Abstract
Heparanase-1 (HPR1), an endoglycosidase that specifically degrades heparan sulfate (HS) proteoglycans, is overexpressed in a variety of malignancies. Our present study sought to determine whether oncogene BRAF and RAS mutations lead to increased HPR1 expression. Reverse transcription-polymerase chain reaction analysis revealed that HPR1 gene expression was increased in HEK293 cells transiently transfected with a mutant BRAF or RAS gene. Flow cytometric analysis revealed that B-Raf activation led to loss of the cell surface HS, which could be blocked by two HPR1 inhibitors: heparin and PI-88. Cotransfection of a BRAF or RAS mutant gene with HPR1 promoter-driven luciferase reporters increased luciferase reporter gene expression in HEK293 cells. Knockdown of BRAF expression in a BRAF-mutated KAT-10 tumor cell line led to the suppression of HPR1 gene expression, subsequently leading to increased cell surface HS levels. Truncational and mutational analyses of the HPR1 promoter revealed that the Ets-relevant elements in the HPR1 promoter were critical for BRAF activation-induced HPR1 expression. Luciferase reporter gene expression driven by a four-copy GA binding protein (GABP) binding site was significantly lower in BRAF siRNA-transfected KAT-10 cells than in the control siRNA-transfected cells. We further showed that BRAF knockdown led to suppression of the expression of the GABPβ, an Ets family transcription factor involved in regulating HPR1 promoter activity. Taken together, our study suggests that B-Raf kinase activation plays an important role in regulating HPR1 expression. Increased HPR1 expression may contribute to the aggressive behavior of BRAF-mutated cancer.
Introduction
Heparanase-1 (HPR1) is an endoglycosidase that specifically degrades heparan sulfate (HS) proteoglycans (HSPGs) [1–4]. HSPGs are heavily present on the cell surface and in the extracellular matrix (ECM) and the basement membrane (BM). HPR1 is overexpressed in a variety of malignancies [1–4]. Breakdown of HSPGs in theBMand ECMleads to the release of many growth factors such as fibroblast growth factor and vascular endothelial growth factor that are trapped in the tumor stroma. These growth factors can promote tumor angiogenesis by stimulating endothelial cell proliferation and migration. In addition, breakdown of the BMand ECMallows tumor cells to invade locally or metastasize to a distant site. Recent studies have shown that HPR1 exerts its many biologic functions independent of its enzymatic activity. For example, HPR1 can enhance cell adhesion [5,6], induce vascular endothelial growth factor expression [7], induce tumor and endothelial cell migration, and induce Akt, p38, and Src phosphorylation [7,8]. HPR1 can induce epidermal growth factor (EGF) receptor phosphorylation and stimulate tumor cell proliferation and growth in an enzymatic activity-independent manner [9]. A conservative, hydrophobic C-terminus domain of HPR1 has been recently identified to mediate these diverse biologic functions [10,11]. HPR1 C-terminus functions as a ligand to bind two potential unknown receptors (a 130- and a 170-kDa protein) to activate the phosphatidylinositol 3-kinase pathway [11]. HPR1 may exert its tumor-promoting effect independent of its enzymatic activity.
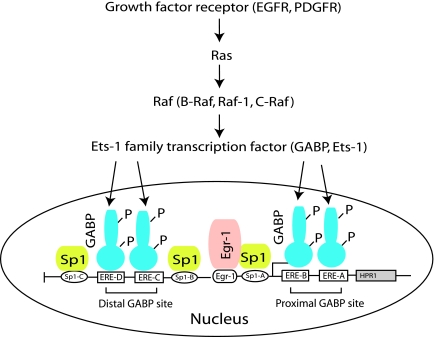
Molecular mechanisms of HPR1 overexpression in a variety of cancers remain poorly understood. We and others have previously characterized the HPR1 promoter [12,13]. Sequence analysis revealed that the TATA-less, GC-rich promoter of the HPR1 gene belongs to the family of housekeeping genes. Three Sp1 sites and four Ets relevant elements (ERE) for two GA binding protein (GABP) binding sites (Figure 7) are located in a 0.3-kb proximal promoter region [13]. GABP and Sp1/Sp3 are two transcription factors that regulate HPR1 basal promoter activity [13]. Later studies demonstrated that Egr-1 is involved in HPR1 gene expression in T cells stimulated by PMAplus ionomycin and in tumor cells [14–16]. Recent studies suggest that increased HPR1 expression in bladder and prostate cancers is largely mediated by HPR1 promoter hypomethylation and Egr-1 overexpression and hyperactivation [16,17] (Figure 7). Because tumor suppressor p53 can negatively regulate HPR1 gene expression [18], p53 gene mutation may also contribute to increased HPR1 expression in a variety of tumors. It is not clear whether oncogene mutation and activation can lead to increased HPR1 expression.
Figure 7.
Signaling pathway and transcription factors involved in HPR1 gene expression in cancer. Mutations and/or overexpression of the growth factor receptor genes such as EGFR and its downstream signaling molecules such as RAS and BRAF genes result in increased expression and/or translocation of the Ets family transcription factors such as GABP and Ets-1. These transcription factors in cooperation with several other transcription factors overexpressed in cancer, such as Sp1 and Egr-1, lead to HPR1 expression in cancer. HPR1 promoter demethylation and p53 mutations also contribute to increased HPR1 gene expression (not shown).
BRAF is an oncogene that is frequently mutated in a variety of malignancies, with the highest frequencies in melanomas and thyroid cancers [19]. We have previously characterized HPR1 expression and BRAF gene mutation in thyroid cancer [20,21]. Interestingly, we found that HPR1 is expressed at relatively low levels in WRO82 and KAT-18 cells, two thyroid tumor cell lines with wild-type BRAF, compared with that in several BRAF mutant tumor cell lines. We hypothesize that BRAF and RAS mutations may be responsible for increased HPR1 expression in cancer. Here we report that HPR1 gene expression was induced in the cells transfected with mutant BRAF and RAS genes and that knockdown of BRAF led to decreased HPR1 expression. GABP, an Ets family transcription factor, was responsible for BRAF mutation-induced HPR1 expression.
Materials and Methods
Plasmids and Cell Lines
pEF6 empty vector, and the vector encoding a wild-type BRAF (pEF6/BRAF), mutant V600E BRAF (pEF6/BRAFV600E), or mutant G12V HRAS (pEF6/HRASG12V), have been previously described [22]. Luciferase reporter constructs driven by various truncated or mutated HPR1 promoters (Figure 5A) and by a four-copy of theGABP binding site derived from human FAS gene have been previously described [13,23]. KAT-10 cells transfected with a small interference (si)RNA vector IMG800 or the vector encoding a BRAF siRNA have been previously reported [24]. KAT-10 cells were previously considered a tumor cell line of thyroid origin but were recently verified as identical to HT29, a human colon cancer cell line [25]. KAT-10 cells were grown in complete RPMI 1640 medium containing 10% fetal bovine serum and 500 µg/ml G418. HEK293, a human kidney embryonic epithelial cell line, was purchased from the American Tissue Culture Collection (Manassas, VA). HEK293 cells were grown in minimum essential medium containing 10% fetal bovine serum, nonessential amino acids, sodium pyruvate, and HEPES buffer.
