Abstract
Urease plays a central role in the pathogenesis of Helicobacter pylori in humans. Maturation of this nickel metalloenzyme in bacteria requires the participation of the accessory proteins UreD (termed UreH in H. pylori), UreF, and UreG which form sequential complexes with the urease apoprotein as well as UreE, a metallochaperone. Here, we describe the crystal structure of C-terminal truncated UreF from H. pylori (residues 1-233), the first UreF structure to be determined, at 1.55 Å resolution using SAD methods. UreF forms a dimer in vitro and adopts an all-helical fold congruent with secondary structure prediction. On the basis of evolutionary conservation analysis, the structure reveals a probable binding surface for interaction with other urease components as well as key conserved residues of potential functional relevance.
Keywords: urease, sequence conservation, ConSurf, protein-protein interactions, dimer, bacteria
INTRODUCTION
Helicobacter pylori is a Gram-negative, microaerophilic bacterium present in the stomachs of approximately half of the human population.1,2 Chronic infection by this microorganism can, in certain individuals, give rise to gastric and duodenal ulcers, gastric adenocarcinoma, and mucosa-associated lymphoid tissue lymphoma. To colonize the stomach and persist in this acidic environment, H. pylori synthesizes prodigious quantities of urease – estimated to account for 6% of the soluble cellular protein.3 Urea that diffuses into the stomach from the bloodstream is hydrolyzed by the cytoplasmic and surface-associated enzyme, resulting in the production of localized concentrations of ammonia and bicarbonate that protect the cells by acting as buffering compounds.4
The crystal structure of H. pylori urease5 reveals it to be a [(UreAB)3]4 supramolecular assembly of two component subunits, where each UreAB catalytic unit contains a buried dinuclear nickel active site. Many other bacteria, fungi, and plants possess nickel-dependent ureases that are related in sequence to that of H. pylori.6 In most bacteria, however, two subunits (also named UreA and UreB) replace the fused UreA of H. pylori and a third subunit, comparable to the H. pylori UreB, is termed UreC. The enzymes from Klebsiella aerogenes and Bacillus pasteurii exhibit (UreABC)3 structures7-9 comparable to one face of the tetrahedral [(UreAB)3]4 structure from H. pylori. In eukaryotes, ureases typically possess a single type of subunit that corresponds to a fusion of the individual bacterial subunits. The structure of the enzyme from jack bean10 reveals a dimer of the trimeric protein [(urease)3]2.
Synthesis of active urease in H. pylori depends on an intricate interplay of numerous accessory proteins.11 Several of these maturation proteins are the products of genes located in the same ureABIEFGH cluster as the subunit genes.12 In addition, hydrogenase accessory proteins HypA and HypB, encoded by hypA and hypB in a separate gene cluster, are needed for activation of both urease and another nickel-containing enzyme, hydrogenase.13 The urease activation functions of only a few of these proteins have been elucidated in H. pylori. UreI is not required for urease synthesis; rather, this H. pylori protein forms a proton-gated urea channel that allows substrate to access the enzyme.14 H. pylori UreE binds one mole of nickel per mole of dimer,15,16 and is proposed to function as the metallochaperone for nickel delivery to urease apoprotein. Despite having a GTP-binding sequence, purified H. pylori UreG possesses negligible GTPase activity.17 A complex between UreE and UreG is stabilized by zinc, but not nickel, ions.16 HypA and HypB were purified from H. pylori and characterized.18 HypA binds stoichiometric amounts of nickel, HypB (containing a GTP-binding motif) hydrolyzes GTP with a turnover number of five min−1, and the two proteins were shown to form a HypA-HypB complex.18 Furthermore, HypA and UreE could be chemically cross-linked, consistent with generation of a HypA-UreE heterodimer.19 Yeast two-hybrid analysis was used to demonstrate UreA-UreA, UreA-UreB, UreA-UreH, UreE-UreG, and UreF-UreH pairwise interactions.20,21 No published reports have thus far described the purification and characterization of UreF or UreH from H. pylori.
Additional insights into the functions and properties of the urease accessory proteins are available from work in other organisms. For example, examination of components of the K. aerogenes ureABCDEFG gene cluster (ureD is an orthologue of H. pylori ureH) expressed in Escherichia coli has been used to generate a model in which UreD, UreF, and UreG bind sequentially22-24 or as a UreDFG complex25 to the urease apoprotein. As shown by in vitro functional studies, fully active urease requires the complete set of accessory proteins, nickel ions, bicarbonate (to form carboxylysine, a bridging ligand of the dinuclear metallocenter), and GTP.26,27 The K. aerogenes urease apoprotein was crystallized28, the metal-binding UreE metallochaperone was extensively characterized29-34 including structural determination35, the UreG GTPase was investigated25, and the UreF protein was purified as a fusion protein with UreE36 or maltose binding protein.37 UreE and UreG orthologues have been examined from other bacteria (e.g.,38-42) with generally similar properties, and the structure of Methanocaldococcus jannaschii HypB, related to UreG, has been elucidated.43 The least understood of the urease accessory proteins are UreD and UreF.
Here, we describe the crystal structure of a truncated form of UreF from H. pylori, henceforth denoted HpUreF, the first UreF to be structurally characterized. Our structure suggests that the protein forms a dimer with a novel all-helical topology and it reveals a probable binding surface for interaction with other urease components based on sequence conservation analysis. We also show that the current model bears little structural resemblance to known GTPase activating proteins.
MATERIALS AND METHODS
Protein Expression and Purification
The ureF gene of H. pylori strain 26695 was amplified from genomic DNA (ATCC) and cloned into a modified pET15b vector (EMD Novagen, Madison, WI) by using standard molecular biology techniques. The resulting vector was transformed into E. coli BL21-CodonPlus (DE3)-RIPL (Stratagene, La Jolla, CA). Cells were grown on selenomethionine medium (Medicilon, Chicago) in 1 L Tunair flasks at 37°C to an OD600 of 1.2, then the temperature was lowered to 16°C and IPTG was added to 0.5 mM. Expression was allowed to proceed overnight, at which point the cells were harvested by centrifugation, flash-frozen in liquid nitrogen, and stored at −80°C.
