Abstract
We previously established an efficient Agrobacterium-mediated transformation system using primary calli derived from mature seeds of the model japonica rice variety Nipponbare. We expected that the shortened tissue culture period would reduce callus browning—a common problem with the indica transformation system during prolonged tissue culture in the undifferentiated state. In this study, we successfully applied our efficient transformation system to Kasalath—a model variety of indica rice. The Luc reporter system is sensitive enough to allow quantitative analysis of the competency of rice callus for Agrobacterium-mediated transformation. We unexpectedly discovered that primary callus of Kasalath exhibits a remarkably high competency for Agrobacterium-mediated transformation compared to Nipponbare. Southern blot analysis and Luc luminescence showed that independent transformation events in primary callus of Kasalath occurred successfully at ca. tenfold higher frequency than in Nipponbare, and single copy T-DNA integration was observed in ~40% of these events. We also compared the competency of secondary callus of Nipponbare and Kasalath and again found superior competency in Kasalath, although the identification and subsequent observation of independent transformation events in secondary callus is difficult due to the vigorous growth of both transformed and non-transformed cells. An efficient transformation system in Kasalath could facilitate the identification of QTL genes, since many QTL genes are analyzed in a Nipponbare × Kasalath genetic background. The higher transformation competency of Kasalath could be a useful trait in the establishment of highly efficient systems involving new transformation technologies such as gene targeting.
Electronic supplementary material
The online version of this article (doi:10.1007/s00299-010-0921-x) contains supplementary material, which is available to authorized users.
Keywords: Agrobacterium, Bacterial density, Genetic transformation, Indica rice, Transformation efficiency
Introduction
Successful Agrobacterium-mediated genetic transformation technology has been reported in various rice cultivars [e.g., Oryza sativa L. ssp. japonica, indica and javanica, and NERICA (O. sativa × O. glaberrima)], and is used widely in the fields of both molecular breeding and functional genomics (Datta et al. 2000; Dong et al. 1996; Ishizaki and Kumashiro 2008; Parkhi et al. 2005; Rashid et al. 1996; Toki et al. 2006). In rice, usually calli derived from mature seeds and immature embryos are used in Agrobacterium-mediated transformation. Routine protocols for highly efficient Agrobacterium-mediated transformation using calli derived from mature seeds have been reported in various cultivars of japonica rice (Hiei and Komari 2008; Nishimura et al. 2006; Toki et al. 2006). For example, it was reported that transgenic plants were obtained using 3-week-old mature seed-derived embryogenic calli in Nipponbare—a model variety of japonica rice—with a transformation efficiency of 40–70% (independent transgenic plants/inoculated callus; (Hiei and Komari 2008). By contrast, transgenic plants were obtained at low frequencies of only 4.6–5.5 and 6.4–7.3% (no. of callus lines regenerated/no. of explants used for co-cultivation) using 2-month-old mature seed-derived embryogenic calli in indica varieties IR64 and IR72, respectively (Kumar et al. 2005). Thus, many indica rice varieties are still recalcitrant to Agrobacterium-mediated transformation using calli derived from mature seeds, and a widely applicable and efficient transformation system has not yet been reported. On the other hand, a protocol for Agrobacterium-mediated transformation using immature embryos has been reported to be applicable to many rice cultivars of both japonica and indica (Hiei and Komari 2008), suggesting that immature embryos are more competent than mature seeds for Agrobacterium-mediated transformation. However, unlike mature seeds, the preparation and manipulation of immature embryos is troublesome because only a limited number of immature embryos can be obtained at the right developmental stage (8–12 days after pollination; (Hiei and Komari 2008). Thus, the establishment of a highly efficient and widely applicable Agrobacterium-mediated transformation using calli derived from mature seeds in indica rice would be of significant benefit to the field.
There are two types of mature seed-derived callus: primary callus, i.e., <10-day-old callus representing proliferation of scutellum; and secondary callus, i.e., >2-week-old callus proliferating from primary callus (Online Resource 1). Primary callus derived from mature seeds is compact and hard, although secondary callus is friable due to vigorous growth. Therefore, antibiotic-resistant secondary calli [Hygromycin (Hyg)-resistant in the case illustrated in Online Resource 1] appearing on primary callus co-cultivated with Agrobacterium can be distinguished clearly and propagated independently. On the other hand, antibiotic-resistant secondary calli are hard to distinguish from secondary callus co-cultivated with Agrobacterium due to the vigorous growth of both transformed and non-transformed cells. Our transformation system using primary callus enables independent transformation events occurring within a single primary callus to be distinguished. In a previous study, we succeeded in obtaining transformed calli of Nipponbare at a frequency of 95–98% [% co-cultured seeds yielding Hyg-tolerant or green fluorescent protein (GFP)-expressing calli] using primary calli, i.e., 5-day-old mature seed-derived embryogenic calli, and transgenic plantlets within a month after the onset of inoculation of mature seeds (Toki et al. 2006). The occurrence of genomic changes at high frequency is associated with plant regeneration from de-differentiated cells (Labra et al. 2001), but a short period of tissue culture helps minimize somaclonal variation. In addition, long periods of culture result in browning, especially in callus of indica rice. Since callus browning results from polyphenol accumulation and cell death, which prevents further proliferation, this is thought to be one of the reasons underlying regeneration failure in this cultivar (Zhao et al. 2009). In our efficient transformation system, somaclonal mutations are thought to occur at lower frequency because the time required for callus induction from mature seeds is shortened (Toki et al. 2006). Regeneration can start before callus browning because of the short period of callus proliferation. Thus, our efficient transformation system using primary callus derived from mature seeds is not only convenient and efficient compared to conventional transformation systems using secondary callus derived from mature seeds and immature embryos, but could also be useful for transformation in indica-type rice.
In this study, we established our efficient transformation system in Kasalath—a model variety of indica-type rice used for quantitative trait locus (QTL) analysis of genes involved in agronomically useful traits. Furthermore, an unexpected finding was that primary callus of Kasalath has remarkably high competency in Agrobacterium infection compared to that of Nipponbare; the number of transformed cells of Kasalath in which T-DNA were expressed transiently and/or stably was much higher than in Nipponbare. Additionally, transformation was achieved in Kasalath under conditions of low density (OD = 10−5) Agrobacterium co-cultivation. We discuss the potential use of this high competency trait to improve gene targeting frequency.
Materials and methods
Plant materials
The indica-type rice Oryza sativa L. cv. Kasalath and the japonica-type rice O. sativa L. cv. Nipponbare were used in this study.
Binary vectors
The binary plasmid vectors used in this study, pCAMBIA1390-Luc [hygromycin phosphotransferase (hpt) expression cassette and luciferase (luc) expression cassette] and pCAMBIA1390-sGFP (hpt expression cassette and sgfp expression cassette Toki et al. 2006) are shown in Fig. 1. The binary vector, pCAMBIA1390 was provided by CAMBIA (Canberra, Australia). To construct pCAMBIA1390-Luc, pCAMBIA1390-sGFP was digested with SalI and BsrGI [blunt-ended with T4 DNA polymerase (Toyobo, Osaka, Japan)], and the SalI-/SacI-digested (blunt-ended with T4 DNA polymerase) luc fragment from the luc expression vector was inserted.
Fig. 1.
