Abstract
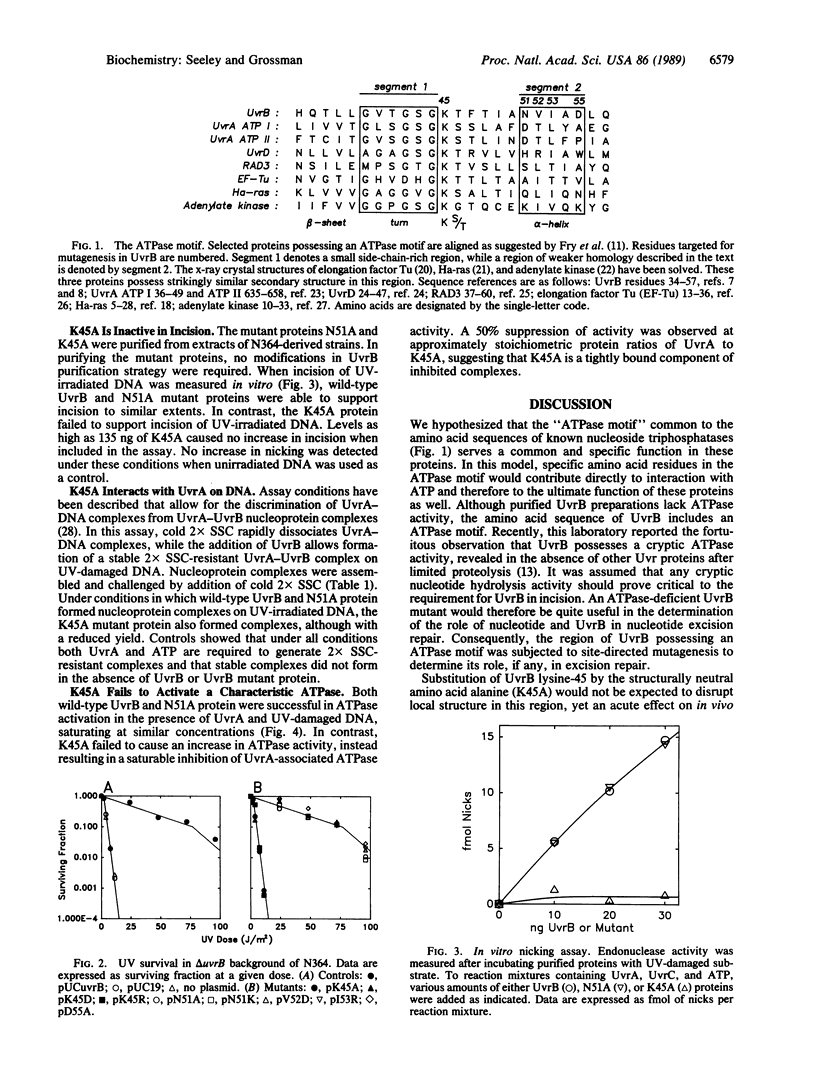
The Escherichia coli UvrB protein possesses an amino acid sequence motif common to many ATPases. The role of this motif in UvrB has been investigated by site-directed mutagenesis. Three UvrB mutants, with amino acid replacements at lysine-45, failed to confer UV resistance when tested in the UV-sensitive strain N364 (delta uvrB), while five other mutants constructed near this region of UvrB confer wild-type levels of UV resistance. Because even the conservative substitution of arginine for lysine-45 in UvrB results in failure to confer UV resistance, we believe we have identified an amino acid side chain in UvrB essential to nucleotide excision repair in E. coli. The properties of two purified mutant UvrB proteins, lysine-45 to alanine (K45A) and asparagine-51 to alanine (N51A), were analyzed in vitro. While the K45A mutant is fully defective in incision of UV-irradiated DNA, K45A is capable of interaction with UvrA in forming an ATP-dependent nucleoprotein complex. The K45A mutant, however, fails to activate the characteristic increase in ATPase activity observed with the wild-type UvrB in the presence of UvrA and DNA. From these results we conclude that there is a second nucleotide-dependent step in incision following initial complex formation, which is defective in the K45A mutant. This experimental approach may prove of general applicability in the study of function and mechanism of other ATPase motif proteins.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arai K., Clark B. F., Duffy L., Jones M. D., Kaziro Y., Laursen R. A., L'Italien J., Miller D. L., Nagarkatti S., Nakamura S. Primary structure of elongation factor Tu from Escherichia coli. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1326–1330. doi: 10.1073/pnas.77.3.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikan E., Kulkarni M. S., Thomas D. C., Sancar A. Sequences of the E. coli uvrB gene and protein. Nucleic Acids Res. 1986 Mar 25;14(6):2637–2650. doi: 10.1093/nar/14.6.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backendorf C., Spaink H., Barbeiro A. P., van de Putte P. Structure of the uvrB gene of Escherichia coli. Homology with other DNA repair enzymes and characterization of the uvrB5 mutation. Nucleic Acids Res. 1986 Apr 11;14(7):2877–2890. doi: 10.1093/nar/14.7.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Caron P. R., Grossman L. Involvement of a cryptic ATPase activity of UvrB and its proteolysis product, UvrB* in DNA repair. Nucleic Acids Res. 1988 Nov 25;16(22):10891–10902. doi: 10.1093/nar/16.22.10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Glynias M. J., Merrick W. C. GTP-binding domain: three consensus sequence elements with distinct spacing. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1814–1818. doi: 10.1073/pnas.84.7.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch P. W., Emmerson P. T. The nucleotide sequence of the uvrD gene of E. coli. Nucleic Acids Res. 1984 Jul 25;12(14):5789–5799. doi: 10.1093/nar/12.14.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry D. C., Kuby S. A., Mildvan A. S. ATP-binding site of adenylate kinase: mechanistic implications of its homology with ras-encoded p21, F1-ATPase, and other nucleotide-binding proteins. Proc Natl Acad Sci U S A. 1986 Feb;83(4):907–911. doi: 10.1073/pnas.83.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay N. J., Walker J. E. Homology between human bladder carcinoma oncogene product and mitochondrial ATP-synthase. Nature. 1983 Jan 20;301(5897):262–264. doi: 10.1038/301262a0. [DOI] [PubMed] [Google Scholar]
- Heil A., Müller G., Noda L., Pinder T., Schirmer H., Schirmer I., von Zabern I. The amino-acid sequence of sarcine adenylate kinase from skeletal muscle. Eur J Biochem. 1974 Mar 15;43(1):131–144. doi: 10.1111/j.1432-1033.1974.tb03393.x. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Hiles I. D., Salmond G. P., Gill D. R., Downie J. A., Evans I. J., Holland I. B., Gray L., Buckel S. D., Bell A. W. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature. 1986 Oct 2;323(6087):448–450. doi: 10.1038/323448a0. [DOI] [PubMed] [Google Scholar]