Cells were thawed on ice and resuspended in binding buffer (50 mM HEPES, pH 7.5, 500 mM NaCl, 5% glycerol, 0.2 mM tris(2-carboxyethyl)phosphine, TCEP) supplemented with 0.5% CHAPS, phenylmethylsulfonylfluoride, and benzamidine. After disruption by sonication and centrifugation at 60,000 g for 40 min, the cell free extracts were applied to a DE-52 column (Ø2.6 × 7 cm) that had been equilibrated with the same buffer and the flow-through fraction was loaded by gravity flow onto a Ni-nitrilotriacetic acid (NTA) column (Qiagen, Germantown, MD). The column was washed with 20 volumes of wash buffer (50 mM HEPES, pH 7.5, 500 mM NaCl, 5% glycerol, 25 mM imidazole, and 0.2 mM TCEP) supplemented with 0.5% CHAPS, followed by 10 volumes of wash buffer. The His6-tagged protein was eluted with the same buffer containing 250 mM imidazole. This sample was concentrated by using a VIVASpin unit (Sartorius NA, Edgewood, NY) and loaded onto a Superdex 200 column (Ø2.6 × 60 cm, GE Healthcare) equilibrated with gel filtration buffer (10 mM HEPES, pH 7.5, 250 mM NaCl, 0.2 mM TCEP). Elution was carried out at a flow rate of 3 mL min−1 at 8°C and HpUreF was eluted as a dimer. The final protein sample was concentrated to 41 mg/ml, divided into 1.5-mg aliquots, flash-frozen, and stored at −80°C.
The protein sample that produced diffracting crystals had two bands with apparent size of 28 and 31 kDa on denaturing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE). All protein samples (selenomethionine-labeled as well as native) demonstrated similar truncation which seemed to occur during expression (and possibly purification). Electrospray ionization (ESI)-time-of-flight (ToF) mass spectrometry (MS) (1 Da error margin) analysis showed the presence of multiple protein species. Major components included the N-terminally His-tagged (MGSSHHHHHHSSGLVPRGSH) full length species (Gly-2′ to Ser-254, 31 kDa on SDS PAGE) and a variety of C-terminal truncations (28 kDa) (peptides ending with any of the four residues from Ala-233 to Gln-236).
Crystallization and Data Collection
Crystallization was performed using a selenomethionine-labeled HpUreF protein sample concentrated to 15 mg ml−1 in a buffer containing 10 mM HEPES at pH 7.5, 250 mM NaCl and 0.2 mM TCEP. Initial screening was carried out by sitting drop vapor diffusion at room temperature using a commercially available sparse-matrix crystallization screening kit (Wizard I from Emerald BioSystems), yielding thick crystals of rectangular shape which appeared after four days in Condition 10 (20% polyethylene glycol monomethylether 2000, TRIS pH 7.0). The crystals belong to the monoclinic space group C2 with unit cell constants of a=135.2 Å, b=89.4 Å, c=66.0 Å, and β=94.0°, containing three molecules per asymmetric unit. A single crystal from an optimized crystallization mixture containing 21% polyethylene glycol monomethylether 2000 and 100 mM Bis-TRIS (pH 6.8) was cryoprotected by soaking in well solution with 18% glycerol (v/v) for 30 s before flash freezing in liquid nitrogen. Diffraction data were collected at 100 K by using synchrotron radiation on beamline 17-ID (IMCA-CAT, Advanced Photon Source, Argonne National Laboratory) at the peak wavelength of the selenium K absorption edge (0.97926 Å). The data were integrated and scaled using HKL2000.44
Structure Determination and Refinement
The structure of HpUreF was determined by using the single-wavelength anomalous dispersion (SAD) method utilizing the anomalous signal from selenium atoms.45,46 Using diffraction data between 29 Å and 2.2 Å, the positions of 20 selenium atoms were found with the program SHELXD47 followed by heavy-atom refinement and phasing using the maximum likelihood-based algorithm as implemented in the autoSHARP program suite.48,49 Phase improvement by density modification generated an interpretable experimental SAD map at 1.55 Å, allowing an initial model (86% complete) to be built using ARP/wARP.50 The model was subsequently improved through alternate cycles of manual building using COOT51 and restrained refinement against a maximum-likelihood target with 5% of the reflections randomly excluded as an Rfree test set. All refinement steps were performed using REFMAC52 in the CCP4 program suite.53 Owing to poorly defined electron density at both N- and C-termini, the final model encompasses residues 27-233 (chain A) and residues 25-233 (chains B, C). The density for a flexible loop region (residues 48–52) in chain C was also missing and therefore not modeled. The final model comprising three molecules of HpUreF and solvent molecules refined to an Rwork of 18.7% and Rfree of 21.1%, including TLS parameterization.54,55 Data collection, phasing and structure-refinement statistics are summarized in Table 1. The Ramachandran plot generated by PROCHECK56 showed excellent stereochemistry overall, with 99.6% of the residues in the most favored (93.6%) and additionally allowed regions (6.1%) with no disallowed residues. Structure figures were prepared using PyMOL.57 Atomic coordinates and structure factors for HpUreF have been deposited in the Protein Data Bank with accession code 3CXN.
TABLE 1.
Data Collection and Refinement Statistics for the Structure of HpUreF
| HpUreF (SeMet, Peak) | |
|---|---|
| Data collection | |
| Space group | C2 |
| Unit cell dimensions a, b, c, β (Å, °) |
135.2, 89.4, 66.0, 94.0 |
| Wavelength, Å | 0.97926 |
| Resolution, Å | 50-1.55 (1.61-1.55)a |
| Rmerge, % | 5.1 (38.3) |
| I / σI) | 34.5 (3.3) |
| Completeness, % | 99.5 (99.1) |
| Multiplicity | 4.2 (4.1) |
| Number of unique reflections |
112962 |
| Refinement | |
| Rwork / Rfree, % | 18.7/21.1 |
| Number of atoms | |
| Protein | 5039 |
| Ligand/ion | 6 |
| Water | 401 |
| Average B-factors, Å2 | |
| Protein | 26.5 |
| Ligand/ion | 54.0 |
| Water | 35.2 |
| R.m.s deviations from ideal | |
| Bond lengths, Å | 0.010 |
| Bond angles, ° | 1.236 |
Values in parentheses correspond to the outermost resolution shell.