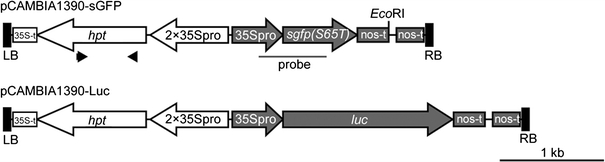
Structure of the T-DNAs used in this study. The gfp and luc expression cassettes were cloned in the binary vector pCAMBIA1390, yielding pCAMBIA1390-sGFP and pCAMBIA1390-Luc, respectively. The gfp and luc genes are under the control of the CaMV 35S promoter (35Spro) and nopaline synthase terminator (nos-t); hpt expression was directed by a duplicated 35S promoter (2 × 35Spro) and the CaMV 35S polyA (35S-t). Black arrowheads in pCAMBIA1390-sGFP indicate the primers used for PCR analysis shown in Fig. 2e. The black bar in pCAMBIA1390-sGFP indicates the region covered by the DIG-labeled gfp probe used for the Southern blot analysis shown in Figs. 2f, 4a and 5b
Agrobacterium-mediated transformation
The binary plasmid vectors described above were transferred into Agrobacterium tumefaciens strain EHA105 by electroporation (Hood et al. 1993) using an E. coli pulser (Bio-Rad, Hercules, CA, USA). A schematic representation of Agrobacterium-mediated transformation using primary callus is shown in Online Resource 1. Dehulled mature seeds, which were sterilized with 70% ethanol and then 2.5% sodium hypochlorite, were inoculated on callus induction N6D medium solidified with 0.4% gelrite [30 g/L sucrose, 0.3 g/L casamino acids, Chu(N6) Medium Salt Mixture (Wako Pure Chemical Industries, Osaka, Japan), 2.878 g/L proline, 2 mg/L glycine, N6-vitamins and 2 mg/L 2,4-dichlorophenoxyacetic acid (2,4-D), pH = 5.8], and cultured at 33°C (light for 10 h)/30°C (dark for 14 h). 7-day-old and 3-week-old calli were used for transformation as primary and secondary calli, respectively. Secondary calli were subcultured on fresh N6D medium for 3 days before the onset of Agrobacterium co-cultivation. Agrobacterium co-cultivation, selection against hygromycin (Hyg) and regeneration of transgenic plants followed Toki et al. (2006). After 3 days of co-cultivation with Agrobacterium at 22–25°C under constant dark on 2N6-AS medium [30 g/L sucrose, 10 g/L glucose, 0.3 g/L casamino acids, Chu(N6) Medium Salt Mixture (Wako Pure Chemical Industries), 2 mg/L glycine, N6-vitamins and 2 mg/L 2,4-D, pH = 5.2], calli were washed with water and carbenicillin (Nakalai tesque, Kyoto, Japan) or meropenem (Wako Pure Chemical Industries) solution, and cultured on N6D medium containing 50 mg/L Hyg (Wako Pure Chemical Industries) and 400 mg/L carbenicillin or 12.5 mg/L meropenem for 3 weeks at 33°C (light for 10 h)/30°C (dark for 14 h). After 10 days of selection, calli were transferred to fresh N6D selection medium. For regeneration, calli growing vigorously on Hyg were transferred to Re-III medium [30 g/L sucrose, 30 g/L sorbitol, 2 g/L casamino acids, MS Medium Salt Mixture (Wako Pure Chemical Industries), 2 mg/L glycine, N6-vitamins, 20 μg/L 1-naphthalene acetic acid and 2 mg/L kinetin, pH = 5.8] with 200 mg/L carbenicillin and 30 mg/L Hyg and cultured for 3 weeks at 30°C under constant light. At 10 days after transfer to Re-III medium, some pieces of vigorously growing or greening callus were transferred to fresh Re-III medium. Shoots arising from callus on Re-III medium were transferred to HF medium [30 g/L sucrose, MS Medium Salt Mixture (Wako Pure Chemical Industries), 2 mg/L glycine and N6-vitamins, pH = 5.8] with 30 mg/L Hyg to allow vigorous growth of roots.
Extraction of rice genomic DNA, PCR analysis and Southern blot analysis
For extraction of genomic DNA, rice seedlings were harvested, immediately frozen in liquid N2 and stored at −80°C. Genomic DNA was extracted from leaves using Nucleon PhytoPure (GE Healthcare, Piscataway, NJ, USA) according to the manufacturer’s protocol. PCR analysis to detect the hpt gene was performed using KOD dash DNA polymerase (Toyobo) with the primer set 5′-ATAGCTGCGCCGATGGTT-3′/5′-CGTCTGCTGCTCCATACAAG-3′. Southern blot analysis was performed according to a standard protocol. Specific DNA probes were prepared using a PCR digoxigenin (DIG) probe synthesis kit (Roche Diagnostics, Basel, Switzerland) according to the manufacturer’s protocol, with the primer sets 5′-GGATTCCATTGCCCAGCTATCTGTC-3′/5′-AGAAGTCGTGCTGCTTCATGTGGTC-3′ for the gfp gene. Southern blot hybridization signals were detected and analyzed using a Lumivision PRO (TAITEC, Saitama, Japan).
Extraction of total RNA and quantitative RT-PCR analysis
Total RNA was extracted from frozen calli with an RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Transcript levels of each gene were measured by real-time quantitative RT-PCR using a Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) and an ABI7300 (Applied Biosystems) according to the manufacturers’ protocols. The gene-specific primers used for quantitative RT-PCR were 5′-CAGAAGAACGGCATCAAGGT-3′ and 5′-CTGGGTGCTCAGGTAGTGGT-3′ for gfp, and 5′-AGGCCAATCGTGAGAAGATGACCCA-3′ and 5′-GTGTGGCTGACACCATCACCAGAG-3′ for OsActin1.
Immunoblot analysis and measurement of GFP fluorescence
Total protein was extracted from frozen calli with extraction buffer (0.1 M sodium phosphate, 0.1% Triton-X and 20% glycerol). Electrophoresis of protein and immunoblot hybridization was performed as described previously (Osakabe et al. 2006). Living colors A.v. (JL-8) monoclonal antibody (Clontech, Mountain View, CA, USA) was used as the antibody against GFP, and recombinant GFP protein (Clontech) was used as a positive control. Signals were detected and analyzed as for Southern blot hybridization.
Observation of GFP fluorescence
GFP fluorescence was observed using a fluorescence microscope with a GFP2 filter (MZ FLIII, Leica Microsystems, Wetzlar, Germany).
Observation of Luc luminescence
Transformed calli were treated with 0.05 mM Beetle Luciferin, Potassium salt (Promega) and kept for 5 min. Luc luminescence images were taken using a high resolution photon counting camera (C2400-77 VIM camera, Hamamatsu Photonics, Hamamatsu, Japan) with a 30 min exposure time, and processed with Aquacosmos (Hamamatsu Photonics).
