biofilm mass by 10 ± 6% (P > 0.05). When mutants impaired in biofilm formation 6H9, 5G4, as well as 8G12, a mutant that forms increased amount of biofilm compared with the wild-type bacterium, were exposed to streptomycin and tetracycline (data not shown), 6H9 and 5G4 mutants did not increase the amount of biofilm (Table 1). The high-biofilm mutant exposure to streptomycin resulted in a small, but consistent, increase in biofilm formation (Table 1).
Table 1.
Effect of streptomycin exposure on biofilm formation by M. avium strains.
| M. avium Strains | No Antibiotics | Biofilm Formation (OD ± SD)a |
||
|---|---|---|---|---|
| Streptomycin (mg/L)b | ||||
| 0.5 | 1 | 2 | ||
| 104 | 0.236 ± 0.026 | 0.156 ± 0.032* | 0.534 ± 0.046* | 0.549 ± 0.041* |
| 101 | 0.243 ± 0.029 | 0.584 ± 0.041* | 0.563 ± 0.041* | 0.545 ± 0.065* |
| A5 | 0.266 ± 0.041 | 0.668 ± 0.036* | 0.681 ± 0.044* | 0.685 ± 0.048* |
| 3362-33 | 0.222 ± 0.032 | 0.508 ± 0.016* | 0.628 ± 0.023* | 0.699 ± 0.037* |
| 3362-34 | 0.278 ± 0.048 | 0.497 ± 0.039* | 0.548 ± 0.062* | 0.591 ± 0.056* |
| A5 | 0.286 ± 0.045 | 0.647 ± 0.051c | 0.670 ± 0.041c | |
| 8G12 | 0.592 ± 0.051 | 0.732 ± 0.056d | 0.744 ± 0.026d | |
| 5G4 | 0.159 ± 0.040a | 0.194 ± 0.024d | 0.211 ± 0.034d | |
| 6H9 | 0.123 ± 0.067a | 0.143 ± 0.042d | 0.173 ± 0.028d | |
P < 0.05 compared with biofilm without exposure to antibiotics.
Biofilm mass was measured by staining the biofilm with crystal violet and determining the optic density of the crystal violet by spectrophotometry.[5, 11]
The results are the mean of three independent experiments.
P < 0.05 compared with A5 without antibiotic exposure.
P > 0.05 compared with the mutant without exposure to antibiotic.
Table 2.
Effect of tetracycline exposure on biofilm formation by M. avium strains.
| M. avium Strains | No Antibiotics | Biofilm Formation (OD ± SD)a |
||
|---|---|---|---|---|
| Tetracycline (mg/L)b | ||||
| 0.5 | 1 | 2 | ||
| 104 | 0.256 ± 0.026 | 0.310 ± 0.042 | 0.316 ± 0.031* | 0.350 ± 0.017* |
| 101 | 0.243 ± 0.029 | 0.314 ± 0.046 | 0.327 ± 0.023* | 0.329 ± 0.036* |
| A5 | 0.266 ± 0.041 | 0.312 ± 0.022 | 0.336 ± 0.039* | 0.342 ± 0.028* |
| 3362-33 | 0.222 ± 0.032 | 0.371 ±0.033* | 0.384 ± 0.040* | 0.388 ± 0.019* |
| 3362-34 | 0.206 ± 0.057 | 0.423 ± 0.042* | 0.457 ± 0.040* | 0.481 ± 0.050* |
Table 3.
Biofilm formation among M. avium strains exposed to ampicillin, moxifloxacin, rifampin and TMP/SMX.
| Biofilm formation (% increase/decrease) at 7 days | |||||
|---|---|---|---|---|---|
| M. avium strain | No antibiotics | ampicillin (10 μg/L) | moxifloxacin (1 μg/L) | TMP/SMX (3 μg/L) | rifampin (1 μg/L) |
| 104 | 0.271 ± 0.032 | 1 ± 0.3 | 0.1 ± 0.02 | 0.2 ± 0.03 | −10 ± 6* |
| 101 | 0.266 ± 0.049 | 0.4 ± 0.1 | 0.2 ± 0.02 | 0.3 ± 0.03 | −7 ± 3* |
| A5 | 0.296 ± 0.057 | 0.3 ± 0.07 | 0.2 ± 0.05 | 0.2 ± 0.08 | −9.2 ± 3* |
| 3362-33 | 0.274 ± 0.037 | 0.2 ± 0.03 | 0.2 ± 0.1 | 0.2 ± 0.05 | −10.1 ± 4* |
| 3362-34 | 0.230 ± 0.025 | 0.1 ± 0.06 | 0.1 ± 0.09 | 0.2 ± 0.06 | −9.6 ± 2* |
Results represent the mean percentage of increase in biofilm formation of two different experiments.