Sequence Conservation
The sequence of HpUreF was used to search for bacterial homologues in the UniProt Knowledgebase by using the program BLAST, available at http://ca.expasy.org/tools/blast. An E-value cutoff of 0.005 was used to ensure inclusion of the distant homologue K. aerogenes, which has emerged as a model system for studying urease accessory proteins. Subsequent removal of all redundant entries from the queried list gave a final result of 224 unique bacterial UreF sequences whereby all entries share less than 90% sequence identity. A multiple sequence alignment of the 224 UreF sequences was performed using the program MUSCLE, available at http://www.ebi.ac.uk/Tools/muscle/index.html, and the results used to calculate the degree of conservation of each residue in HpUreF using the ConSurf server,58-60 available at http://consurf.tau.ac.il. Conservation scores were also mapped onto the protein structure by the server using a representative coloring scheme. The JPRED server,61 available at http://www.compbio.dundee.ac.uk/www-jpred, was utilized for secondary structure prediction calculations.
Supplemental Data
Supplemental data include two figures and can be found with the article online.
RESULTS
Overall Structure
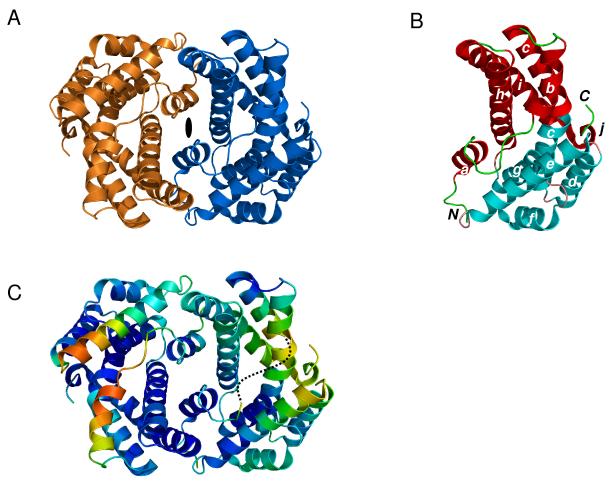
The structure of a C-terminal truncated HpUreF (residues 1-233) was determined to a resolution of 1.55 Å by using selenomethionine enrichment and SAD methods (Table 1). The crystallographic asymmetric unit contains three copies of the protein (denoted chains A, B, and C), and shows HpUreF to be a homodimer consistent with the observed migration position during gel filtration chromatography. In the present crystal form, chains A and C form a dimer while a second dimer is composed of chain B and its nearest symmetry equivalent (B’) (Figure 1A). The protomers in each dimer are related by a non-crystallographic (A and C) and crystallographic (B and B’) two-fold symmetry axis, respectively. Clear electron density allowed for structural characterization of residues 25-233 of the total 254 amino acids in this H. pylori urease accessory protein.
Figure 1.
A: Ribbon representation of the HpUreF dimer viewed down the crystallographic twofold axis (designated by solid oval). The two protein subunits are depicted in gold and blue, respectively. B: Tertiary structure of a protomer chain with helices labeled (a to j) from the N to C termini and color-coded to depict the two individual domains. The dimerization and insertion domains are shown in red (with green loops) and cyan (with salmon loops), respectively. C: Dimer comprising chains A (left) and C (right) color-coded according to temperature factors to highlight the most flexible parts of the structure. The dashed line denotes a disordered segment in chain C (residues 48–52), which was not included in the final model owing to poor electron density. Temperature factors are depicted on a scale going from blue to red to indicate increasing values of B-factors, respectively.
Superposition of the three polypeptide chains in the asymmetric unit revealed only minor differences (root-mean-square deviations for Cα atoms ~ 0.4 Å; data not shown). Discussion will be limited to chain B unless indicated otherwise. The HpUreF protomer adopts an all-helical fold that consists of a bundle of nine α-helices arranged in an antiparallel fashion (Figure 1B), terminated at the carboxy-terminal end by a short 310 helix (helix j). A pronounced kink of approximately 35° in the middle of a 35-residue central helix (helix c) further defines two domains within the overall structure. A domain involved in dimerization of HpUreF (denoted D domain henceforth and shown in red in Figure 1B) is highlighted by a 4-helix bundle comprising helix b (residues 50-60), the N-terminal portion of the kinked helix c (residues 65-81), helix h (residues 173-197) and helix i (residues 200-225). Additional flanking by an N-terminal helix a (residues 30-40), linked to helix b through a 9-residue loop, as well as a short C-terminal 310 helix j (residues 226-230) completes the description of the D domain. The a – b loop also represents the longest loop in the compact structure and the only one not employed as structural turns in the contiguous helix-turn-helix topology. Insertion of a 5-helix bundle between helix c and helix h introduces a second domain in the protomer structure (henceforth denoted I for insertion domain and shown in cyan in Figure 1B). The I domain is composed of the C-terminal portion of the kinked helix c (residues 82-98), helix d (residues 100-113), helix e (residues 117-138), helix f (residues 143-154), and helix g (residues 158-171). With the exception of a buried hydrophobic residue Met-138 (selenomethionine) at the dimer interface, the I domain participates minimally in the dimerization of HpUreF.
A six-helix sandwich grouped as two helical triads related by a two-fold symmetry axis defines the dimer interface. Helix a forms a helical triad with helix h’ and the N-terminal section of the kinked helix i’ from the opposing subunit. Within each triad, dimer interactions are characterized by a network of intersubunit hydrophobic, polar as well as ion pair interactions, supporting the notion that isolated HpUreF is a dimer. Multiple contacts define the interactions between helices a and h’ beginning with a salt bridge that is established at the base of helix a between Glu-32 and Arg-179′ (3.1 Å). Residue Asp-40 hydrogen bonds to Tyr-183′ (2.8 Å) as well as Ser-187′ (2.8 Å) through side-chain interactions. Intersubunit interactions also are observed between helices a and i’ featuring hydrogen bonds between the side-chain of Gln-37 and Gln-212′ (3.3 Å, side-chain) as well as Gln-205′ (2.8 Å, side-chain; 3.3 Å, main-chain carbonyl). Furthermore, the carboxylate of Asp-30 hydrogen bonds to the side-chain of Asn-216′ (2.9 Å). In addition to polar contacts, hydrophobic interactions are also present at the dimer interface with several residues buried in hydrophobic pockets throughout the interface including Phe-33, Leu-36 and Leu-209 from helices a and i as well as Val-42, Phe-43, and Pro-44 from the a – b loop. Notably, a total surface area of ~1640 Å2 is buried at the HpUreF dimer interface, corresponding to 15% of the accessible surface area of each protomer.