Results
Efficient transformation in Kasalath
First, we applied our efficient transformation system using primary calli to Kasalath. To test the ability of callus formation in Kasalath, mature seeds were inoculated on N6D medium and grown for 7 days at 30–33°C; the rate of callus induction from mature seeds was about 75% (n = 640; cf. ~ 90% in Nipponbare, n = 160). The fresh weight of 7-day-old callus of Kasalath was 2.77 ± 0.22 (gFW/100 calli; average ± SD), which was similar to that of Nipponbare [2.61 ± 0.51 (gFW/100calli)] (Fig. 2a). On the other hand, the fresh weight of 3-week-old secondary calli of Kasalath was 9.01 ± 1.78 (gFW/100 primary calli; average ± SD), which was about half that of Nipponbare [18.2 ± 1.55 (gFW/100 primary calli)]. To see if transformed plants could be obtained using primary calli derived from mature seeds of Kasalath, 7-day-old calli were infected with Agrobacterium harboring pCAMBIA1390-sGFP vector (Fig. 1). Following a 3-week selection on 50 mg/L Hyg, Hyg-tolerant calli were observed from almost all calli lines tested (Fig. 2b; Table 1). After selection, calli propagated on selection medium were transferred to regeneration medium containing 30 mg/L Hyg. After 3 weeks of regeneration, shoots and roots emerged from about half of the Hyg-tolerant callus lines (Fig. 2c; Table 1) and regenerated plants were obtained successfully (Fig. 2d).
Fig. 2.
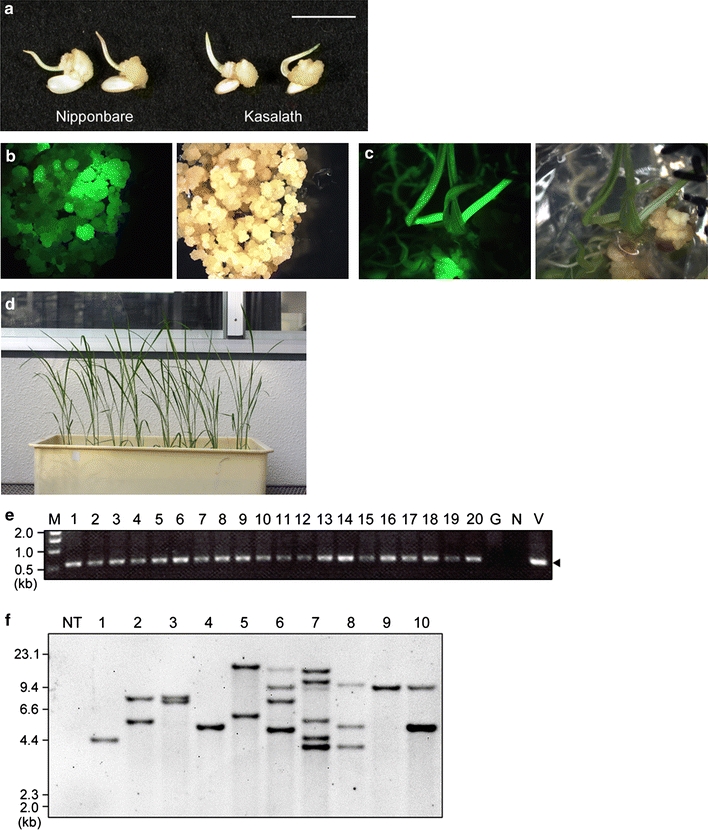
Agrobacterium-mediated transformation in primary calli of Kasalath. a Rice calli induced on N6D medium from mature seeds of Nipponbare (left) and Kasalath (right) for 7 days. Bar 1 cm. b Transformed calli 21 days after the onset of selection on Hyg. Left panel, under blue light; right panel, under white light. c Regenerated shoots 24 days after the onset of regeneration. Left panel, under blue light; right panel, under white light. d Transgenic Kasalath plants in the T0 generation. e An example of PCR analysis of the presence of the hpt gene in T0 plants. The arrowhead indicates the 0.59 kb band corresponding to the hpt gene of the T-DNA. M DNA size marker, 1–20 genomic DNA extracted from transformed T0 plants (lane 1–4 4 independent regenerated plantlets in Exp.3 shown in Table 1, lane 5–20 16 independent regenerated plantlets in Exp.4), G genomic DNA extracted from a non-transformant; N no template; V binary vector pCAMBIA1390-sGFP as template. f Southern blot analysis of 1.5 μg of EcoRI-digested genomic DNA extracted from T0 plants using the gfp probe shown in Fig. 1. Lanes: NT non-transformant, 1–10 transgenic T0 plants
Table 1.
Transformation efficiency of Kasalath and Nipponbare
| No. of calli infected with Agrobacterium (A) | No. of lines successful in clonal propagation (B) | No. of lines successfully regenerated (C) | Clonal propagation efficiency (B/A) (%)a | Regeneration efficiency (C/B) (%)b | |
|---|---|---|---|---|---|
| Kasalath-Exp.1 | 120 | 119 | 48 | 99.1 | 40.3 |
| Kasalath-Exp.2 | 124 | 121 | 81 | 97.6 | 66.9 |
| Kasalath-Exp.3 | 124 | 119 | 70 | 96.0 | 58.8 |
| Kasalath-Exp.4 | 121 | 118 | 65 | 97.5 | 55.1 |
| Nipponbarec | 96 | 88 | 80 | 91.7 | 91.0 |
aPrimary callus derived from one seed was counted as one line. Clonal propagation efficiency was calculated as the percentage of the number of clonal lines successfully propagated compared to the number of Agrobacterium-infected calli
bRegeneration efficiency was calculated as the ratio of regenerated plant lines in which T-DNA insertion was confirmed to the number of clonal lines successfully propagated. Significant difference between Nipponbare and Kasalath at P < 0.01, as determined by t test
cAs reported by Saika and Toki (2009b)
PCR analysis and Southern blot analysis were performed to check whether T-DNA was inserted into the rice genome in regenerated plants (T0 plants). PCR analysis using genomic DNA extracted from 16 independent regenerated plantlets in each experiment demonstrated that T-DNA had inserted into the genome of all regenerated plantlets tested (Fig. 2e). Southern blot analysis was performed to confirm the number of T-DNA copies integrated into the rice genome of transgenic rice plants. Transgenic plants of Kasalath contained single (Fig. 2f, lanes 1, 4 and 9) or multiple (lanes 2, 3, 5–8, 10) hybridizing bands. On average, 2.4 T-DNA copies were integrated in Kasalath (n = 10; Fig. 2f). Previously, we reported that, on average, 2.1 copies of T-DNA are integrated in rice plants of Nipponbare (Nakamura et al. 2007), i.e., comparable to the integrated T-DNA copy number in transgenic plants of Kasalath. GFP fluorescence was observed in 67 of 94 T1 plants (the progeny of line No.1 in Fig. 2f), which fit a 3:1 ratio (χ 2 = 0.70; P > 0.05). These results show that transgenes in transformed Kasalath are stably inherited to the T1 generation in a Mendelian manner. Thus, stable integration of T-DNA in the T0 generation and inheritance to the T1 generation were confirmed in transformed rice plants of Kasalath.
Transformation frequency in primary and secondary calli of Nipponbare and Kasalath
In a previous report, we calculated the transformation efficiency of Nipponbare (Saika and Toki 2009b). We counted one primary callus derived from one seed as one line, and calculated clonal propagation efficiency as the ratio of the number of clonal lines successfully propagated to the number of Agrobacterium-infected calli under our experimental conditions, regardless of statistical significance (Table 1). However, more Hyg-resistant secondary calli were apparently propagated from Agrobacterium co-cultivated primary calli in Kasalath compared to Nipponbare (Fig. 3a). This suggested the possibility that higher numbers of independent cells were successfully transformed in Kasalath than in Nipponbare. To confirm this, we first compared the number of cells transformed with pCAMBIA1390-sGFP (shown in Fig. 1) emitting green fluorescence in Kasalath primary calli with that in Nipponbare. Interestingly, the number of cells in which GFP signals were observed in Kasalath was found to be much higher than that in Nipponbare after 8 days of selection (Fig. 3b; Online Resource 2a). We repeated the transformation with another vector, pCAMBIA1304, which harbors a gfp-gus expression cassette. As with GFP, the number of GUS-staining cells in Kasalath was higher than in Nipponbare (Online Resource 2b), confirming that the difference in the number of GFP-positive cells in transformed calli between Nipponbare and Kasalath shown in Fig. 3b and Online Resource 2a was not caused by differences in the spectral qualities of GFP due to cellular conditions.