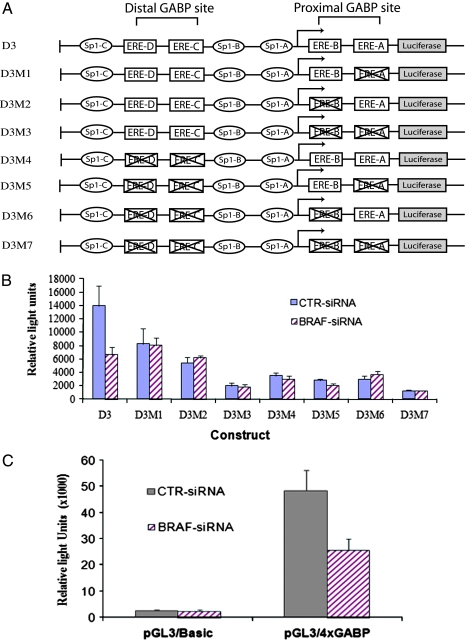
Figure 5.
Evidence that the GABP binding site in the HPR1 promoter is critical for BRAF-mediated HPR1 gene regulation. (A) Schematic illustration of a panel of the HPR1 promoter-driven luciferase reporter constructs. ERE sites marked with a cross sign denote those with altered nucleotides can no longer be bound by Ets transcription factors. (B) Control or BRAF siRNA-transfected KAT-10 cells were transiently transfected with the luciferase reporter constructs as shown in (A) or with 4xGABP binding site-driven luciferase reporter construct (C). After incubating for 48 hours, the cells were harvested and analyzed for luciferase activity.
Luciferase Reporter Gene Expression
HEK293 cells seeded in a 24-well plate were transfected with pGL3/basic, a 0.7-kb or 3.5-kbHPR1 promoter-driven luciferase reporter gene plus pEF6 empty vector, pEF6/BRAF, pEF6/BRAFV600E, and pEF6/HRASG12V by using FuGENE6 (Roche Applied Science, Indianapolis, IN) following the manufacturer's instructions (Figure 1B). An internal control plasmid, pCMV/SPORT, which encodes a β-galactosidase gene, was included in the transfection mixture. Control or BRAF siRNA-transfected KAT-10 cells were similarly transfected with the luciferase reporter gene driven by a 0.3-, 0.7-, or 3.5-kb HPR1 promoter (pGL3/HPR-0.3, pGL3/HPR1-0.7, or pGL3/HPR-3.5; Figure 4), by a 0.3-kb HPR1 promoter with various mutated ERE sites (Figure 5, A and B), or by a four-copy GABP binding site (Figure 5C). Luciferase reporter gene without a HPR1 promoter (pGL3/Basic) was included as a negative control. After incubating for 48 hours, cells were harvested and analyzed for luciferase activity by using the luciferin substrate and reading in a TECAN plate reader (Phenix Research Products, Hayward, CA). The relative light unit in each sample was normalized against the β-galactosidase activity measured by a colorimetric assay as previously reported [26]. The means ± SDs of the data in triplicate from one experiment are presented. The experiment was repeated at least once with similar results.
Figure 1.
B-Raf kinase activation induces HPR1 expression. (A) Induction of HPR1 gene expression. HEK293 cells were transiently transfected with the pEF6 empty vector or the vector encoding wild-type BRAF, BRAFV600E, or HRASG12V. Total RNA was extracted and analyzed for HPR1 mRNA expression by RT-PCR. (B) Induction of HPR1 promoter-driven luciferase reporter gene expression. HEK293 cells were cotransfectedwith the empty vector or the vector encoding wild-type BRAF, BRAFV600E, or HRASG12V plus control luciferase reporter or the luciferase reporter gene driven by a 0.7- or 3.5-kb HPR1 promoter. pCMV/SPORT, which encodes a β-galactosidase gene, was included as an internal control. After incubating for 48 hours, the cells were harvested and analyzed for luciferase activity. The relative luciferase activity was calculated. One representative example in triplicate from three experiments with similar results is presented.
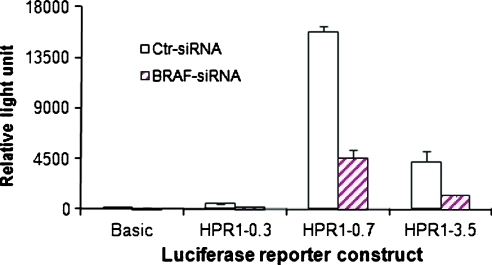
Figure 4.
Decreased HPR1 promoter activity in BRAF siRNA-transfected KAT-10 cellls. Control or BRAF siRNA-transfected KAT-10 cells were transiently transfected with the luciferase reporter gene expression driven by a 0.3-, 0.7-, or 3.5-kb HPR1 promoter. pGL3/Basic luciferase reporter construct was included as a negative control. After incubating for 48 hours, the cells were harvested and analyzed for luciferase activity.
Flow Cytometric Analysis
HEK293 cells were seeded in six-well plates. On 40% confluence, the cells were transfected with pEF6 empty vector, pEF6/BRAF, pEF6/BRAFV600E, and pEF6/HRASG12V. After incubating for 24 hours, the cells were left untreated or treated with two HPR1 inhibitors, heparin or PI-88 (50 µg/ml). After incubating for another 24 hours, the cells were harvested and used for the preparation of single cell suspensions. Single cell suspensions of HEK293 and KAT-10 cells were incubated with an anti-HS mAb (0.5 µg/sample; clone HepSS; Seikagaku Corp, Chuo-ku, Tokyo, Japan) or mouse immunoglobulin M (IgM) as an isotype control for 30 minutes at 4°C, followed by incubation with fluorescein isothiocyanate-labeled goat antimouse IgM (5 µl/sample) for 30 minutes at 4°C. To increase the fluorescence intensity, the cells were further stained with a fluorescein isothiocyanate-labeled rabbit antigoat IgG (5 µl/sample) for 30 minutes at 4°C. Cell surface HS levels were analyzed by fluorescence-activated cell sorter (FACS) in a Becton Dickinson flow cytometer.
HPR1 Activity Assay
HPR1 activity in cell lysates was measured by using a novel ELISA as previously described [27–30]. HPR1 activity in cell lysates was calculated based on a standard curve of serially diluted purified platelet HPR1 (starting at 1:200) [31] at a concentration of 1 µl of HPR1 with the activity of degrading 0.133 µg of heparan sulfate per hour at 37°C in HPR1 buffer. HPR1 activity was designated as per 100 U capable of degrading 1 ng of heparan sulfate at 37°C per hour in HPR1 buffer.