P < 0.05 compared with biofilm formation without antibiotic exposure.
Biofilm-related genes
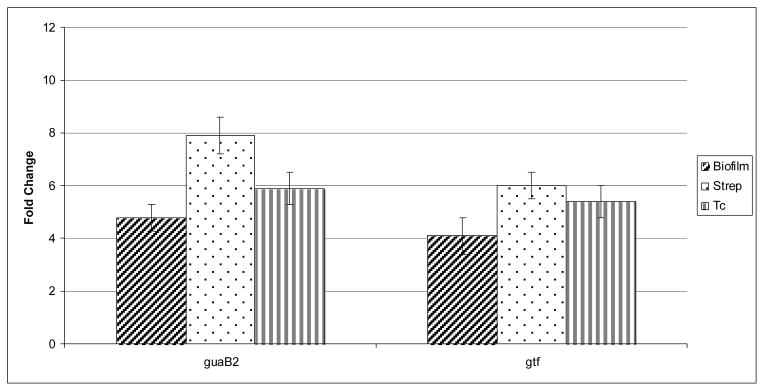
In order to examine whether exposure to streptomycin or tetracycline induces biofilm-related gene regulation in M. avium, MAC 104 biofilm was exposed to 1 mg/L of each antibiotic and the expression of guaB2 and gtf was monitored. As shown in Figure 1, in agreement with previously published data, both genes were expressed during biofilm formation. Gene expression was, however, significantly greater when antibiotics were applied to biofilms.
Figure 1.
Real-Time PCR showing the fold increase, compared to the bacteria not exposed to antibiotics, in the expression of guaB2 and gtf genes. Biofilm (diagonal stripes rectangle) = no antibiotic treatment; Strep = in presence of 1 mg/L of streptomycin; Tc (vertical stripe rectangle) = in presence of 1 mg/L of tetracycline. The assay was run as described in Materials and Methods.
*P < 0.05 for the comparison between biofilm and biofilm exposed to streptomycin.
We established the MIC of clarithromycin and ethambutol against M. avium strains, M. avium 104, 101, A5, 3362-33, 3362-34, and the 5G4 and 6H9 mutants. All the strains had MIC of 2 mg/L for clarithromycin, except the 3362-33 strain, for which the MIC was 4 mg/L. Similarly, the MICs of ethambutol did not vary significantly, with all the strains, except for 101 and A5 (MIC 2 mg/L), having inhibitory concentration at 1 mg/L.
Planktonic and sessile subpopulations
To examine the MICs of the compounds against two subpopulations of M. avium in biofilm, planktonic and sessile, both populations were purified and bacteria were kept at 4°C until the assay. In addition, the two populations were cultured in 7H9 broth for two passages of 10 days before determination of MIC. Table 4 shows that sessile bacteria had MIC of clarithromycin 5-fold greater than the planktonic bacteria, and a MIC of ethambutol 4-fold greater than for the planktonic bacteria. Sessile bacteria passed twice in 7H9 broth reverted the phenotype and had MICs similar to planktonic M. avium.
Table 4.
Minimum inhibitory concentration (MIC) of clarithromycin and ethambutol to planktonic and sessile subpopulations of M. avium.
| Subpopulation of M. avium 104 | MIC |
|
|---|---|---|
| Clarithromycin | Ethambutol | |
| planktonica | 4 mg/L | 2 mg/L |
| sessilea | 64 mg/Lb | 16 mg/Lb |
| planktonicc passed in medium | 4 mg/L | 2 mg/L |
| sessilec passed in medium | 4 mg/L | 2 mg/L |
Planktonic and sessile subpopulations were collected as described in Materials and Methods. Bacteria were kept at 4°C to maintain the phenotype, then placed in 7H9 broth with different concentrations of antibiotics for 48 h. The inoculum was determined by plating onto 7H10 agar without antibiotics.