Fig. 3.
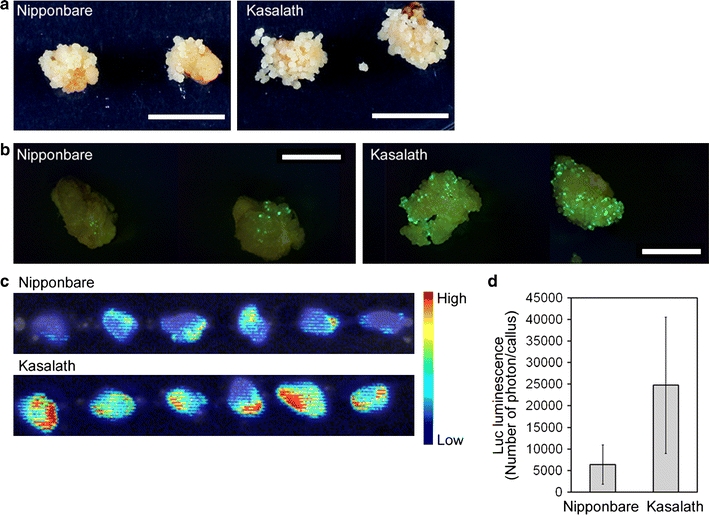
Transformed primary calli of Nipponbare and Kasalath. a Transformed callus 14 days after the onset of selection. Hyg-tolerant cells were grown from all transformed callus of Kasalath, whereas only some transformed callus of Nipponbare grew. b Representative images of green fluorescence (viewed under blue light) emitted from callus transformed with pCAMBIA1390-sGFP 8 days after the onset of selection. Bar 5 mm. Other images are shown in Online Resource 2a. c Luminescence emitted from callus transformed with pCAMBIA1390-Luc just after the onset of selection. Luminescence intensity is shown by the false color scale. d Graphical representation of the Luc luminescence intensity emitted from the transformed Nipponbare or Kasalath calli shown in Fig. 3c. The y axis shows luminescence level (photon) per callus. Data are presented as mean ± SD (n = 75). Significant difference between Nipponbare and Kasalath at P < 0.01 as determined by t test
Two stages in Agrobacterium-mediated transformation have been proposed: (1) transient expression of foreign genes on T-DNA that is not integrated into the plant genome (early stage), and (2) stable expression of foreign genes on T-DNA that is successfully integrated into the plant genome (late stage). In previous reports of transformed calli of Nipponbare, GFP signals were derived from transient expression of a foreign gene on T-DNA that was not integrated into the rice genome after 1–2 days of Agrobacterium elimination, with cells that have been stably transformed with T-DNA starting growth via cell division and cell expansion 4 days after Agrobacterium elimination (Toki et al. 2006). We observed transformed cells in the early transformation stage just after the onset of selection. Since the auto-fluorescence of rice callus can interfere with the observation of GFP fluorescence emitted from transformed cells, we used the Luc luminescence system, in which background emitted from rice callus can be reduced. Luc luminescence from transformed calli in Kasalath was found to be higher than that in Nipponbare just after the onset of selection (Fig. 3c, d). These results suggested that callus of Kasalath contains more transformed cells than that of Nipponbare in later stages because of the larger number of transformed cells of Kasalath at the early stage.
Southern blot analysis using EcoRI-digested or non-digested genomic DNA was performed to estimate the integrated T-DNA copy number in transformed callus. Comparison of the band pattern detected by hybridization with a GFP probe between EcoRI-digested or non-digested genomic DNA showed that the high molecular weight bands observed in non-digested DNA (indicated by a black arrowhead; Fig. 4a) were not detected following digestion with EcoRI, suggesting that they represented T-DNA integrated into the rice genome. Moreover, only the upper band of two bands observed in non-digested DNA (white arrowheads; Fig. 4a) was detected following digestion with EcoRI, suggesting that this band represented the 10.2 kb binary vector derived from survival of Agrobacterium in rice callus. Southern blot analysis using a GFP probe showed that the intensity of bands corresponding to T-DNA integrated into the genome of Kasalath was higher than that of Nipponbare (Fig. 4a). In addition, we examined expression levels of the gfp gene in transformed callus of Nipponbare and Kasalath at both early (just after onset of selection) and late (7 days after onset of selection) stages of transformation. The levels of gfp mRNA and GFP protein accumulation in transformed callus of Kasalath were higher than those in Nipponbare at both timepoints (Fig. 4b, c). These results again support the finding that callus of Kasalath contains more transformed cells than that of Nipponbare.
Fig. 4.
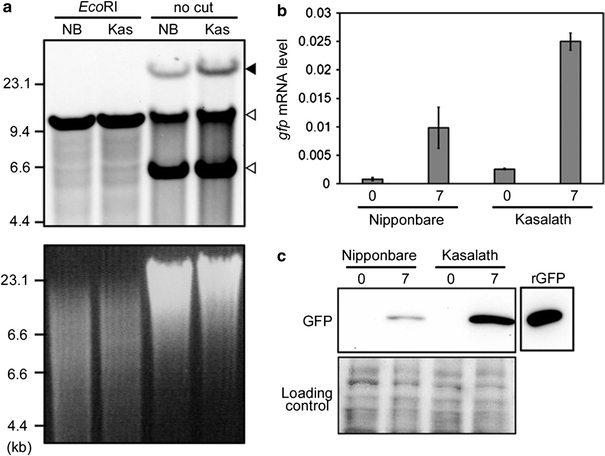
Molecular analysis of transformed calli of Nipponbare and Kasalath. a Integrated T-DNA copy number in transformed callus of Nipponbare (NB) and Kasalath (Kas). Southern blot analysis using a GFP probe (shown in Fig. 1) and 5 μg of genomic DNA extracted from transformed primary calli 3 days after the onset of Agrobacterium co-cultivation and digested with EcoRI or non-digested (no cut). Upper panel Southern blot analysis using GFP probe. Lower panel ethidium bromide (EtBr) staining of genomic DNA as a loading control. Black and white arrowheads show the bands corresponding to T-DNA integrated into rice genome and binary vector in Agrobacterium, respectively. b gfp mRNA levels in transformed calli. For qRT-PCR, 5 ng of total RNA extracted from Agrobacterium-co-cultivated calli was used as a template. All mRNA levels were normalized to the OsActin1 level as a control. Data are the mean ± SD of three separate PCR analyses 0 and 7 days after the onset of Hyg selection. c Immuno-blot analysis of GFP protein expression in Agrobacterium co-cultivated calli. 15 μg of total soluble proteins extracted from transformed calli and 10 ng recombinant GFP protein (rGFP) were separated by SDS-PAGE and analyzed with an anti-GFP antibody. Upper panel western blot analysis. Lower panel membrane stained with MemCode Reversible Protein Stain Kit after blotting as a loading control 0 and 7 days after the onset of Hyg selection
Careful observation of transformed calli under blue light showed that GFP spots were dispersed over the entire surface of transformed primary callus, suggesting that transformation events occurred independently in each GFP positive cell (Fig. 3b; Online Resource 2a). We checked whether independent transformation events in Kasalath occurred with higher frequency than in Nipponbare. As shown schematically in Fig. 5a, every piece of secondary callus grown under Hyg selection was propagated and some of them were used for molecular analysis. The number of emerging Hyg-resistant secondary calli per primary callus co-cultivated with Agrobacterium in Kasalath was over eightfold higher than that in Nipponbare (Table 2). Southern blot analysis showed that almost all the Hyg-resistant secondary calli isolated were derived from independent transformation events in Nipponbare and Kasalath, and there was no significant difference between the number of T-DNA copies integrated into the genome of isolated Hyg-resistant secondary calli between Nipponbare and Kasalath (Table 2; Fig. 5b). Moreover, single copy T-DNA integration was observed to have occurred in ~40% of isolated secondary calli of both Nipponbare and Kasalath (Fig. 5c). Thus, we confirmed that successful independent transformation events in Kasalath occur at a frequency over eightfold higher than in Nipponbare.