Reverse Transcription-Polymerase Chain Reaction
Total RNA was extracted from control or BRAF siRNA-transfected KAT-10 cells and HEK293 cells transfected with the pEF6 empty vector, pEF6/BRAF, pEF6/BRAFV600E, and pEF6/HRASG12V using TRIzol (Invitrogen, Camarillo, CA) and quantified in a spectrophotometer by ultraviolet absorption. Complementary DNA was synthesized by using AMV reverse transcriptase with 500 ng of total RNA and oligo(dT) priming. Polymerase chain reaction (PCR) was conducted by using Taq DNA polymerase (Invitrogen) with a pair of primers listed in Table 1. The PCR conditions for amplifying HPR1 and GAPDH are an initial denaturation of 2 minutes at 94°C, followed by 32 cycles of denaturation for 45 seconds at 94°C, annealing for 1 minute at 55°C, and extension for 1 minute at 72°C. PCR products were separated on a 1.5% agarose gel by electrophoresis and visualized by ethidium bromide staining. HPR1, BRAF, and GABPα and β expressions in KAT-10 cells were also quantified by real-time reverse transcription (RT)-PCR with primers listed in Table 1. Complementary DNAwas synthesized as above used for amplifying these genes in triplicate after a 1:10 dilution. PCR in a 25-µl volume was conducted by using cyber green mixture and ran initially at 50°C for 2 minutes and at 95°C for 10 minutes followed by 40 cycles of (95°C for 15 seconds and 60°C for 1 minute). PCR was analyzed on Applied Biosystems 7300 (Foster City, CA). The relative Ct value was calculated according to the standard formula 2Δ/ΔCt (Δ/ΔCt = ΔCt value of sample - ΔCt value of control). ΔCt value of each sample is the Ct value for BRAF siRNA-transfected cells normalized to 18S ribosomal RNA (rRNA), whereas ΔCt value of the control is the Ct value for control siRNA-transfected cells normalized to 18S rRNA. The relative expression of these genes from one representative of two or more independent experiments with similar results is presented.
Table 1.
Primers Used to Amplify HPR1, BRAF, GABPα, GABPβ, GAPDH, and 18S rRNA.
| Gene | Size (bp) | Forward | Reverse |
| HPR1 | 587 | 5′-TTCGATCCCAAGAAGG-AATCAAC-3′ | 5′GTAGTGATGCCATGTAACTGAAT-C-3′ |
| GAPDH | 527 | 5′TGAAGGTCGGAGTCAACGGATTTGGTC-3′ | 5′ATGGACTGTGGTCATGAGTCCTTCCACG-3′ |
| HPR1 | 86 | 5′-GCTAGAGCTCTCGACTCTC-3′ | 5′-GCAGGCTTCGAGCGCAGC-3′ |
| BRAF | 199 | 5′-TTGAACACCACCCAATACCA-3′ | 5′-GTCTCGTTGCCCAAATTGAT-3′ |
| GABPα | 116 | 5′-ACGATGGGGACATGATTTGT-3′ | 5′-TTCTGTGACCAAACGGTTCA-3′ |
| GABPβ | 121 | 5′-GTGTGAGCAGAGATGCCAGA-3′ | 5′-TCCTTTGCATTGACATCAGC-3′ |
| 18S rRNA | 101 | 5′-ATGCTCTTAGCTGAGTGTCCCG-3 | 5′-ATTCCTAGCTGCGGTATCCAGG-3 |
Western Blot
KAT-10 cells were lysed in NP-40 lysis buffer (50mMTris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 5 mM EDTA, 10 µg/ml aprotinin, 10 µg/ml leupeptinin, and 1 mM phenylmethylsulfonyl fluoride). Cell lysates were incubated on ice for 30 minutes and then spun down at 15,000 rpm for 15 minutes at 4°C. After electrophoresis and transfer to nitrocellulose membranes, B-Raf, Sp1, Sp3, Egr-1, phorspho-ERK, ERK, and GABPα and β were detected by their specific antibodies followed by horseradish peroxidase-conjugated goat antimouse or rabbit IgG and SuperSignal Western Pico enhanced chemiluminescence substrate (Pierce Chemical Co, Rockford, IL). These antibodies include rabbit anti-B-Raf, Sp1, Sp3, Egr-1 IgG(Santa Cruz Biotechnology, Inc, San Diego, CA), rabbit anti-phospho ERK and ERK IgG (Cell Signaling Technology, Danvers, MA), and rabbit anti-GABPα and β antisera (kindly provided by Dr. U. A. Rapp, University of Würzburg, Würzburg, Germany).
Immunofluorescence Staining of GABPα and β
KAT-10 cells grown on coverslips were washed three times with cold PBS and fixed with methanol at -20°C for 5 minutes. Coverslips were blocked with 5% normal goat serum for 30 minutes at room temperature. GABPα and β were detected by rabbit anti-GABPα or β antisera, followed by fluorescein-conjugated goat antirabbit IgG. The coverslips were mounted with 50% glycerin in PBS containing antifade reagent 1,4-diazabicyclo[2.2.2]octane (25 mg/ml) and 4,6-diamidino-2-phenylindole (0.5 mg/ml; Sigma Chemical Co, St Louis, MO). GABPα or β expression was examined under a fluorescence microscope. The pictures were taken with a digital camera attached in an Olympus BX41TF fluorescence microscope (Leeds Precision Instruments, Inc, Minneapolis, MN).
Statistical Analysis
The differences in the expression of HPR1, BRAF, GABPα or β, and HPR1 activity were statistically analyzed by using an unpaired Student t test. P < .05 was considered statistically significant.
Results
Induction of HPR1 Expression by Activation of B-Raf Kinase
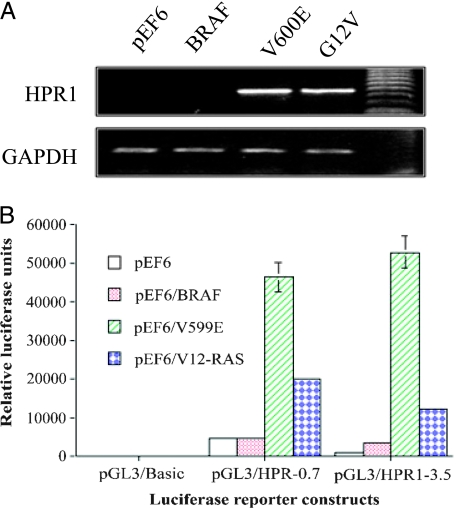
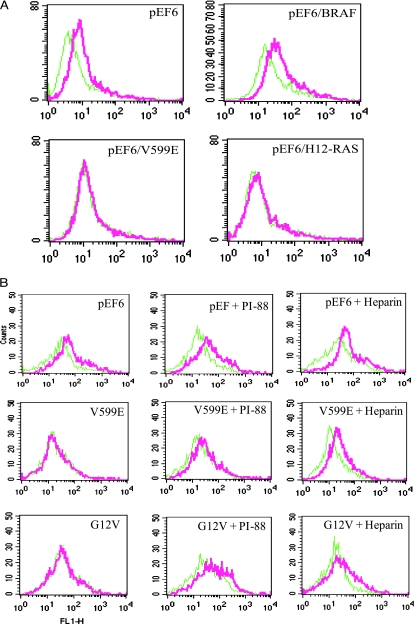
We first tested if RAS and BRAF gene mutations led to increased HPR1 gene expression. HPR1 messenger RNA (mRNA) levels in HEK293 cells transiently transfected with the pEF6 empty vector or the vector encoding wild-type BRAF, BRAFV600E, or RASG12V were analyzed by semiquantitative RT-PCR. As shown in Figure 1A, HPR1 mRNA was undetectable in HEK293 cells transfected with the pEF6 empty vector or the vector encoding wild-type BRAF but was induced in the cells transfected with a constitutively active BRAFV600E mutant or HRASG12V genes. A 0.7-kb HPR1 promoter-driven luciferase reporter gene expression was dramatically increased in HEK293 cells cotransfected with pEF6/BRAFV600E or pEF6/HRASG12V but not in the cells cotransfected with pEF6 or pEF6/BRAF (Figure 1B). Expression of a 3.5-kb HPR1 promoter-driven luciferase reporter gene was also slightly increased in HEK293 cells cotransfected with the pEF6 encoding wild-type BRAF but was dramatically increased in the cells cotransfected by pEF6/BRAFV600E and pEF6/HRASG12V. Induction of HPR1 promoter-driven luciferase expression by HRASV12 was less potent than BRAFV600E mutant, although the former is much more potent in activating the MAP kinase pathway [22].