P < 0.05 compared t the planktonic subpopulation.
Planktonic and sessile subpopulations were cultured twice on 7H10 agar at 37°C for 10 days before the determination of MIC.
To determine whether planktonic phenotype and the biofilm (sessile) phenotype would be able to infect mice equally, mice were exposed to bacteria (planktonic and sessile) via aerosol, and at 7 days and 5 weeks after infection, mice were harvested. The results (Table 5) show that infection with both strains were similar in the number of cfu in lungs and spleen, suggesting that both phenotypes are able to establish infection equally.
Table 5.
Infection of C57BL/6 mice with planktonic and sessile M. avium strain 104.
| Bacteriaa | cfu/lung and spleen |
|||
|---|---|---|---|---|
| spleenb | lungb | |||
| Day 7 | Week 5 | Day 7 | Week 5 | |
| planktonic | 6.1 ± 1 × 104 | 3 ± 2 × 107 | 2 ± 0.9 × 105 | 7.1 ± 1.8 × 108 |
| sessile | 8.7 ± 0.7 × 104 | 5.1 ± 0.8 × 107 | 3.3 ± 1.1 × 105 | 9.6 ± 0.6 × 108 |
inoculum: Planktonic: 1.4 × 108; Sessile: 1.6 × 108
P values were not significant between the experimental groups at all the time points.
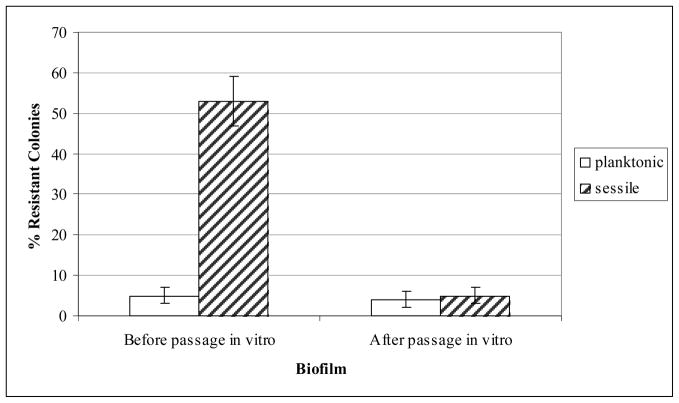
Sessile bacteria showed increased resistance to antibiotics. To investigate whether the phenotype is stable and represents a selection of a subpopulation of M. avium, sessile and planktonic clones were plated on 7H10 or 7H10 with clarithromycin at 20 mg/L, and the percent of resistant colonies was quantified. As shown in Figure 2, the resistant phenotype for clarithromycin is only observed in the first passage but is lost in subsequent passages.
Figure 2.
Selection of resistant and susceptible bacteria. Planktonic and sessile subpopulations were plated onto 7H10 agar. The 7H10 agar plates contained 20 mg/L of clarithromycin. The figure shows how many of the original colonies were resistant to antibiotics.
DISCUSSION
M. avium is an environmental pathogen that causes lung disease in patients with underlying pulmonary conditions [14]. M. avium is believed to be acquired primarily from the environment, more frequently from soil and water sources [15, 16]. It has been demonstrated that the bacterium can be isolated from biofilms on sauna walls and urban water systems [15, 16]. More recently, it has been shown that a large number of M. avium is commonly cultured from household shower heads [17]. Therefore, individuals are exposed frequently to the bacterium, and those with risk factors may develop disease.
M. avium’s ability to form biofilm appears to be a required characteristic for causing pulmonary infection in mice. Mutant strains with deficiency in developing biofilm had impaired ability to cause disease when delivered to the mice by aerosol [6].
Environmental bacteria are likely to compete with other bacteria for nutrients and survival. Many bacteria are known to produce antimicrobials that are lethal against other environmental organisms. Natural evolution would support the development of defense mechanisms, which many times is represented by biofilm formation, in order to defend against environmental bacteria-generated products. Current opinion is that many of these products released into the environment would work as signaling molecules for metabolic pathways in other organisms [18].