Fig. 5.
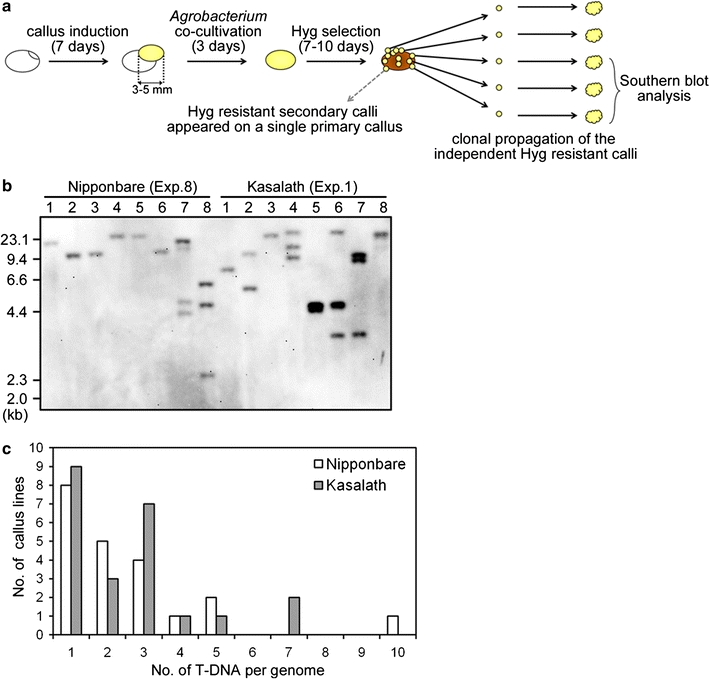
Independent transformation events in primary callus. a Schematic representation of Hyg-tolerant callus isolation. 7-day-old primary callus derived from mature seeds of Nipponbare or Kasalath was co-cultivated with Agrobacterium harboring a binary vector, pCAMBIA1390-sGFP for 3 days. After Agrobacterium-infection, washed calli were grown under Hyg pressure for 7–10 days. The resulting Hyg-tolerant callus pieces were isolated and propagated independently. To confirm whether independent transformation events had occurred successfully, some lines were chosen randomly and used for Southern blot analysis. b Southern blot analysis of 1 μg of EcoRI-digested genomic DNA extracted from Hyg-tolerant calli isolated individually from one primary callus using the gfp probe shown in Fig. 1. Lanes 8 callus lines of Nipponbare-Exp.8 and Kasalath-Exp.1 (Table 2). c T-DNA copy number in isolated clonal propagated calli. The x and y axes show T-DNA copy number and the frequency of transformed callus lines, respectively
Table 2.
Independent transformation events from one primary callus
| No. of Hyg-resistant secondary callus lines observed on a single primary callus | No. of Hyg-resistant callus lines analyzeda | Average copy no. of T-DNAa | |
|---|---|---|---|
| Nipponbare-Exp.1 | 3 | 3/3 | 1.3 |
| Nipponbare-Exp.2 | 4 | 4/4 | 2 |
| Nipponbare-Exp.3 | 5 | 4/5 | 4.2 |
| Nipponbare-Exp.4 | 9 | n.d. | n.d. |
| Nipponbare-Exp.5 | 2 | n.d. | n.d. |
| Nipponbare-Exp.6 | 3 | n.d. | n.d. |
| Nipponbare-Exp.7 | 4 | 4/4 | 3.5 |
| Nipponbare-Exp.8 | 8 | 6/8 | 1.8 |
| Nipponbare-Exp.9 | 8 | n.d. | n.d. |
| Nipponbare-Exp.10 | 8 | n.d. | n.d. |
| Average | 5.4 ± 2.6b | 2.6 ± 2.1 | |
| Kasalath-Exp.1 | >50c | 8/8 | 2 |
| Kasalath-Exp.2 | 42 | 7/7 | 3.7 |
| Kasalath-Exp.3 | 41 | 8/8 | 2.1 |
| Kasalath-Exp.4 | >50c | n.d. | n.d. |
| Kasalath-Exp.5 | >50c | n.d. | n.d. |
| Average | 46.6 ± 4.7b | 2.6 ± 1.8 |
aIndependent transformation events and T-DNA copy numbers were determined by Southern blot analysis as shown in Fig. 5b
bSignificant difference between Nipponbare and Kasalath at P < 0.01, as determined by t test
cCalculated as 50
Furthermore, we determined the transformation frequency in secondary callus of Nipponbare and Kasalath using the Luc reporter system. Similar to the results from primary calli, Luc luminescence from transformed secondary calli in Kasalath was found to be much higher than that in Nipponbare at both 0 and 7 days after selection (Fig. 6). This result suggested that Kasalath callus is highly competent for Agrobacterium-mediated transformation compared to Nipponbare, regardless of whether primary or secondary callus is used.
Fig. 6.
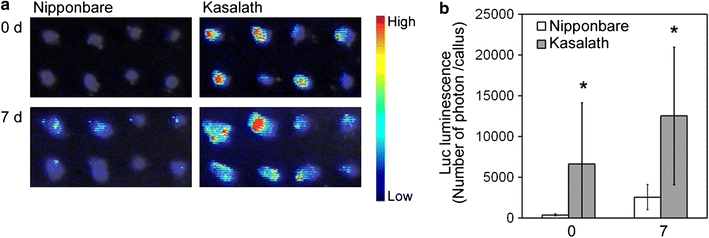
Transformed secondary calli of Nipponbare and Kasalath. a Luminescence emitted from secondary callus transformed with pCAMBIA1390-Luc 0 or 7 days after the onset of Hyg selection. Luminescence intensity is indicated by the false color scale. b Graphic representation of the Luc luminescence intensity emitted from the transformed Nipponbare (white bars) or Kasalath (gray bars) calli shown in a. The y axis shows luminescence level (photons) per callus. Data are presented as mean ± SD (n = 40). The experiments were performed twice using different samples. Similar results were obtained in another experiment. Asterisks indicate significant difference between Nipponbare and Kasalath at P < 0.01 as determined by t test
Agrobacterium density in co-cultivation with callus of Kasalath
The density of Agrobacterium in co-cultivation with rice callus is considered an important factor affecting transformation efficiency. We next examined the effect of low bacterial density in co-cultivation on transformation efficiency in Kasalath. Primary callus of Nipponbare and Kasalath were transformed with various densities of Agrobacterium harboring pCAMBIA1390-Luc (Fig. 1). In the early transformation stage, transformation frequency was estimated as Luc luminescence intensity. Dilution of Agrobacterium density to 10−1 to 10−5 reduced the luminescence intensity in both Nipponbare and Kasalath (Fig. 7a, b). However, luminescence intensities from transformed calli of Kasalath were found to be consistently higher than those of Nipponbare at any bacterial density in co-cultivation (Fig. 7a, b). Similar results were observed at later transformation stages in the evaluation of Hyg-tolerant cells (Fig. 7c; Table 3). All calli of Kasalath were transformed successfully (cf. 10–25% in Nipponbare) when the Agrobacterium density was adjusted to OD = 10−3 in co-cultivation (Table 3). Surprisingly, some calli of Kasalath gave rise successfully to transformed cells even at very low density (OD = 10−5) of Agrobacterium co-cultivation (Table 3).