B-Raf Kinase Activation Leads to Accelerated Cell Surface HS Degradation
We next tested whether increased HPR1 expression bymutant BRAF or HRAS leads to the degradation of HSPG. HEK293 cells were transiently transfected with pEF6, pEF6/BRAF, pEF6/BRAFV600E, or pEF6/HRASV12. As shown in Figure 2A, the cell surface HS expression was abrogated in the cells transfected with mutant BRAF or RAS gene but not in those transfected with the pEF6 empty vector or pEF6/BRAF (Figure 2A). Two HPR1 inhibitors, PI-88 or heparin (50 µg/ml each), were able to restore the cell surface HS levels of HEK293 cells transfected with pEF6/BRAFV600E or pEF6/HRASG12V (Figure 2B). Similar results were obtained in 10/56A cells, immortalized human glomerular epithelial cells (data not shown). These observations suggest that downregulation of HS expression in transfected cells is due to increased degradation of HS by HPR1.
Figure 2.
B-Raf kinase activation induces cell surface HS degradation. (A) HEK293 cells transiently transfected with the indicated vectors. After incubating for 48 hours, the cells were harvested and analyzed for cell surface HS levels by FACS. (B) HPR1 inhibitors restored cell surface HS levels. Cells transiently transfected with the empty vector or the vector encoding BRAFV600E or HRASG12V in the absence or presence of PI-88 or heparin (50 µg/ml) for 48 hours were analyzed for cell surface HS levels by FACS with anti-HSmAb. Green line indicates IgM control; red line, anti-HS mAb.
Inhibition of HPR1 Gene Expression in a BRAF-Suppressed Tumor Cell Line
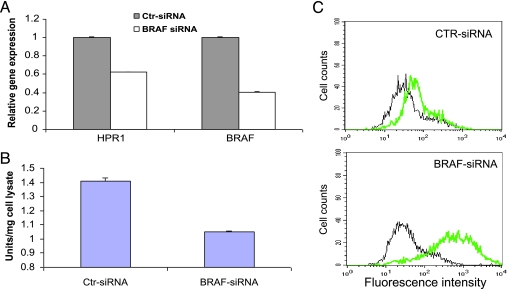
To verify that B-Raf kinase activation plays a critical role in inducing HPR1 gene expression, we tested if BRAF knockdown leads to decreased HPR1 gene expression. Knockdown of BRAF expression in a BRAF-mutated KAT-10 cell line by stable transfection of BRAF siRNA has been previously reported [24]. RT-PCR analysis revealed that BRAF knockdown reduced BRAF and HPR1 mRNA levels by 60% and 38%, respectively (Figure 3A). HPR1 enzymatic activity in the cell lysates of BRAF siRNA-transfected KAT-10 cells was reduced by 30% compared with that in control siRNA-transfected cells (Figure 3B). However, the cell surface HSlevels were dramatically increased in BRAF siRNA-transfected KAT 10 cells compared with that in control siRNA-transfected KAT-10 cells (Figure 3C). These observations strongly suggest that B-Raf kinase activation leads to increased HPR1 expression.
Figure 3.
BRAF gene knockdown leads to decreased HPR1 expression and increased cell surface HS levels. (A) Real-time RT-PCR analysis of HPR1 and BRAF mRNA. Total RNA was extracted from KAT-10 cells and analyzed for HPR1 and BRAF mRNA by real-time RT-PCR. One of two experiments in triplicate with similar results is shown. (B) HPR1 enzymatic activity. Cell lysates of KAT-10 cells transfected with control or BRAF siRNA were analyzed for HPR1 activity by ELISA. (C) FACS analysis of the cell surface HS levels. Single cell suspensions of KAT-10 cells transfected with control or BRAF siRNA were analyzed for cell surface HS levels by FACS. Black line indicates IgM control; green line, anti-HS mAb.
HPR1 Promoter Activity Is Decreased in BRAF-Suppressed KAT-10 Cells
Our cotransfection experiment in Figure 1 suggests that mutant BRAF or HRAS leads to increased HPR1 promoter activation. To corroborate this observation, we tested whether HPR1 promoter activity was decreased in BRAF-suppressed KAT-10 cells. The luciferase reporter gene driven by a 0.3-, 0.7-, or 3.5-kb HPR1 promoter was introduced into control or BRAF siRNA-transfected KAT-10 cells. As shown in Figure 4, luciferase reporter gene expression driven by 0.3-, 0.7-, and 3.5-kb HPR1 promoter were at least two-fold higher in control than BRAF siRNA-transfected KAT-10 cells. These observations suggest that down-regulation of HPR1 gene expression in BRAF-suppressed KAT-10 cells is mediated by suppression of HPR1 promoter activity.
Identification of a GABP-Binding Site as a Critical cis-Responsive Element Responsible for BRAF-Induced HPR1 Gene Expression
We and others have previous shown that several transcription factors, mainly Sp1, the Ets family transcription factors, and Egr-1 (Figure 7) play an important role in regulating basal and inducible HPR1 promoter activity [12–16]. GABP, a member of the Ets family transcription factors, can be activated by B-Raf kinase. We tested whether elimination of the Ets transcription factor binding site in the HPR1 promoter could abrogate BRAF mutation-induced HPR1 promoter activity. A panel of HPR1 promoter constructs with the ERE sites being mutated (Figure 5A) was introduced into control or BRAF siRNA-transfected KAT-10 cells. Consistent with our previous observations [13], mutation of any one of the four ERE sites significantly reduced HPR1 promoter activity (Figure 5B). Luciferase gene expression driven by a 0.3-kb HPR1 promoter in control siRNA-transfected KAT-10 cells was about two-fold higher than that in BRAF siRNA-transfected KAT-10 cells. Elimination of any of the four ERE sites in this 0.3-kb HPR1 promoter led to a comparable HPR1 promoter activity in control or in BRAF siRNA-transfected KAT-10 cells. These observations suggest that the presence of all ERE sites in the HPR1 promoter is required for mediating BRAF mutation-induced HPR1 promoter activation.