We show in this study that M. avium responds with an increase of biofilm formation when exposed to streptomycin and tetracycline at sub-inhibitory concentration, but not when incubated with ampicillin, TMP/SMX, moxifloxacin or rifampin. Tetracycline had less accentuated effect against some of the strains tested. The finding was substantiated by the enhanced expression of M. avium biofilm-related genes upon exposure to antibiotics. Interestingly, ampicillin is derived from Penicillium and rifampin from Nocardia. The importance of the observation is, in fact, that many patients with chronic lung conditions are treated for infections caused by many pathogens with antibiotics, such as aminoglycosides or tetracyclines [19]. Therefore, there is a possibility that, in case M. avium would be colonizing an individual receiving antibiotic, either for prophylaxis or therapy, it would potentially result in the production of increased amounts of biofilm and further establishment of the infection.
In terms of existence and evolution, host infection is a consequence of M. avium survival in the environment. M. avium utilizes many of the features necessary to overcome environmental challenges to infect and persist in the host. Among them, genes associated to amoeba invasion [20] and genes linked to biofilm formation [12]. Here we show that an iatrogenic action, antibiotic use, reproduces the environmental conditions of mixed biofilms, in which the bacterial structure becomes more resistant to outside-inflicted damage. Therefore, specific consideration should be given when a decision is made to administer antimicrobials to patients with chronic lung conditions in which the presence of M. avium is a possibility. Future studies will be needed to evaluate if the effect of antibiotics in vivo-formed biofilm follows similar dynamics.
Once in a particular environment, biofilms are made of at least two populations of bacteria, planktonic and sessile. The results obtained confirmed what many other groups working with different pathogens have already observed. While the sessile phenotype is resistant to antibiotics, the planktonic bacteria are susceptible. Such resistance has been described as associated with the organization of the biofilm, but when biofilm was dispersed, bacteria still expressed a significant resistant phenotype. The mechanism for resistance (or reduced susceptibility) is unknown; however, the presence of biofilm matrix and its impact on bacterial phenotype, upregulation of bacterial antibiotic pump expression, for example, can be factors influencing the phenotype.
Many studies in other systems have looked at planktonic phenotypes of pathogens. Recent work has evaluated the susceptibility of the planktonic population form of Mycobacterium abscessus and determined that it is susceptible to antibiotics [10]. There is, however, no information about the detachment of planktonic bacteria from biofilms and the mechanisms of that detachment.
In summary, M. avium forms increasing amounts of biofilm in presence of antibiotics such as streptomycin and tetracycline, which stimulate biofilm-related gene expression in the bacterium. Once formed, biofilms are made of two distinct populations of bacteria, sessile, the more resistant phenotype, and planktonic, a susceptible phenotype. The role of clinically used antibiotics in stimulating signal transduction and metabolic pathways in bacteria is not well known and should be studied further in mycobacteria.
Acknowledgments
We are indebted to Denny Weber for help editing and preparing the manuscript. Thank you to Chris Lambros for reviewing the manuscript, and we thank Nima Motamedi for the technical help.
FUNDING
This work was supported in part by the National Institutes of Health (contract number AI-25468 to L.Y.) and the National Institutes of Health (grant number AI043199 to L.E.B.).
Footnotes
TRANSPARENCY DECLARATIONS
None to declare.