The molecular surface of the dimer is a lozenge-shaped “bowl” whereby the bottom face is delineated by a large flat surface patch while a prominent cavity characterizes the top face. In spite of its compactness, as further evidenced by the paucity of extended internal loops, the HpUreF dimer displays a significant amount of main chain flexibility. As gleaned from the B-factor distribution in all three independent protein chains, mobility appears to be greatest along the rim of the bowl at the periphery of the dimer, away from the interface (Figure 1C). The dimer comprising chains A and C, whose average main-chain temperature factors are 25.8 and 29.6 Å2, respectively, display considerably less rigidity than the B-B’ dimer (20.7 Å2). Curiously, helix b and a portion of the long loop exhibit the highest degree of flexibility in all cases. For instance, dynamic disorder precluded modeling of residues Tyr-48 to Phe-52 in chain C. Similarly, in chains A and B, the highest temperature factors are found in helix b.
Mapping of Conserved Residues
The propensity for UreF to interact with UreH/UreD and other accessory proteins in the maturation of urease has been demonstrated for H. pylori and K. aerogenes, where several protein complexes involving this member have been elucidated from the latter organism.22,24-27,36,62,63 In an effort to uncover potential binding sites to support these protein-protein interactions in the HpUreF structure, we identified conserved residues from a broad analysis of 224 bacterial UreF sequences and mapped the assigned conservation levels (1 to 9) onto the three-dimensional structure using the Consurf server58-60 (Figure 2). The result revealed a large clustering of conserved residues covering the top face of the HpUreF dimer and extending into the dimer interface (Figure 2B). In contrast, only weakly or non-conserved residues occupy the rest of the dimer surface.
Figure 2.
A: Multiple sequence alignment of HpUreF and selected bacterial homologues with shading to highlight only the most conserved residues (conservation scores 6–9). The degree of conservation as assigned by the ConSurf server is shown by the different levels of shading. The observed (labeled) and predicted secondary structure elements for HpUreF are indicated below the sequence alignment. B: Mapping of conservation scores onto the molecular surface of the HpUreF dimer. Three representative views of the dimer are shown, varying in the amount of rotation along the vertical axis: top [same orientation as in Fig. 1(A)], side (~90° rotation from front view), and bottom (~108° rotation from front view). Conservation scores were calculated for one polypeptide chain using the ConSurf server and projected on the dimeric structure. The degree of conservation for each residue is depicted by the indicated coloring scheme, where the amino acids are colored maroon to white to turquoise as the extent of conservation decreases.
The finding of a large conserved cavity on the dimer surface presents a potential binding site for urease-related protein-protein interactions. Of potential significance, the highly flexible region of HpUreF, which includes helix b and the preceding floppy loop (residues 41-59), corresponds to a very well conserved block in the sequence (Figure 2A). Within the a – b loop, the presence of two highly conserved and conformationally influential residues, Pro-44 and Gly-46, suggests a possible link between conformation of this loop, which ultimately controls the movement of helix b, and protein function. It is noteworthy to recognize that Pro-44, whose main-chain carbonyl is hydrogen-bonded to the guanidinium side-chain of a conserved Arg-130, is fully conserved among all bacterial UreF sequences considered in this study (Supplementary Figure S1). Similarly, Gly-46 is also conserved in all bacterial species with the exception of two distant UreF homologues with 22% sequence identity and belonging to the Roseobacter lineage, a family of marine bacteria, where a mutation to Ser is observed at this position. A number of residues with the highest conservation level assigned are found in helix b. At the N-terminus of the helix, residue His-50 is oriented towards the core of the protomer, forming a salt bridge with a highly conserved Glu-85 (2.9 Å) from helix c as well as a hydrogen bond to the side-chain of a weakly conserved Gln-185 (2.5 Å) from helix h. In spite of being part of a flexible region, His-50 may play an essential role in maintaining the structural integrity of the protein with substitutions to other aromatic residues observed in UreF homologues (Figure 2A). The remaining helix b residues possessing the highest assigned conservation level are all solvent-exposed and these include Ser-51, Gly-53, Glu-55, and Tyr-57. It is intriguing to note that the side chains of Ser-51 and Glu-55 are both oriented towards the conserved cavity, forming a part of its surface. Moreover, the conservation of these residues in bacteria is extremely high. The acidic Glu-55 is completely conserved in the present set of bacterial sequences (Figure S1). Similarly, Ser-51 is a fully conserved residue except for the UreF of a marine cyanobacterium strain with 21% overall sequence identity, where substitution to Ala at this position is observed (Figure S1). Among residues occupying the dimer cavity, Lys-195 represents another highly conserved site, with some homologous UreF sequences substituting this site with an equivalently charged Arg (Figure S1). As part of helix h in the D domain, Lys-195 and its counterpart Lys-195′, separated by a distance of 11.5 Å within the dimer, are situated curiously at the mouth of the binding cavity above the dimer interface. On the basis of high residue conservation and placement in the dimer cavity, we speculate that the surface-exposed Ser-51, Glu-55 and Lys-195 represent potential sites of interaction with urease-related components, but further investigation is warranted to confirm their functional significance.