Fig. 7.
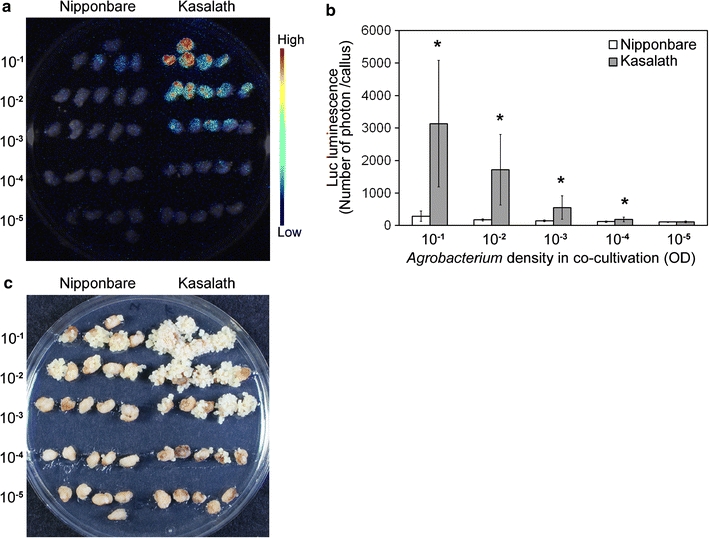
Effect of Agrobacterium density on transformation frequency. a Luminescence emitted from callus transformed with pCAMBIA1390-Luc just after the onset of Hyg selection. Luminescence intensity is indicated by the false color scale. b Graphical representation of the Luc luminescence intensity emitted from the transformed Nipponbare (white bars) or Kasalath (gray bars) calli shown in a. The x and y axes show Agrobacterium density in co-cultivation and luminescence level (photon) per callus, respectively. Data are presented as mean ± SD (n = 10). The experiments were performed twice using different samples. Similar results were obtained in both experiments. Asterisks indicate significant difference between Nipponbare and Kasalath in each Agrobacterium density at P < 0.01 as determined by t test. c Hyg-tolerant cells 14 days after onset of selection. 10−1 to 10−5 Agrobacterium density in co-cultivation with rice callus (OD600). The frequencies of Hyg-tolerant cells are listed in Table 3
Table 3.
Effect of Agrobacterium density on the transformation frequency of Nipponbare and Kasalath
| Transformation frequencya | Agrobacterium density (OD600) | ||||
|---|---|---|---|---|---|
| 10−1 | 10−2 | 10−3 | 10−4 | 10−5 | |
| Nipponbare-Exp.1 | 9/10 | 10/10 | 1/10 | 1/10 | 0/10 |
| Kasalath-Exp.1 | 9/10 | 10/10 | 10/10 | 6/10 | 1/10 |
| Nipponbare-Exp.2 | 8/16 | 9/16 | 4/16 | 0/16 | 0/16 |
| Kasalath-Exp.2 | 16/16 | 16/16 | 16/16 | 12/16 | 3/16 |
aTransformation frequency was defined as the number of calli that could produce Hyg-resistant secondary calli/number of primary calli inoculated with Agrobacterium
Discussion
Fast and efficient transformation in Kasalath
In this study, we successfully produced transgenic plants of Kasalath using primary callus derived from mature seeds (Fig. 2). To date, transformation experiments using secondary callus derived from mature seeds and callus derived from immature embryos of Kasalath have been reported (Du et al. 2009; Hiei and Komari 2008; Nishimura et al. 2006; Prodhan et al. 2008; Yamaya et al. 2002). However, there are no reports of transformation using primary callus of Kasalath derived from mature seeds.
Genetic information on Kasalath, such as end sequencing and chromosomal mapping of bacterial artificial chromosome clones, as well as a high density linkage map comparing Kasalath with Nipponbare and Koshihikari are already available (Ebitani et al. 2005; Harushima et al. 1998; Katagiri et al. 2004). Moreover, QTL analysis between Kasalath and Koshihikari has shown that Kasalath has alleles with positive effects for 14 agronomic and morphological traits (Madoka et al. 2008). Thus, Kasalath has been used frequently for the isolation of genes controlling useful agronomic traits in forward genetics, and could be used to improve agronomic traits including nutrient and water uptake, and yield in japonica rice. Our rapid and efficient transformation system could help to research functional genomics and could be applied to molecular breeding in Kasalath.
Luc—a highly-sensitive transformation monitoring system
As described above, T-DNA integration steps can be divided into two main stages: transient expression of double-stranded T-DNA in the nucleus at the early stage and stable expression of T-DNA integrated into the plant genome at the late stage. In this study, we were able to visualize and sequentially observe independent transformation events in transformed cells using GFP and the Luc system (Figs. 3, 6). This is the first report of sequential comparison of transformation processes between rice cultivars using visual markers. We applied our transformation system to both primary and secondary callus (Figs. 3, 6; Online Resource 2). However, in our high-speed transformation system, transformed primary callus proved more suitable for sequential observation than secondary callus because of the rapid change in shape of the latter due to the vigorous growth of both transformed and non-transformed cells. Indeed, this might explain why a high transformation frequency in Kasalath has not been reported to date.
This method could be also useful in analyzing details of the mechanism of T-DNA integration into the plant genome. Forward and reverse genetic approaches have succeeded in isolating over 125 genes involved in Agrobacterium-mediated transformation in Arabidopsis (Zhu et al. 2003), some of which have been characterized. In yeast, non-homologous end joining (NHEJ)—the mechanism used to repair DNA double strand breaks—plays a crucial role in Agrobacterium T-DNA integration into the genome. However, the mechanism of T-DNA integration via NHEJ in plants remains unclear, although analyses of T-DNA integration and transformation efficiency have been performed using NHEJ mutants such as atligIV and atku80 (Gelvin 2008). We are currently establishing a transformation monitoring system to analyze the molecular mechanisms of T-DNA integration by visualizing transient and stable expressions of T-DNA. The mechanism of NHEJ in T-DNA integration in plants could be elucidated using such a system.