To verify that BRAF mutation-induced HPR1 gene expression is indeed mediated by the GABP binding sites present in the HPR1 promoter, we tested whether luciferase reporter gene driven by a four-copy GABP binding site is higher in control than in BRAF siRNA-transfected KAT-10 cells. As shown in Figure 5C, there was no difference in the luciferase activity in control and BRAF siRNA-transfected KAT-10 cells transfected with a pGL3/Basic reporter construct. However, luciferase gene expression driven by the 4xGABP binding site was almost two-fold higher in control than in BRAF siRNA-transfected KAT-10 cells. This observation strongly suggests that the transcriptional activity of the Ets family transcription factors is reduced in BRAF-suppressed KAT-10 cells.
Down-regulation of GABPβ Expression in BRAF-Suppressed KAT-10 Cells
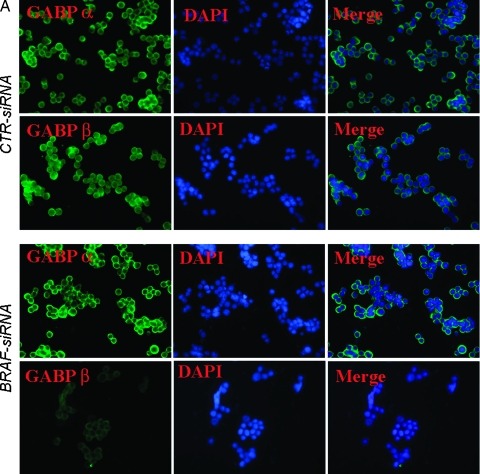
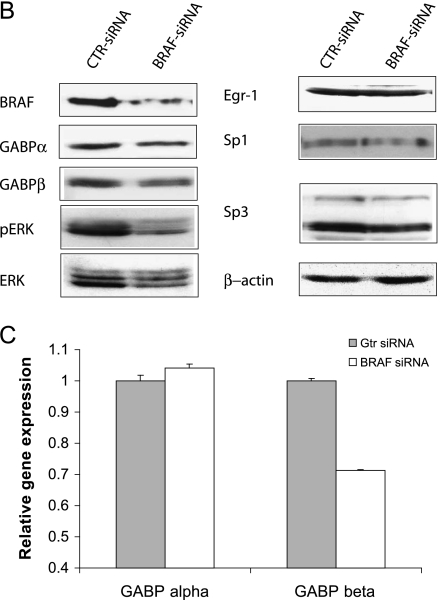
Several Ets family transcription factors are upregulated in various malignancies [32,33], probably because of oncogene mutation and activation. Here we tested whether decreased HPR1 promoter activity in BRAF-suppressed KAT-10 cells was due to decreased GABP expression or activation. We first conducted immunofluorescence (IF) staining to determine whether there was a difference in GABP nucleus translocation between control and BRAF-suppressed KAT-10 cells. As shown in Figure 6A, both GABPα and β were extensively stained in the cytoplasm and nuclei of control siRNA-transfected KAT-10 cells. The signals for GABPβ but not GABPα were much weaker in both the cytoplasm and nucleus in BRAF siRNA-transfected cells than in control siRNA-transfected cells. To verify that GABPβ expression was downregulated in BRAF-suppressed KAT-10 cells, Western blot analysis was conducted to compare GABPα and β expression. GABPβ was expressed at a significantly lower level in BRAF siRNA-transfected than in control siRNA-transfected KAT-10 cells (Figure 6B). ERK phosphorylation and BRAF expression were dramatically reduced in BRAF siRNA-transfected KAT-10 cells compared with those in control siRNA-transfected KAT-10 cells. The levels of Sp1, Sp3, total ERK, and Egr-1 expression were not reduced or were only marginally reduced in BRAF siRNA-transfected KAT-10 cells. To further verify the reduction of GABPβ in BRAF siRNA-transfected cells, we conducted real-time RT-PCR and found that GABPβ mRNA levels were 30% lower in BRAF siRNA-transfected cells than in control siRNA-transfected KAT-10 cells. However, there was no difference in GABPα mRNA levels between BRAF siRNA- and control siRNA-transfected KAT-10 cells. These observations suggest that suppression of BRAF expression leads to downregulation of GABPβ expression, subsequently leading to decreased HPR1 promoter activation and gene expression.
Figure 6.
Down-regulation of GABPβ in BRAF-suppressed KAT-10 cells. (A) IF staining of GABPα and β. Control or BRAF siRNA-transfected KAT-10 cells were stained with an anti-GABPα and β rabbit IgG, followed by staining with fluorescein-conjugated goat antirabbit IgG. A normal rabbit antiserum was included as a negative control. No nonspecific signal was presented (photograph not shown). (B) Western blot analyses of the signaling molecules and transcription factors involved in regulating HPR1 gene expression. Cell lysates from control or BRAF siRNA-transfected KAT-10 cells were analyzed for the expression of several transcription factors and signaling molecules by their specific antibodies. (C) Real-time RT-PCR analyses of GABPα and β expression. Total RNA was extracted fromKAT-10 cells transfected with BRAF or control siRNA expression vector and analyzed for GABPα and β by real-time RT-PCR. One of two experiments in triplicate with similar results is shown.
Discussion
The molecular mechanisms of increased HPR1 expression in a variety of malignancies are poorly understood. Increased HPR1 expression in prostate and bladder cancers is mediated by increased Egr-1 expression and HPR1 promoter hypomethylation [16,17]. p53 mutation may also contribute to increased HPR1 expression because wild-type p53 can negatively regulate HPR1 expression [18]. In the present study, we provide several lines of evidence that RAS or BRAF gene mutations are responsible for increased HPR1 expression: 1) mutant RAS or BRAF genes were able to directly activate the HPR1 promoter and to induce HPR1 gene expression; 2) increased HPR1 expression in mutant BRAF or RAS-transfected HEK293 cells led to the loss of cell surface HS, which could be blocked by two HPR1 inhibitors; 3) knockdown of BRAF gene expression in BRAF-mutated KAT-10 cells led to decreased HPR1 expression and increased cell surface HS levels; and 4) mechanistic studies demonstrated that GABP, a critical transcription factor involved in regulating HPR1 gene expression, was downregulated when BRAF expression was suppressed. These observations collectively suggest that increased HPR1 expression in a wide range of tumors could be caused in part by mutation of oncogenes such as BRAF, RAS, or their upstream growth factor receptors such as EGFR (Figure 7).
The Ets transcription factor family has more than 30 members that bind to the core DNA sequence GGA (A/T). GABP is a unique member of the Ets family because its α and β subunits form a heterodimer complex that often binds tandem ERE sites separated by approximately 10 to 30 bp. A recent genome-wide GABP occupancy study indicates that many GABP binding locations contain two ERE sites [34]. There are two sets of tandem ERE sites within the proximal region of the HPR1 promoter (Figure 7) [13]. Mutation of any one of the ERE sites should eliminate the binding of GABP to one set of tandem ERE sites. In addition, GABP cooperates with its neighboring Sp1, so that disruption of one set of GABP binding site leads to a significant reduction of HPR1 promoter activity [13]. This may partially explain that, although one set of the GABP binding sites in the HPR1 remains functional, there was no significant difference in luciferase reporter gene expression driven by the HPR1 promoter in control and BRAF siRNA-transfected KAT-10 cells.