DISCUSSION
Over the past several decades, much has been learned about the metal dependence and function of the nickel-containing metalloenzyme urease. Of particular interest is the finding that maturation of bacterial ureases typically requires the presence of UreD (or UreH in H. pylori), UreE, UreF, and UreG accessory proteins.6 UreD, UreF, and UreG form sequential complexes with the urease apoprotein,22-24 but the precise functions of these accessory proteins in activating urease remain to be characterized. A role in nickel delivery to the UreDFG-urease apoprotein complex has been attributed to UreE, a so-called metallochaperone whose structure has been determined.35,41 The recognition of a nucleotide-binding motif (P-loop) in the UreG sequence, the abolishment of urease activity when this site is mutated, and in vitro GTP-dependent activation of the UreDFG-urease apoprotein complex all support the notion that UreG serves as a GTPase.25-27 Whereas both UreE and UreG are soluble proteins that have been extensively investigated,15,17,38-42 progress in the study of UreD and UreF has been hindered by their insolubility for proteins from the microorganisms thus far studied, and no structural studies have been described for these proteins from any species. Encouragingly, the translational fusions of K. aerogenes UreF with the maltose binding protein37 or with UreE36 were soluble and subsequently isolated, and the latter was shown to be able to facilitate urease activation in vivo. In the present study, ureF from H. pylori was heterologously expressed in E. coli and the corresponding protein, fortunately shown to be soluble, was purified. Adventitious limited proteolysis provided a fortuitous route to a soluble and crystallizable form of HpUreF.
The overall HpUreF structure adopts an all-helical fold for the crystallographically observed residues 25-233, representing the first structural characterization of UreF from any species. As shown in Figure 2A, the positions of all helices and connecting loops correlate well with the predicted JPRED model for the secondary structure. The latter may also be invoked to ascertain the extent of secondary structure for the unmodeled portions at the N and C termini, where lack of electron density or limited proteolysis precluded their complete modeling in the current crystal form. For the N-terminal region, examination of the crystal packing revealed the presence of large channels oriented along the [001] crystal direction in the vicinity of the first modeled residue in each protein chain, providing the possibility that crystallographic disorder could account for the missing peptide residues 1-24 as well as the uncleaved N-terminal polyhistidine tag that was preserved throughout purification of the protein. The calculated secondary structure, which shows this region to be mainly unstructured, lends additional support for this notion. Significantly, this N-terminal region is not present in UreF of many microorganisms and is unlikely to be functionally important. Using the same analysis, the C-terminal region in all protein chains present in the crystal was found to be restrictively packed against a neighboring molecule in the crystal lattice, consequently presenting insufficient volume to fully account for the missing residues 234-254. This result necessarily implicates a C-terminally truncated form of the protein to be present in the current crystal form. Thus, consistent with observed bands in the denaturing gel, we deduce that a 28-kDa protein species of HpUreF is the most likely fragment to give rise to the crystals. Furthermore, on the basis of predicted secondary structure, we also infer the existence of an additional surface-exposed and flexible “helix k” in a complete structure model of full-length HpUreF which would include the protease-sensitive segment bounded by residues 234-254 (Figure 2A). The C-terminal region is present and highly conserved in UreF proteins from many species, so it is likely to be functionally important; indeed, this region of the K. aerogenes protein was shown to be essential for generating an activation complex.36
Mapping of residue conservation onto the structure revealed a large conserved cavity on the dimer surface. A second region containing conserved residues is the dimer interface. Here, the hydrogen-bonded pairs Asp-40/Ser187′ and Gln-37/Gln-205′, with the highest levels of conservation, represent the most essential residues involved in dimerization of purified HpUreF. These residues could be highly conserved simply as a means to retain appropriate interactions between the protomers. Alternatively, these residues might be critical to interactions between other urease components and this interface region of UreF. In this regard we note three lines of evidence that the UreF-UreF interface is disrupted in the UreDF-urease apoprotein complex. First, chemical cross-linking studies of the purified K. aerogenes UreDF-UreABC complex (recall that UreA of H. pylori corresponds to a fusion of these UreA and UreB subunits) failed to identify any UreF-UreF cross-links, but did lead to cross-linking of UreF and UreB.62 Secondly, the isolated K. aerogenes UreDF-UreABC complex was shown by a combination of native and denaturing polyacrylamide gel electrophoretic analyses with densitometry to be a mixture of species that was interpreted as the trimeric urease apoprotein binding from zero to a maximum of three UreDF units; i.e. (UreDF)0-3-(UreABC)3, inconsistent with UreF-UreF interactions in all samples.22 Third, small-angle x-ray scattering measurements and modeling, again carried out with K. aerogenes UreDF-UreABC, were consistent with both UreD and UreF binding to the vertices of the trimeric apoprotein;63 a situation that precludes retention of the UreF-UreF interface. Additional evidence for disruption of the UreF-UreF interface is available from studies involving the translational fusion of K. aerogenes UreE and UreF.36 The UreEF fusion protein, in which UreF was functional as shown by its ability to activate urease in the cell, eluted as a monomer when examined by gel filtration chromatography in comparison to globular protein standards. In sum, substantial evidence exists to demonstrate that the UreF-UreF dimer interface is not fixed, at least for the K. aerogenes protein. Thus, we suggest that the UreF-UreF interface residues indicated above may be critical to requisite interactions between UreF and another urease protein component.
UreF is likely to interact with UreG within the UreDFG-urease apoprotein complex. Adhering to the idea that UreG is a GTPase, a recent study64 described a potential evolutionary link between UreF and a class of GAPs. The investigators used a comparative modeling approach to create a partial homology model of B. pasteurii UreF (BpUreF) derived by a threading algorithm search that uncovered two model templates of GTPase-activating factors represented by the structures of the rgRGS domain of H. sapiens p115RhoGEF (PDB 1IAP, E-value = 4.21) and the RGS-like domain of H. sapiens PDZ-RhoGEF (PDB 1HTJ). While both structures share an all-helical assignment for the secondary structure, a comparison of the proposed homology model for BpUreF (27% sequence identity) and the experimentally determined HpUreF structure revealed no obvious resemblance (Z < 1.5 in DALI pairwise comparisons with PDB 1IAP and PDB 1HTJ).