References
- 1.Costerton W, Veeh R, Shirtliff M, Pasmore M, Post C, Ehrlich G. The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest. 2003;112:1466–1477. doi: 10.1172/JCI20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ebihara T, Sasaki H. Image in clinical medicine. Bronchiectasis with mycobacterium avium complex infection. N Engl J Med. 2002;346:1372. doi: 10.1056/NEJMicm010899. [DOI] [PubMed] [Google Scholar]
- 3.Fujita J, Ohtsuki Y, Suemitsu I, et al. Pathological and radiological changes in resected lung specimens in mycobacterium avium intracellulare complex disease. Eur Respir J. 1999;13:535–540. doi: 10.1183/09031936.99.13353599. [DOI] [PubMed] [Google Scholar]
- 4.Martinez A, Torello S, Kolter R. Sliding motility in mycobacteria. J Bacteriol. 1999;181:7331–7338. doi: 10.1128/jb.181.23.7331-7338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter G, Wu M, Drummond DC, Bermudez LE. Characterization of biofilm formation by clinical isolates of mycobacterium avium. J Med Microbiol. 2003;52:747–752. doi: 10.1099/jmm.0.05224-0. [DOI] [PubMed] [Google Scholar]
- 6.Yamazaki Y, Danelishvili L, Wu M, et al. The ability to form biofilm influences mycobacterium avium invasion and translocation of bronchial epithelial cells. Cell Microbiol. 2006;8:806–814. doi: 10.1111/j.1462-5822.2005.00667.x. [DOI] [PubMed] [Google Scholar]
- 7.Griffith DE, Brown BA, Cegielski P, Murphy DT, Wallace RJ., Jr Early results (at 6 months) with intermittent clarithromycin-including regimens for lung disease due to mycobacterium avium complex. Clin Infect Dis. 2000;30:288–292. doi: 10.1086/313644. [DOI] [PubMed] [Google Scholar]
- 8.Griffith DE, Brown BA, Murphy DT, Girard WM, Couch L, Wallace RJ., Jr Initial (6-month) results of three-times-weekly azithromycin in treatment regimens for mycobacterium avium complex lung disease in human immunodeficiency virus-negative patients. J Infect Dis. 1998;178:121–126. doi: 10.1086/515597. [DOI] [PubMed] [Google Scholar]
- 9.Reich JM, Johnson RE. Mycobacterium avium complex pulmonary disease. Incidence, presentation, and response to therapy in a community setting. Am Rev Respir Dis. 1991;143:1381–1385. doi: 10.1164/ajrccm/143.6.1381. [DOI] [PubMed] [Google Scholar]
- 10.Greendyke R, Byrd TF. Differential antibiotic susceptibility of mycobacterium abscessus variants in biofilms and macrophages compared to that of planktonic bacteria. Antimicrob Agents Chemother. 2008;52:2019–2026. doi: 10.1128/AAC.00986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter G, Young LS, Bermudez LE. A subinhibitory concentration of clarithromycin inhibits mycobacterium avium biofilm formation. Antimicrob Agents Chemother. 2004;48:4907–4910. doi: 10.1128/AAC.48.12.4907-4910.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamazaki Y, Danelishvili L, Wu M, Macnab M, Bermudez LE. Mycobacterium avium genes associated with the ability to form a biofilm. Appl Environ Microbiol. 2006;72:819–825. doi: 10.1128/AEM.72.1.819-825.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harriff MJ, Danelishvili L, Wu M, et al. Mycobacterium avium genes mav_5138 and mav_3679 are transcriptional regulators that play a role in invasion of epithelial cells, in part by their regulation of cipa, a putative surface protein interacting with host cell signaling pathways. J Bacteriol. 2009;191:1132–1142. doi: 10.1128/JB.01359-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aksamit TR. Mycobacterium avium complex pulmonary disease in patients with pre-existing lung disease. Clin Chest Med. 2002;23:643–653. doi: 10.1016/s0272-5231(02)00022-9. [DOI] [PubMed] [Google Scholar]
- 15.Torvinen E, Lehtola MJ, Martikainen PJ, Miettinen IT. Survival of mycobacterium avium in drinking water biofilms as affected by water flow velocity, availability of phosphorus, and temperature. Appl Environ Microbiol. 2007;73:6201–6207. doi: 10.1128/AEM.00828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Reyn CF, Maslow JN, Barber TW, Falkinham JO, 3rd, Arbeit RD. Persistent colonisation of potable water as a source of mycobacterium avium infection in aids. Lancet. 1994;343:1137–1141. doi: 10.1016/s0140-6736(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 17.Falkinham JO, 3rd, Iseman MD, de Haas P, van Soolingen D. Mycobacterium avium in a shower linked to pulmonary disease. J Water Health. 2008;6:209–213. doi: 10.2166/wh.2008.032. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman LR, D’Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 19.Chodosh S. Treatment of acute exacerbations of chronic bronchitis: State of the art. Am J Med. 1991;91:87S–92S. doi: 10.1016/0002-9343(91)90317-q. [DOI] [PubMed] [Google Scholar]
- 20.Danelishvili L, Wu M, Stang B, et al. Identification of mycobacterium avium pathogenicity island important for macrophage and amoeba infection. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11038–11043. doi: 10.1073/pnas.0610746104. [DOI] [PMC free article] [PubMed] [Google Scholar]