GABP is one of the transcription factors that are involved in regulating HPR1 gene expression [13]. Our present study demonstrates that GABPβ expression was downregulated in BRAF-suppressed KAT-10 cells, as analyzed by Western blot and real-time RT-PCR. IF staining revealed that the signals for GABPβ expression were reduced in both cytoplasm and nucleus. It is likely that the reduced presence of GABPβ in nucleus may not be due to its inability to translocate into nucleus. Nevertheless, because the HPR1 promoter- and 4xGABP binding site-driven luciferase reporter gene expression was significantly lower in BRAF siRNA than in control siRNA-transfected KAT-10 cells, this suggests that GABP plays an important role in mediating mutant BRAF-induced HPR1 expression. It should be noted that BRAF gene mutation may induce the expression of other Ets family members that also contribute to HPR1 expression. For example, activation of the MAP kinase pathway leads to increased Ets2 expression and phosphorylation, subsequently leading to increased matrix metalloproteinase 9 expression [35–37].
Previous studies have shown that GABP can be phosphorylated by Raf-1 kinase [38,39]. Fromm and Burden [40] showed that GABPα can be phosphorylated at threonine 280 and activated by ERK and JNK kinases in neuregulin-1-stimulated muscle cells. However, a recent study suggests that GABPα phosphorylation is not essential for neuregulin-1-induced acetylcholine receptor expression [41]. Although it is likely that MAP kinase activation due to BRAF gene mutation may lead to increased phosphorylation of GABPα, increased GABPβ expression in BRAF-mutated cell lines may play a dominant role in mediating BRAF mutation-induced HPR1 expression.
It has been well documented that several transcription factors of the Ets family, in particular Ets-1, Ets-2, and PEA, are overexpressed in a variety of malignancies such as breast and ovarian cancers. GABP is ubiquitously expressed in normal tissues. GABP plays a critical role in regulating the expression of BRCA1 and prolactin, both of which are involved in tumorigenesis [42–44]. Our present study suggests the possibility that GABPβ expression can be upregulated in a variety of cancers due to the activation of the MAP kinase pathway. GABP and Sp1, another housekeeping transcription factor that is upregulated in a variety of cancers [45], can cooperate to induce HPR1 expression in cancer (Figure 7). Egr-1 is another transcription factor involved in regulating HPR1 gene expression [12–15]. Egr-1 overexpression in cancer may also contribute to increased HPR1 expression (Figure 7). Our present study shows that there was no significant difference in Sp1 and Egr-1 expression in control and BRAF siRNA-transfected KAT-10 cells. In addition, when the GABP binding sites in the HPR1 promoter were mutated, there was no difference in luciferase gene expression in control or BRAF siRNA vector-transfected KAT-10 cells. These observations suggest that Sp1 and Egr-1 do not contribute to mutant B-Raf kinase-mediated increase of HPR1 expression. Because we did not analyze the expression of other Ets family member, it remains unknown if other Ets transcript factors may also required for mutant BRAF-induced HPR1 expression.
BRAF-mutated thyroid cancers and melanomas tend to be more aggressive than those without BRAF mutation [19,46]. B-Raf kinase activation due to the BRAF gene mutations leads to the up-regulation of several molecules that are involved in tumor metastasis [19,46]. For example, BRAFV600E mutation has been shown to regulate melanoma metastasis by increasing fibronectin expression and by promoting interaction of melanoma cells with neutrophils to facilitate extravasation across the endothelial cell lining as well as enhancing proliferation in the lung microenvironment [47,48]. Inhibition of these processes leads to a decrease of lung metastasis of BRAF-mutated melanoma cells by four- to five-fold [49]. BRAF gene mutation also leads to the upregulation of matrix metalloproteinase expression in thyroid cancer and melanoma [50,51]. Our present study demonstrates that BRAF mutation led to up-regulation of HPR1, an endoglycosidase that is involved in tumor angiogenesis and metastases. This observation is consistent with a prior study showing that sorafenib, an inhibitor of B-Raf kinase and other growth factor receptor, blocks vascular development in malignant melanomas in a xenograft model [52]. It is anticipated that sorafenib treatment may lead to a significant decrease in HPR1 expression, subsequently suppressing tumor angiogenesis. These studies collectively suggest that up-regulation of HPR1 and other molecules due to BRAF gene mutation may act in concert to promote tumor angiogenesis and metastases.
Acknowledgments
The authors thank R. Marais (The Institute of Cancer Research, Signal Transduction Team, Cancer Research UK Centre for Cell and Molecular Biology, London, United Kingdom) for kindly providing the pEF6 vectors, U. A. Rapp (University of Würzburg, Würzburg, Germany) for anti-GABPα and β rabbit antisera, and Jeffrey Platt (Transplantation Biology Program, University of Michigan) for kindly providing purified platelet HPR1.
Footnotes
This work was supported in part by grants from the Thyroid Research Advisory Council and the ThyCa Foundation to X.X.
References
- 1.Vlodavsky I, Friedmann Y, Elkin M, Aingorn H, Atzmon R, Ishai-Michaeli R, Bitan M, Pappo O, Peretz T, Michal I, et al. Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nat Med. 1999;5:793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- 2.Vlodavsky I, Friedmann Y. Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J Clin Invest. 2001;108:341–347. doi: 10.1172/JCI13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hulett M, Freeman C, Hamdorf B, Baker R, Harris M, Parish C. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis. Nat Med. 1999;5:803–809. doi: 10.1038/10525. [DOI] [PubMed] [Google Scholar]
- 4.Parish CR, Freeman C, Hulett MD. Heparanase: a key enzyme involved in cell invasion. Biochim Biophys Acta. 2001;1471:M99–M108. doi: 10.1016/s0304-419x(01)00017-8. [DOI] [PubMed] [Google Scholar]
- 5.Zetser A, Bashenko Y, Miao HQ, Vlodavsky I, Ilan N. Heparanase affects adhesive and tumorigenic potential of human glioma cells. Cancer Res. 2003;63:7733–7741. [PubMed] [Google Scholar]