A search for structural neighbors to HpUreF failed to identify meaningful matches when using the Secondary Structure Matching (SSM) algorithm.65 Unexpectedly, a homology search using DALI66 yielded the citrate synthase (CS) fold family as the top match, with the Thermotoga maritima protein (TmCS) producing the best overall result (Z = 7.2, PDB 2P2W). Despite sharing a sequence identity of only 8%, a DALI superposition revealed that the arrangement of eight of the nine α-helices (a-h) in the HpUreF fold is remarkably similar in its topology to a portion of the TmCS fold that comprises the entire small domain as well as a part of the large domain (Supplementary Figure S2A). Moreover, the I domain in HpUreF is similar to the small domain in TmCS that is considered also to be an insertion into the large domain of the CS fold (SCOP nomenclature). Citrate binds in a cleft between the small and large domains of CS, where the catalytic His-211TmCS stabilizes acetyl-coenzyme A enolization in the enzymatic reaction; the highly conserved His-50HpUreF is similarly positioned, but is more likely involved in domain stabilization. Further evidence for functional divergence of these proteins in spite of their common fold is provided by a comparison of their dimerization surfaces which do not coincide.
Of potential functional relevance, the homology search revealed a weak but intriguing structural similarity between the D domain, particularly the bundle defined by helices a, b, c, h, and i, and the GAP domains of SynGAP (Z = 4.4, PDB 3BXJ)67 (Figure S2B), neurofibromin (Z = 3.6, PDB 1NF1),68 p120GAP (Z = 3.5, PDB 1WER),69 and IQGAP1 (Z = 3.0, PDB 3FAY).70 Interestingly, the catalytic interface in each of these structures coincided with the dimer interface in HpUreF. The catalytic Arg fingers (e.g., Arg-470SynGAP) and invariant Lys (e.g., Lys-613SynGAP) required for substrate binding are lacking in HpUreF. Similarly, the catalytic Thr-1046 of IQGAP1 is replaced by Asp-40HpUreF. Although a weak match was also noted for the guanine nucleotide exchange factor catalytic domain of SopE from Salmonella typhimurium (Z = 2.9, PDB 1GZS),71 no functional parallels were apparent in spite of a similar arrangement in a three-helix bundle common to both structures.
This study presents the first report of a structure for the C-terminal truncated UreF urease accessory protein. It is only distantly related structurally to GAPs, so the attractive hypothesis that UreF functions as a GAP acting upon UreG appears unlikely. In line with an alternate role that has been proposed for UreF in the UreABC-UreDF complex, it could also be hypothesized that Lys-195 prevents nickel binding to the noncarboxylated urease apoprotein by regulating the sequential incorporation of bicarbonate into the active site before nickel can gain access. Mapping of residue conservation onto the three-dimensional structure has allowed identification of other potentially essential residues in the activity of UreF, providing a basis for further exploration of its function by mutagenesis studies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Aiping Dong for his assistance with data collection and Jodi Boer for generating an initial sequence alignment and WebLogo plots that were superseded by the more detailed sequence analysis reported here. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with the Center for Advanced Radiation Sources at the University of Chicago. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. These studies were supported by the Ontario Research and Development Challenge Fund (99-SEP-0512 to E.F.P.) and National Institutes of Health grant DK45686 (to R.P.H.).
REFERENCES
- 1.Kusters JG, Van Vliet AHM, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atherton JC. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annu Rev Pathol Mech Dis. 2006;1:63–96. doi: 10.1146/annurev.pathol.1.110304.100125. [DOI] [PubMed] [Google Scholar]
- 3.Hu L-T, Mobley HLT. Purification and N-terminal analysis of urease from Helicobacter pylori. Infect Immun. 1990;58:992–998. doi: 10.1128/iai.58.4.992-998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stingl K, Altendorf K, Bakker EP. Acid survival of Helicobacter pylori: how does urease activity trigger cytoplasmic pH homeostasis? Trends Microbiol. 2002;10:70–74. doi: 10.1016/s0966-842x(01)02287-9. [DOI] [PubMed] [Google Scholar]
- 5.Ha N-C, Oh S-T, Sung JY, Cha K-A, Lee MH, Oh B-H. Supramolecular assembly and acid resistance of Helicobacter pylori urease. Nat Struct Biol. 2001;8:505–509. doi: 10.1038/88563. [DOI] [PubMed] [Google Scholar]
- 6.Carter EL, Flugga N, Boer JL, Mulrooney SB, Hausinger RP. Interplay of metal ions and urease. Metallomics. 2009;1:207–221. doi: 10.1039/b903311d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benini S, Rypniewski WR, Wilson KS, Miletti S, Ciurli S, Mangani S. A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels. Structure. 1999;7:205–216. doi: 10.1016/S0969-2126(99)80026-4. [DOI] [PubMed] [Google Scholar]
- 8.Jabri E, Carr MB, Hausinger RP, Karplus PA. The crystal structure of urease from Klebsiella aerogenes. Science. 1995;268:998–1004. [PubMed] [Google Scholar]
- 9.Pearson MA, Michel LO, Hausinger RP, Karplus PA. Structure of Cys319 variants and acetohydroxamate-inhibited Klebsiella aerogenes urease. Biochemistry. 1997;36:8164–8172. doi: 10.1021/bi970514j. [DOI] [PubMed] [Google Scholar]
- 10.Sheridan L, Wilmont CM, Cromie KD, van der Logt P, Phillips SEV. Crystallization and preliminary X-ray structure determination of jack bean urease with a bound antibody fragment. Acta Crystallogr D Biol Crystallogr. 2002;58:374–376. doi: 10.1107/s0907444901021503. [DOI] [PubMed] [Google Scholar]