Application of highly efficient transformation to new technologies
In a previous study, we succeeded in obtaining transformed calli with high efficiency (95–98%) by use of primary calli in Nipponbare (Toki et al. 2006). This level of efficiency is thought to be adequate to produce transgenic plants for functional genomics of genes of interest because we succeeded in clonal propagation of at least one cluster of successfully transformed cells in almost all co-cultivated seeds (Toki et al. 2006). However, for some purposes, such as the large-scale identification of gene function using transgenic plants, visual selection under no antibiotic pressure, and gene modification by gene targeting (GT) via homologous recombination (HR), much higher transformation efficiencies, i.e., many more transformed cells per co-cultivated callus, are required. Targeted gene modification by GT via HR has been reported in rice (Endo et al. 2007; Saika and Toki 2009a; Terada et al. 2007; Terada et al. 2002; Yamauchi et al. 2009). The efficiency of obtaining cells in which GT had successfully occurred was very low because the efficiency of HR between the targeted genomic locus and the donor DNA was quite low, and the ratio of random integration of donor DNA into the genome in higher plants is of the order of 10−3 to 10−6 (Iida and Terada 2005). Both improving the efficiency of HR and increasing the number of cells in which donor DNAs are integrated into plant nuclear DNA would lead to increased GT efficiency (Saika and Toki 2009a). In this study, we showed that the number of independent transformation events in each primary callus of Kasalath was ca. 10 times greater than that in Nipponbare, and that single copy of T-DNA integration was observed in ~40% of these events (Table 2; Fig. 5). This highly efficient transformation of Kasalath suggests that this variety could be used to improve the efficiency of some of these new transformation techniques.
Transformation efficiency is thought to be determined by the efficiency of tissue culture and the ability to accept a transgene via Agrobacterium; both these characteristics are regulated by both genetic and physiological factors. It has been shown that IR64 and IR72 are recalcitrant to tissue culture compared to Nipponbare and Kasalath, with consequent low transformation efficiencies. On the other hand, Kasalath showed higher competency for Agrobacterium-mediated transformation compared to Nipponbare in spite of the fact that the tissue culture performance of both cultivars were comparable. Thus, Kasalath is a good candidate cultivar for the isolation of genes involved in high competency for Agrobacterium-mediated transformation. The isolation and characterization of genes involved in this trait would be of interest not only from the point of view of basic research into the rice–Agrobacterium interaction, but also in terms of improving transformation technology. We are currently performing genetic analysis to isolate and characterize the genes involved in this trait using our highly sensitive transformation monitoring system.
Agrobacterium density and transformation frequency
Agrobacterium density in co-cultivation affects transformation efficiency. Under our experimental conditions, we succeeded in clonal propagation of Hyg-resistant cells at a low frequency of 10–20% even at very low bacterial density (OD = 10−5) in co-cultivation (Fig. 7c; Table 3). It has been suggested that Agrobacterium induces browning and programmed cell death in non-host plant cells. Inhibition of cell death as well as efficient T-DNA transfer can have a dramatic impact on transformation efficiency (Khanna et al. 2007). Moreover, overgrowth of Agrobacterium is thought to cause a decrease in transformation efficiency. On the contrary, a high density of Agrobacterium in co-cultivation enhanced transformation frequency (Fig. 7). Thus, high competency to Agrobacterium-mediated transformation observed in Kasalath could be applied to the transformation of any plant hyper-susceptible or resistant to Agrobacterium infection. Our results suggest that transformation efficiency could be improved if survival of Agrobacterium after the onset of selection can be prevented by the use of appropriate culture conditions and efficient antibiotics. It is possible that cultivation under high temperature conditions (32–33°C) also stimulates removal of Agrobacterium in rice (Toki et al. 2006). In addition, application of meropenem—a β-lactam antibiotic—was reported to improve transformation efficiency without the need for modification of protocols in many plants (Ogawa and Mii 2007).
Conclusion
In conclusion, we have succeeded in obtaining regenerated transformed plants of the rice indica variety Kasalath via Agrobacterium-mediated transformation using primary callus derived from mature seeds. Furthermore, Kasalath is shown to be more competent for Agrobacterium co-cultivation and subsequent clonal propagation than Nipponbare. This higher transformation competency of Kasalath could be exploited in the application of new transformation technologies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank M. Yano for providing Kasalath seeds, Y. Niwa for providing sGFP, A. Baba for providing pCAMBIA1390-sGFP and, K. Osakabe and S. Nonaka for providing the luc expression vector. We also thank K. Osakabe, K. Abe, M. Endo, S. Nonaka and N. Ohtsuki for stimulating discussion, and K. Amagai, R. Aoto, C. Furusawa, A. Nagashii, E. Ozawa and F. Suzuki for technical help. This work was supported financially by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan to H.S. and S.T., and a grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan to H.S. This work was also supported by a Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) grant to S.T. and the Budget for Nuclear Research of the Ministry of Education, Culture, Sports, Science and Technology, based on screening and counseling by the Atomic Energy Commission to S.T.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- GFP
Green fluorescent protein
- hpt
Hygromycin phosphotransferase
- hyg
Hygromycin
- Luc
Luciferase
References
- Datta K, Koukolikova-Nicola Z, Baisakh N, Oliva N, Datta SK. Agrobacterium-mediated engineering for sheath blight resistance of indica rice cultivars from different ecosystems. Theor Appl Genet. 2000;100:832–839. doi: 10.1007/s001220051359. [DOI] [Google Scholar]
- Dong JJ, Teng WM, Buchholz WG, Hall TC. Agrobacterium-mediated transformation of Javanica rice. Mol Breed. 1996;2:267–276. doi: 10.1007/BF00564204. [DOI] [Google Scholar]
- Du B, Zhang WL, Liu BF, Hu J, Wei Z, Shi ZY, He RF, Zhu LL, Chen RZ, Han B, He GC. Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc Natl Acad Sci USA. 2009;106:22163–22168. doi: 10.1073/pnas.0912139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebitani T, Takeuchi Y, Nonoue Y, Yamamoto T, Takeuchi K, Yano M. Construction and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of indica rice cultivar ‘Kasalath’ in a genetic background of japonica elite cultivar ‘Koshihikari’. Breed Sci. 2005;55:65–73. doi: 10.1270/jsbbs.55.65. [DOI] [Google Scholar]