- 6.Goldshmidt O, Zcharia E, Cohen M, Aingorn H, Cohen I, Nadav L, Katz BZ, Geiger B, Vlodavsky I. Heparanase mediates cell adhesion independent of its enzymatic activity. FASEB J. 2003;17:1015–1025. doi: 10.1096/fj.02-0773com. [DOI] [PubMed] [Google Scholar]
- 7.Zetser A, Bashenko Y, Edovitsky E, Levy-Adam F, Vlodavsky I, Ilan N. Heparanase induces vascular endothelial growth factor expression: correlation with p38 phosphorylation levels and Src activation. Cancer Res. 2006;66:1455–1463. doi: 10.1158/0008-5472.CAN-05-1811. [DOI] [PubMed] [Google Scholar]
- 8.Cohen-Kaplan V, Naroditsky I, Zetser A, Ilan N, Vlodavsky I, Doweck I. Heparanase induces VEGF C and facilitates tumor lymphangiogenesis. Int J Cancer. 2008;123:2566–2573. doi: 10.1002/ijc.23898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen-Kaplan V, Doweck I, Naroditsky I, Vlodavsky I, Ilan N. Heparanase augments epidermal growth factor receptor phosphorylation: correlation with head and neck tumor progression. Cancer Res. 2008;68:10077–10085. doi: 10.1158/0008-5472.CAN-08-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai NS, Simizu S, Morisaki D, Muroi M, Osada H. Requirement of the conserved, hydrophobic C-terminus region for the activation of heparanase. Exp Cell Res. 2008;314:2834–2845. doi: 10.1016/j.yexcr.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Fux L, Feibish N, Cohen-Kaplan V, Gingis-Velitski S, Feld S, Geffen C, Vlodavsky I, Ilan N. Structure-function approach identifies a COOH-terminal domain that mediates heparanase signaling. Cancer Res. 2009;69:1758–1767. doi: 10.1158/0008-5472.CAN-08-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu WC, Liu YN, Kang BB, Chen JH. Trans-activation of heparanase promoter by ETS transcription factors. Oncogene. 2003;22:919–923. doi: 10.1038/sj.onc.1206201. [DOI] [PubMed] [Google Scholar]
- 13.Jiang P, Kumar A, Parrillo JE, Dempsey LA, Platt JL, Prinz RA, Xu X. Cloning and characterization of the human heparanase-1 (HPR1) gene promoter: role of GA-binding protein and Sp1 in regulating HPR1 basal promoter activity. J Biol Chem. 2002;277:8989–8998. doi: 10.1074/jbc.M105682200. [DOI] [PubMed] [Google Scholar]
- 14.de Mestre AM, Rao S, Hornby JR, Soe-Htwe T, Khachigian LM, Hulett MD. Early growth response gene 1 (EGR1) regulates heparanase gene transcription in tumor cells. J Biol Chem. 2005;280:35136–35147. doi: 10.1074/jbc.M503414200. [DOI] [PubMed] [Google Scholar]
- 15.de Mestre AM, Khachigian LM, Santiago FS, Staykova MA, Hulett MD. Regulation of inducible heparanase gene transcription in activated T cells by early growth response 1. J Biol Chem. 2003;278:50377–50385. doi: 10.1074/jbc.M310154200. [DOI] [PubMed] [Google Scholar]
- 16.Ogishima T, Shiina H, Breault JE, Terashima M, Honda S, Enokida H, Urakami S, Tokizane T, Kawakami T, Ribeiro-Filho LA, et al. Promoter CpG hypomethylation and transcription factor EGR1 hyperactivate heparanase expression in bladder cancer. Oncogene. 2005;24:6765–6772. doi: 10.1038/sj.onc.1208811. [DOI] [PubMed] [Google Scholar]
- 17.Ogishima T, Shiina H, Breault JE, Tabatabai L, Bassett WW, Enokida H, Li LC, Kawakami T, Urakami S, Ribeiro-Filho LA, et al. Increased heparanase expression is caused by promoter hypomethylation and up-regulation of transcriptional factor early growth response-1 in human prostate cancer. Clin Cancer Res. 2005;11:1028–1036. [PubMed] [Google Scholar]
- 18.Baraz L, Haupt Y, Elkin M, Peretz T, Vlodavsky I. Tumor suppressor p53 regulates heparanase gene expression. Oncogene. 2006;25:3939–3947. doi: 10.1038/sj.onc.1209425. [DOI] [PubMed] [Google Scholar]
- 19.Michaloglou C, Vredeveld LC, Mooi WJ, Peeper DS. BRAF(E600) in benign and malignant human tumours. Oncogene. 2008;27:877–895. doi: 10.1038/sj.onc.1210704. [DOI] [PubMed] [Google Scholar]
- 20.Xu X, Quiros RM, Gattuso P, Ain KB, Prinz RA. High prevalence of BRAF gene mutation in papillary thyroid carcinomas and thyroid tumor cell lines. Cancer Res. 2003;63:4561–4567. [PubMed] [Google Scholar]
- 21.Xu X, Quiros RM, Maxhimer JB, Jiang P, Marcinek R, Ain KB, Platt JL, Shen J, Gattuso P, Prinz RA. Inverse correlation between heparan sulfate composition and heparanase-1 gene expression in thyroid papillary carcinomas: a potential role in tumor metastasis. Clin Cancer Res. 2003;9:5968–5979. [PubMed] [Google Scholar]
- 22.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 23.Li XR, Chong AS, Wu J, Roebuck KA, Kumar A, Parrillo JE, Rapp UR, Kimberly RP, Williams JW, Xu X. Transcriptional regulation of Fas gene expression by GA-binding protein and AP-1 in T cell antigen receptor·CD3 complex-stimulated T cells. J Biol Chem. 1999;274:35203–35210. doi: 10.1074/jbc.274.49.35203. [DOI] [PubMed] [Google Scholar]
- 24.Liu D, Liu Z, Condouris S, Xing M. BRAF V600E maintains proliferation, transformation, and tumorigenicity of BRAF-mutant papillary thyroid cancer cells. J Clin Endocrinol Metab. 2007;92:2264–2271. doi: 10.1210/jc.2006-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schweppe RE, Klopper JP, Korch C, Pugazhenthi U, Benezra M, Knauf JA, Fagin JA, Marlow L, Copland JA, Smallridge RC, et al. Deoxyribonucleic acid profiling analysis of 40 human thyroid cancer cell lines reveals cross-contamination resulting in cell line redundancy and misidentification. J Clin Endocrinol Metab. 2008;93:4331–4341. doi: 10.1210/jc.2008-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eustice DC, Feldman PA, Colberg-Poley AM, Buckery RM, Neubauer RH. A sensitive method for the detection of beta-galactosidase in transfected mammalian cells. Biotechniques 11, 1991;739–740:742–743. [PubMed] [Google Scholar]
- 27.Xu X, Ding J, Rao G, Shen J, Prinz RA, Rana N, Dmowski WP. Estradiol induces heparanase-1 expression and heparan sulphate proteoglycan degradation in human endometrium. Hum Reprod. 2007;22:927–937. doi: 10.1093/humrep/del483. [DOI] [PubMed] [Google Scholar]
- 28.Xu X, Rao G, Quiros RM, Kim AW, Miao HQ, Brunn GJ, Platt JL, Gattuso P, Prinz RA. In vivo and in vitro degradation of heparan sulfate (HS) proteoglycans by HPR1 in pancreatic adenocarcinomas. Loss of cell surface HS suppresses fibroblast growth factor 2-mediated cell signaling and proliferation. J Biol Chem. 2007;282:2363–2373. doi: 10.1074/jbc.M604218200. [DOI] [PubMed] [Google Scholar]
- 29.Maxhimer JB, Somenek M, Rao G, Pesce CE, Baldwin D, Jr, Gattuso P, Schwartz MM, Lewis EJ, Prinz RA, Xu X. Heparanase-1 gene expression and regulation by high glucose in renal epithelial cells: a potential role in the pathogenesis of proteinuria in diabetic patients. Diabetes. 2005;54:2172–2178. doi: 10.2337/diabetes.54.7.2172. [DOI] [PubMed] [Google Scholar]