- 11.Maier RJ, Benoit S, Seshadri S. Nickel-binding and accessory proteins facilitating Ni-enzyme maturation in Helicobacter pylori. BioMetals. 2007;20:655–664. doi: 10.1007/s10534-006-9061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cussac V, Ferrero RL, Labigne A. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J Bacteriol. 1992;174:2466–2473. doi: 10.1128/jb.174.8.2466-2473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olson JW, Mehta NS, Maier RJ. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol Microbiol. 2001;39:176. doi: 10.1046/j.1365-2958.2001.02244.x. [DOI] [PubMed] [Google Scholar]
- 14.Weeks DL, Eskandar S, Scott DR, Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. 2000;287:482–485. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]
- 15.Benoit S, Maier RJ. Dependence of Helicobacter pylori urease activity on the nickel-sequestering ability of the UreE accessory protein. J Bacteriol. 2003;185:4787–4795. doi: 10.1128/JB.185.16.4787-4795.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellucci M, Zambelli B, Musiani F, Turano P, Ciurli S. Helicobacter pylori UreE, a urease accessory protein: specific Ni(2+)- and Zn(2+)-binding properties and interaction with its cognate UreG. Biochem J. 2009;422(1):91–100. doi: 10.1042/BJ20090434. [DOI] [PubMed] [Google Scholar]
- 17.Mehta N, Benoit S, Maier RJ. Roles of conserved nucleotide-binding domains in accessory proteins, HypB and UreG, in the maturation of nickel-enzymes required for efficient Helicobacter pylori colonization. Microb Pathog. 2003;35:229–234. doi: 10.1016/s0882-4010(03)00151-7. [DOI] [PubMed] [Google Scholar]
- 18.Mehta NS, Olson JW, Maier RJ. Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. J Bacteriol. 2003;185:726–734. doi: 10.1128/JB.185.3.726-734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benoit SL, Mehta N, Weinberg MV, Maier C, Maier RJ. Interaction beween the Helicobacter pylori accessory proteins HypA and UreE is needed for urease maturation. Microbiology. 2007;153:1474–1482. doi: 10.1099/mic.0.2006/003228-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rain J-C, Selig L, de Reuse H, Battaglia V, Reverdy C, Simon S, Lenzen G, Petel F, Wojcik J, Schächter V, Chemama Y, Labigne A, Legrain P. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 21.Voland P, Weeks DL, Marcus EA, Prinz C, Sachs G, Scott D. Interactions among the seven Helicobacter pylori proteins encoded by the urease gene cluster. Am J Physiol Gastrointest Liver Physiol. 2003;284:G96–G106. doi: 10.1152/ajpgi.00160.2002. [DOI] [PubMed] [Google Scholar]
- 22.Moncrief MBC, Hausinger RP. Purification and activation properties of UreD-UreF-urease apoprotein complexes. J Bacteriol. 1996;178:5417–5421. doi: 10.1128/jb.178.18.5417-5421.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park I-S, Carr MB, Hausinger RP. In vitro activation of urease apoprotein and role of UreD as a chaperone required for nickel metallocenter assembly. Proc Natl Acad Sci USA. 1994;91:3233–3237. doi: 10.1073/pnas.91.8.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park I-S, Hausinger RP. Evidence for the presence of urease apoprotein complexes containing UreD, UreF, and UreG in cells that are competent for in vivo enzyme activation. J Bacteriol. 1995;177:1947–1951. doi: 10.1128/jb.177.8.1947-1951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moncrief MBC, Hausinger RP. Characterization of UreG, identification of a UreD-UreF-UreG complex, and evidence suggesting that a nucleotide-binding site in UreG is required for in vivo metallocenter assembly of Klebsiella aerogenes urease. J Bacteriol. 1997;179:4081–4086. doi: 10.1128/jb.179.13.4081-4086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soriano A, Colpas GJ, Hausinger RP. UreE stimulation of GTP-dependent urease activation in the UreD-UreF-UreG-urease apoprotein complex. Biochemistry. 2000;39:12435–12440. doi: 10.1021/bi001296o. [DOI] [PubMed] [Google Scholar]
- 27.Soriano A, Hausinger RP. GTP-dependent activation of urease apoprotein in complex with the UreD, UreF, and UreG accessory proteins. Proc Natl Acad Sci USA. 1999;96:11140–11144. doi: 10.1073/pnas.96.20.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jabri E, Karplus PA. Structures of the Klebsiella aerogenes urease apoprotein and two active-site mutants. Biochemistry. 1996;35:10616–10626. doi: 10.1021/bi960424z. [DOI] [PubMed] [Google Scholar]
- 29.Brayman TG, Hausinger RP. Purification, characterization, and functional analysis of a truncated Klebsiella aerogenes UreE urease accessory protein lacking the histidine-rich carboxyl terminus. J Bacteriol. 1996;178:5410–5416. doi: 10.1128/jb.178.18.5410-5416.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colpas GJ, Brayman TG, McCracken J, Pressler MA, Babcock GT, Ming L-J, Colangelo CM, Scott RA, Hausinger RP. Spectroscopic characterization of metal binding by Klebsiella aerogenes UreE urease accessory protein. J Biol Inorg Chem. 1998;3:150–160. [Google Scholar]
- 31.Colpas GJ, Brayman TG, Ming L-J, Hausinger RP. Identification of metal-binding residues in the Klebsiella aerogenes urease nickel metallochaperone, UreE. Biochemistry. 1999;38:4078–4088. doi: 10.1021/bi982435t. [DOI] [PubMed] [Google Scholar]
- 32.Grossoehme NE, Mulrooney SB, Hausinger RP, Wilcox DE. Thermodynamics of Ni2+, Cu2+, and Zn2+ binding to urease metallochaperone UreE. Biochemistry. 2007;46:10506–10516. doi: 10.1021/bi700171v. [DOI] [PubMed] [Google Scholar]
- 33.Lee MH, Pankratz HS, Wang S, Scott RA, Finnegan MG, Johnson MK, Ippolito JA, Christianson DW, Hausinger RP. Purification and characterization of Klebsiella aerogenes UreE protein: a nickel-binding protein that functions in urease metallocenter assembly. Protein Sci. 1993;2:1042–1052. doi: 10.1002/pro.5560020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colpas GJ, Hausinger RP. In vivo and in vitro kinetics of metal transfer by the Klebsiella aerogenes urease nickel metallochaperone, UreE. J Biol Chem. 2000;275:10731–10737. doi: 10.1074/jbc.275.15.10731. [DOI] [PubMed] [Google Scholar]
- 35.Song HK, Mulrooney SB, Huber R, Hausinger RP. Crystal structure of Klebsiella aerogenes UreE, a nickel-binding metallochaperone for urease activation. J Biol Chem. 2001;276:49359–49364. doi: 10.1074/jbc.M108619200. [DOI] [PubMed] [Google Scholar]