- Husain I., Van Houten B., Thomas D. C., Sancar A. Sequences of Escherichia coli uvrA gene and protein reveal two potential ATP binding sites. J Biol Chem. 1986 Apr 15;261(11):4895–4901. [PubMed] [Google Scholar]
- Jurnak F. Structure of the GDP domain of EF-Tu and location of the amino acids homologous to ras oncogene proteins. Science. 1985 Oct 4;230(4721):32–36. doi: 10.1126/science.3898365. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Milman G. Expression plasmid containing the lambda PL promoter and cI857 repressor. Methods Enzymol. 1987;153:482–491. doi: 10.1016/0076-6879(87)53073-7. [DOI] [PubMed] [Google Scholar]
- Oh E. Y., Claassen L., Thiagalingam S., Mazur S., Grossman L. ATPase activity of the UvrA and UvrAB protein complexes of the Escherichia coli UvrABC endonuclease. Nucleic Acids Res. 1989 Jun 12;17(11):4145–4159. doi: 10.1093/nar/17.11.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E. Y., Grossman L. Characterization of the helicase activity of the Escherichia coli UvrAB protein complex. J Biol Chem. 1989 Jan 15;264(2):1336–1343. [PubMed] [Google Scholar]
- Oh E. Y., Grossman L. Helicase properties of the Escherichia coli UvrAB protein complex. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3638–3642. doi: 10.1073/pnas.84.11.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinstein J., Brune M., Wittinghofer A. Mutations in the nucleotide binding loop of adenylate kinase of Escherichia coli. Biochemistry. 1988 Jun 28;27(13):4712–4720. doi: 10.1021/bi00413a020. [DOI] [PubMed] [Google Scholar]
- Reynolds P., Higgins D. R., Prakash L., Prakash S. The nucleotide sequence of the RAD3 gene of Saccharomyces cerevisiae: a potential adenine nucleotide binding amino acid sequence and a nonessential acidic carboxyl terminal region. Nucleic Acids Res. 1985 Apr 11;13(7):2357–2372. doi: 10.1093/nar/13.7.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Rupp W. D. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell. 1983 May;33(1):249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- Schulz G. E., Elzinga M., Marx F., Schrimer R. H. Three dimensional structure of adenyl kinase. Nature. 1974 Jul 12;250(462):120–123. doi: 10.1038/250120a0. [DOI] [PubMed] [Google Scholar]
- Sigal I. S., Gibbs J. B., D'Alonzo J. S., Temeles G. L., Wolanski B. S., Socher S. H., Scolnick E. M. Mutant ras-encoded proteins with altered nucleotide binding exert dominant biological effects. Proc Natl Acad Sci U S A. 1986 Feb;83(4):952–956. doi: 10.1073/pnas.83.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Higgins D., Prakash L., Prakash S. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 1988 Oct;7(10):3263–3269. doi: 10.1002/j.1460-2075.1988.tb03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. C., Levy M., Sancar A. Amplification and purification of UvrA, UvrB, and UvrC proteins of Escherichia coli. J Biol Chem. 1985 Aug 15;260(17):9875–9883. [PubMed] [Google Scholar]
- Yeung A. T., Mattes W. B., Grossman L. Protein complexes formed during the incision reaction catalyzed by the Escherichia coli UvrABC endonuclease. Nucleic Acids Res. 1986 Mar 25;14(6):2567–2582. doi: 10.1093/nar/14.6.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung A. T., Mattes W. B., Oh E. Y., Grossman L. Enzymatic properties of purified Escherichia coli uvrABC proteins. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6157–6161. doi: 10.1073/pnas.80.20.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung A. T., Mattes W. B., Oh E. Y., Yoakum G. H., Grossman L. The purification of the Escherichia coli UvrABC incision system. Nucleic Acids Res. 1986 Nov 11;14(21):8535–8556. doi: 10.1093/nar/14.21.8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos A. M., Tong L., Milburn M. V., Matias P. M., Jancarik J., Noguchi S., Nishimura S., Miura K., Ohtsuka E., Kim S. H. Three-dimensional structure of an oncogene protein: catalytic domain of human c-H-ras p21. Science. 1988 Feb 19;239(4842):888–893. doi: 10.1126/science.2448879. [DOI] [PubMed] [Google Scholar]