- 30.Quiros RM, Rao G, Plate J, Harris JE, Brunn GJ, Platt JL, Gattuso P, Prinz RA, Xu X. Elevated serum heparanase-1 levels in patients with pancreatic carcinoma are associated with poor survival. Cancer. 2006;106:532–540. doi: 10.1002/cncr.21648. [DOI] [PubMed] [Google Scholar]
- 31.Ihrcke NS, Parker W, Reissner KJ, Platt JL. Regulation of platelet heparanase during inflammation: role of pHand proteinases. J Cell Physiol. 1998;175:255–267. doi: 10.1002/(SICI)1097-4652(199806)175:3<255::AID-JCP3>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 32.Turner DP, Watson DK. ETS transcription factors: oncogenes and tumor suppressor genes as therapeutic targets for prostate cancer. Expert Rev Anticancer Ther. 2008;8:33–42. doi: 10.1586/14737140.8.1.33. [DOI] [PubMed] [Google Scholar]
- 33.de Launoit Y, Baert JL, Chotteau-Lelievre A, Monte D, Coutte L, Mauen S, Firlej V, Degerny C, Verreman K. The Ets transcription factors of the PEA3 group: transcriptional regulators in metastasis. Biochim Biophys Acta. 2006;1766:79–87. doi: 10.1016/j.bbcan.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 34.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fowles LF, Martin ML, Nelsen L, Stacey KJ, Redd D, Clark YM, Nagamine Y, McMahon M, Hume DA, Ostrowski MC. Persistent activation of mitogen-activated protein kinases p42 and p44 and ets-2 phosphorylation in response to colony-stimulating factor 1/c-fms signaling. Mol Cell Biol. 1998;18:5148–5156. doi: 10.1128/mcb.18.9.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trimboli AJ, Cantemir-Stone CZ, Li F, Wallace JA, Merchant A, Creasap N, Thompson JC, Caserta E, Wang H, Chong JL, et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature. 2009;461:1084–1091. doi: 10.1038/nature08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCarthy SA, Chen D, Yang BS, Garcia Ramirez JJ, Cherwinski H, Chen XR, Klagsbrun M, Hauser CA, Ostrowski MC, McMahon M. Rapid phosphorylation of Ets-2 accompanies mitogen-activated protein kinase activation and the induction of heparin-binding epidermal growth factor gene expression by oncogenic Raf-1. Mol Cell Biol. 1997;17:2401–2412. doi: 10.1128/mcb.17.5.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmeyer A, Avots A, Flory E, Weber CK, Serfling E, Rapp UR. The GABP-responsive element of the inerleukin-2 enhancer is regulated by JNK/SAPK-activating pathways in T lymphocytes. J Biol Chem. 1998;273:10112–10119. doi: 10.1074/jbc.273.17.10112. [DOI] [PubMed] [Google Scholar]
- 39.Flory E, Hoffmeyer A, Smola U, Rapp UR, Bruder JT. Raf-1 kinase targets GA-binding protein in transcriptional regulation of the human immunodeficiency virus type 1 promoter. J Virol. 1996;70:2260–2268. doi: 10.1128/jvi.70.4.2260-2268.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fromm L, Burden SJ. Neuregulin-1-stimulated phosphorylation of GABP in skeletal muscle cells. Biochemistry. 2001;40:5306–5312. doi: 10.1021/bi002649m. [DOI] [PubMed] [Google Scholar]
- 41.Herndon CA, Fromm L. Neuregulin-1 induces acetylcholine receptor transcription in the absence of GABPαlpha phosphorylation. J Neurosci Res. 2008;86:982–991. doi: 10.1002/jnr.21563. [DOI] [PubMed] [Google Scholar]
- 42.Atlas E, Stramwasser M, Mueller CR. A CREB site in the BRCA1 proximal promoter acts as a constitutive transcriptional element. Oncogene. 2001;20:7110–7114. doi: 10.1038/sj.onc.1204890. [DOI] [PubMed] [Google Scholar]
- 43.Hoare S, Copland JA, Wood TG, Jeng YJ, Izban MG, Soloff MS. Identification of a GABP alpha/beta binding site involved in the induction of oxytocin receptor gene expression in human breast cells, potentiation by c-Fos/c-Jun. Endocrinology. 1999;140:2268–2279. doi: 10.1210/endo.140.5.6710. [DOI] [PubMed] [Google Scholar]
- 44.Schweppe RE, Melton AA, Brodsky KS, Aveline LD, Resing KA, Ahn NG, Gutierrez-Hartmann A. Purification and mass spectrometric identification of GA-binding protein (GABP) as the functional pituitary Ets factor binding to the basal transcription element of the prolactin promoter. J Biol Chem. 2003;278:16863–16872. doi: 10.1074/jbc.M213063200. [DOI] [PubMed] [Google Scholar]
- 45.Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 46.Xing M. BRAF mutation in papillary thyroid cancer: pathogenic role, molecular bases, and clinical implications. Endocr Rev. 2007;28:742–762. doi: 10.1210/er.2007-0007. [DOI] [PubMed] [Google Scholar]
- 47.Gaggioli C, Robert G, Bertolotto C, Bailet O, Abbe P, Spadafora A, Bahadoran P, Ortonne JP, Baron V, Ballotti R, et al. Tumor-derived fibronectin is involved in melanoma cell invasion and regulated by V600E B-Raf signaling pathway. J Invest Dermatol. 2007;127:400–410. doi: 10.1038/sj.jid.5700524. [DOI] [PubMed] [Google Scholar]
- 48.Liang S, Sharma A, Peng HH, Robertson G, Dong C. Targeting mutant (V600E) B-Raf in melanoma interrupts immunoediting of leukocyte functions and melanoma extravasation. Cancer Res. 2007;67:5814–5820. doi: 10.1158/0008-5472.CAN-06-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma A, Tran MA, Liang S, Sharma AK, Amin S, Smith CD, Dong C, Robertson GP. Targeting mitogen-activated protein kinase/extracellular signal-regulated kinase kinase in the mutant (V600E) B-Raf signaling cascade effectively inhibits melanoma lung metastases. Cancer Res. 2006;66:8200–8209. doi: 10.1158/0008-5472.CAN-06-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huntington JT, Shields JM, Der CJ, Wyatt CA, Benbow U, Slingluff CL, Jr, Brinckerhoff CE. Overexpression of collagenase 1 (MMP-1) is mediated by the ERK pathway in invasive melanoma cells: role of BRAF mutation and fibroblast growth factor signaling. J Biol Chem. 2004;279:33168–33176. doi: 10.1074/jbc.M405102200. [DOI] [PubMed] [Google Scholar]
- 51.Mesa C, Jr, Mirza M, Mitsutake N, Sartor M, Medvedovic M, Tomlinson C, Knauf JA, Weber GF, Fagin JA. Conditional activation of RET/PTC3 and BRAFV600E in thyroid cells is associated with gene expression profiles that predict a preferential role of BRAF in extracellular matrix remodeling. Cancer Res. 2006;66:6521–6529. doi: 10.1158/0008-5472.CAN-06-0739. [DOI] [PubMed] [Google Scholar]
- 52.Sharma A, Trivedi NR, Zimmerman MA, Tuveson DA, Smith CD, Robertson GP. Mutant V599EB-Raf regulates growth and vascular development of malignant melanoma tumors. Cancer Res. 2005;65:2412–2421. doi: 10.1158/0008-5472.CAN-04-2423. [DOI] [PubMed] [Google Scholar]