- 36.Kim JK, Hausinger RP. The UreEF fusion protein provides a soluble and functional form of the UreF urease accessory protein. J Bacteriol. 2006;188:8413–8420. doi: 10.1128/JB.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim KY, Yang CH, Lee MH. Expression of the recombinant Klebsiella aerogenes UreF protein as a MalE fusion. Arch Pharm Res. 1999;22:274–278. doi: 10.1007/BF02976362. [DOI] [PubMed] [Google Scholar]
- 38.Sriwanthana B, Island MD, Maneval D, Mobley HLT. Single-step purification of Proteus mirabilis urease accessory protein UreE, a protein with a naturally occurring histidine tail, by nickel chelate affinity chromatography. J Bacteriol. 1994;176:6836–6841. doi: 10.1128/jb.176.22.6836-6841.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zambelli B, Musiani F, Savini M, Tucker P, Ciurli S. Biochemical studies on Mycobacterium tuberculosis UreG and comparative modeling reveal structural and functional conservation among the bacterial UreG family. Biochemistry. 2007;46:3171–3182. doi: 10.1021/bi6024676. [DOI] [PubMed] [Google Scholar]
- 40.Zambelli B, Turano P, Musiani F, Neyroz P, Ciurli S. Zn2+-linked dimerization of UreG from Helicobacter pylori, a chaperone involved in nickel trafficking and urease activation. Proteins. 2009;74:222–239. doi: 10.1002/prot.22205. [DOI] [PubMed] [Google Scholar]
- 41.Remaut H, Safarof N, Ciurli S, Van Beeumen J. Structural basis for Ni2+ transport and assembly of the urease active site by the metallo-chaperone UreE from Bacillus pasteurii. J Biol Chem. 2001;276:49365–49370. doi: 10.1074/jbc.M108304200. [DOI] [PubMed] [Google Scholar]
- 42.Zambelli B, Stola M, Musiani F, De Vriendt K, Samyn B, Devreese B, Van Beeumen J, Dikiy A, Bryant DA, Ciurli S. UreG, a chaperone in the urease assembly process, is an intrinsically unstructured GTPase that specifically binds Zn2+ J Biol Chem. 2005;280:4684–4695. doi: 10.1074/jbc.M408483200. [DOI] [PubMed] [Google Scholar]
- 43.Gasper R, Scrima A, Wittinghofer A. Structural insights into HypB, a GTP-binding protein that regulates metal binding. J Biol Chem. 2006;281:27492–27502. doi: 10.1074/jbc.M600809200. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.Dauter Z, Dauter M, Dodson E, Jolly SAD. Acta Crystallogr D Biol Crystallogr. 2002;58:494–506. doi: 10.1107/s090744490200118x. [DOI] [PubMed] [Google Scholar]
- 46.Dodson E. Is it jolly SAD? Acta Crystallogr D Biol Crystallogr. 2003;59:1958–1965. doi: 10.1107/s0907444903020936. [DOI] [PubMed] [Google Scholar]
- 47.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 48.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 49.de la Fortelle E, Bricogne G. Maximum-Likelihood Heavy-Atom Parameter Refinement for Multiple Isomorphous Replacement and Multiwavelength Anomalous Diffraction Methods. Meth Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 50.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 53.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 54.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 55.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Meth Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 56.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 57.DeLano WL. The PYMOL Molecular Graphic System. DeLano Scientific LLC; San Carlos, CA: 2002. [Google Scholar]
- 58.Armon A, Graur D, Ben-Tal N. ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J Mol Biol. 2001;307:447–463. doi: 10.1006/jmbi.2000.4474. [DOI] [PubMed] [Google Scholar]
- 59.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 60.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang Z, Kuchar J, Hausinger RP. Chemical crosslinking and mass spectrometric identification of sites of interaction for UreD, UreF, and urease. J Biol Chem. 2004;279:15305–15313. doi: 10.1074/jbc.M312979200. [DOI] [PubMed] [Google Scholar]
- 63.Quiroz-Valenzuela S, Sukuru SCK, Hausinger RP, Kuhn LA, Heller WT. The structure of urease activation complexes examined by flexibility analysis, mutagenesis, and small-angle X-ray scattering. Arch Biochem Biophys. 2008;480:51–57. doi: 10.1016/j.abb.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salomone-Stagni M, Zambelli B, Musiani F, Ciurli S. A model-based proposal for the role of UreF as a GTPase-activating protein in the urease active site biosynthesis. Proteins. 2007;68:749–761. doi: 10.1002/prot.21472. [DOI] [PubMed] [Google Scholar]
- 65.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 66.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pena V, Hothorn M, Eberth A, Kaschau N, Parret A, Gremer L, Bonneau F, Ahmadian MR, Scheffzek K. The C2 domain of SynGAP is essential for stimulation of the Rap GTPase reaction. EMBO Rep. 2008;9:350–355. doi: 10.1038/embor.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scheffzek K, Ahmadian MR, Wiesmuller L, Kabsch W, Stege P, Schmitz F, Wittinghofer A. Structural analysis of the GAP-related domain from neurofibromin and its implications. EMBO J. 1998;17:4313–4327. doi: 10.1093/emboj/17.15.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scheffzek K, Lautwein A, Kabsch W, Ahmadian MR, Wittinghofer A. Crystal structure of the GTPase-activating domain of human p120GAP and implications for the interaction with Ras. Nature. 1996;384:591–596. doi: 10.1038/384591a0. [DOI] [PubMed] [Google Scholar]
- 70.Kurella VB, Richard JM, Parke CL, Lecour LF, Jr., Bellamy HD, Worthylake DK. Crystal Structure of the GTPase-activating Protein-related Domain from IQGAP1. J Biol Chem. 2009;284:14857–14865. doi: 10.1074/jbc.M808974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buchwald G, Friebel A, Galan JE, Hardt WD, Wittinghofer A, Scheffzek K. Structural basis for the reversible activation of a Rho protein by the bacterial toxin SopE. EMBO J. 2002;21:3286–3295. doi: 10.1093/emboj/cdf329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.