- Endo M, Osakabe K, Ono K, Handa H, Shimizu T, Toki S. Molecular breeding of a novel herbicide-tolerant rice by gene targeting. Plant J. 2007;52:157–166. doi: 10.1111/j.1365-313X.2007.03230.x. [DOI] [PubMed] [Google Scholar]
- Gelvin SB. Function of host proteins in the Agrobacterium-mediated plant transformation process. In: Tzfira T, Citovsky V, editors. Agrobacterium: from biology to biotechnology. New York: Springer; 2008. pp. 483–522. [Google Scholar]
- Harushima Y, Yano M, Shomura P, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin SY, Antonio BA, Parco A, Kajiya H, Huang N, Yamamoto K, Nagamura Y, Kurata N, Khush GS, Sasaki T. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics. 1998;148:479–494. doi: 10.1093/genetics/148.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Komari T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat Protoc. 2008;3:824–834. doi: 10.1038/nprot.2008.46. [DOI] [PubMed] [Google Scholar]
- Hood EE, Gelvin SB, Melchers LS, Hoekema A. New Agrobacterium helper plasmids for gene-transfer to plants. Transgenic Res. 1993;2:208–218. doi: 10.1007/BF01977351. [DOI] [Google Scholar]
- Iida S, Terada R. Modification of endogenous natural genes by gene targeting in rice and other higher plants. Plant Mol Biol. 2005;59:205–219. doi: 10.1007/s11103-005-2162-x. [DOI] [PubMed] [Google Scholar]
- Ishizaki T, Kumashiro T. Genetic transformation of NERICA, interspecific hybrid rice between Oryza glaberrima and O.sativa, mediated by Agrobacterium tumefaciens. Plant Cell Rep. 2008;27:319–327. doi: 10.1007/s00299-007-0465-x. [DOI] [PubMed] [Google Scholar]
- Katagiri S, Wu JZ, Ito YK, Karasawa W, Shibata M, Kanamori H, Katayose Y, Namiki N, Matsumoto T, Sasaki T. End sequencing and chromosomal in silico mapping of BAC clones derived from an indica rice cultivar, Kasalath. Breed Sci. 2004;54:273–279. doi: 10.1270/jsbbs.54.273. [DOI] [Google Scholar]
- Khanna HK, Paul JY, Harding RM, Dickman MB, Dale JL. Inhibition of Agrobacterium-induced cell death by antiapoptotic gene expression leads to very high transformation efficiency of banana. Mol Plant Microbe Interact. 2007;20:1048–1054. doi: 10.1094/MPMI-20-9-1048. [DOI] [PubMed] [Google Scholar]
- Kumar KK, Maruthasalam S, Loganathan M, Sudhakar D, Balasubramanian P. An improved Agrobacterium-mediated transformation protocol for recalcitrant elite indica rice cultivars. Plant Mol Biol Rep. 2005;23:67–73. doi: 10.1007/BF02772648. [DOI] [Google Scholar]
- Labra M, Savini C, Bracale M, Pelucchi N, Colombo L, Bardini M, Sala F. Genomic changes in transgenic rice (Oryza sativa L.) plants produced by infecting calli with Agrobacterium tumefaciens. Plant Cell Rep. 2001;20:325–330. doi: 10.1007/s002990100329. [DOI] [Google Scholar]
- Madoka Y, Kashiwagi T, Hirotsu N, Ishimaru K. Indian rice “Kasalath” contains genes that improve traits of Japanese premium rice “Koshihikari”. Theor Appl Genet. 2008;116:603–612. doi: 10.1007/s00122-007-0693-z. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Hakata M, Amano K, Miyao A, Toki N, Kajikawa M, Pang J, Higashi N, Ando S, Toki S, Fujita M, Enju A, Seki M, Nakazawa M, Ichikawa T, Shinozaki K, Matsui M, Nagamura Y, Hirochika H, Ichikawa H. A genome-wide gain-of-function analysis of rice genes using the FOX-hunting system. Plant Mol Biol. 2007;65:357–371. doi: 10.1007/s11103-007-9243-y. [DOI] [PubMed] [Google Scholar]
- Nishimura A, Aichi I, Matsuoka M. A protocol for Agrobacterium-mediated transformation in rice. Nat Protoc. 2006;1:2796–2802. doi: 10.1038/nprot.2006.469. [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Mii M. Meropenem and moxalactam: novel beta-lactam antibiotics for efficient Agrobacterium-mediated transformation. Plant Sci. 2007;172:564–572. doi: 10.1016/j.plantsci.2006.11.003. [DOI] [Google Scholar]
- Osakabe K, Abe K, Yoshioka T, Osakabe Y, Todoriki S, Ichikawa H, Hohn B, Toki S (2006) Isolation and characterization of the RAD54 gene from Arabidopsis thaliana. Plant J 48:827–842 [DOI] [PubMed]
- Parkhi V, Rai M, Tan J, Oliva N, Rehana S, Bandyopadhyay A, Torrizo L, Ghole V, Datta K, Datta SK. Molecular characterization of marker-free transgenic lines of indica rice that accumulate carotenoids in seed endosperm. Mol Genet Genomics. 2005;274:325–336. doi: 10.1007/s00438-005-0030-7. [DOI] [PubMed] [Google Scholar]
- Prodhan SH, Hossain A, Nagamiya K, Komamine A, Morishima H. Improved salt tolerance and morphological variation in indica rice (Oryza sativa L.) transformed with a catalase gene from E. coli. Plant Tissue Cult Biotechnol. 2008;18:57–63. [Google Scholar]
- Rashid H, Yokoi S, Toriyama K, Hinata K. Transgenic plant production mediated by Agrobacterium in Indica rice. Plant Cell Rep. 1996;15:727–730. doi: 10.1007/BF00232216. [DOI] [PubMed] [Google Scholar]
- Saika H, Toki S. Towards a highly efficient gene targeting system in higher plants. JARQ. 2009;43:81–85. doi: 10.1007/s10817-009-9127-8. [DOI] [Google Scholar]
- Saika H, Toki S. Visual selection allows immediate identification of transgenic rice calli efficiently accumulating transgene products. Plant Cell Rep. 2009;28:619–626. doi: 10.1007/s00299-009-0671-9. [DOI] [PubMed] [Google Scholar]
- Terada R, Urawa H, Inagaki Y, Tsugane K, Iida S. Efficient gene targeting by homologous recombination in rice. Nat Biotechnol. 2002;20:1030–1034. doi: 10.1038/nbt737. [DOI] [PubMed] [Google Scholar]
- Terada R, Johzuka-Hisatomi Y, Saitoh M, Asao H, Iida S. Gene targeting by homologous recombination as a biotechnological tool for rice functional genomics. Plant Physiol. 2007;144:846–856. doi: 10.1104/pp.107.095992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006;47:969–976. doi: 10.1111/j.1365-313X.2006.02836.x. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Johzuka-Hisatomi Y, Fukada-Tanaka S, Terada R, Nakamura I, Iida S. Homologous recombination-mediated knock-in targeting of the MET1a gene for a maintenance DNA methyltransferase reproducibly reveals dosage-dependent spatiotemporal gene expression in rice. Plant J. 2009;60:386–396. doi: 10.1111/j.1365-313X.2009.03947.x. [DOI] [PubMed] [Google Scholar]
- Yamaya T, Obara M, Nakajima H, Sasaki S, Hayakawa T, Sato T. Genetic manipulation and quantitative-trait loci mapping for nitrogen recycling in rice. J Exp Bot. 2002;53:917–925. doi: 10.1093/jexbot/53.370.917. [DOI] [PubMed] [Google Scholar]
- Zhao LN, Zhou HJ, Lu LX, Liu L, Li XH, Lin YJ, Yu SB. Identification of quantitative trait loci controlling rice mature seed culturability using chromosomal segment substitution lines. Plant Cell Rep. 2009;28:247–256. doi: 10.1007/s00299-008-0641-7. [DOI] [PubMed] [Google Scholar]
- Zhu YM, Nam J, Humara JM, Mysore KS, Lee LY, Cao HB, Valentine L, Li JL, Kaiser AD, Kopecky AL, Hwang HH, Bhattacharjee S, Rao PK, Tzfira T, Rajagopal J, Yi HC, Veena, Yadav BS, Crane YM, Lin K, Larcher Y, Gelvin MJK, Knue M, Ramos C, Zhao XW, Davis SJ, Kim SI, Ranjith-Kumar CT, Choi YJ, Hallan VK, Chattopadhyay S, Sui XZ, Ziemienowicz A, Matthysse AG, Citovsky V, Hohn B, Gelvin SB (2003) Identification of Arabidopsis rat mutants. Plant Physiol 132:494–505 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


