Abstract
In this review we provide a systematic analysis of transcriptomic signatures derived from 42 breast cancer gene expression studies, in an effort to identify the most relevant breast cancer biomarkers using a meta-analysis method. Meta-data revealed a set of 117 genes that were the most commonly affected ranging from 12% to 36% of overlap among breast cancer gene expression studies. Data mining analysis of transcripts and protein-protein interactions of these commonly modulated genes indicate three functional modules significantly affected among signatures, one module related with the response to steroid hormone stimulus, and two modules related to the cell cycle. Analysis of a publicly available gene expression data showed that the obtained meta-signature is capable of predicting overall survival (P < 0.0001) and relapse-free survival (P < 0.0001) in patients with early-stage breast carcinomas. In addition, the identified meta-signature improves breast cancer patient stratification independently of traditional prognostic factors in a multivariate Cox proportional-hazards analysis.
Keywords: breast cancer, biomarkers, gene expression signatures
Introduction
Development of effective tools such as DNA microarrays for monitoring gene expression on a large scale has resulted in the discovery of gene networks and regulatory pathways in various tumor processes. In this respect, global gene expression in breast cancer has been profiled extensively over the last decade, which allowed the identification of breast cancer molecular subtypes and the development of prognostic and predictive gene signatures, resulting in an improved understanding of the heterogeneity of breast cancer.
In pioneering work, Perou et al used cDNA arrays to test the expression of approximately 8,000 genes in samples from 42 breast cancer patients1. This first report suggested that primary breast carcinomas could be classified into specific ‘intrinsic subtypes’ distinguished by particular gene expression patterns. These data were confirmed and extended by Sorlie et al., who investigated the clinical usefulness of the breast cancer subtypes identified by screening for correlations between gene expression patterns and clinically relevant parameters. They demonstrated that classification of tumors based on gene expression patterns could be used as a prognostic marker with respect to overall and relapse-free survival in a subset of patients who had received uniform therapy.2,3 The five subtypes identified (Luminal A, Luminal B, Basal-like, ERBB2 positive/ER negative and normal breast-like) represent different biological entities and might originate from different cell types. One of the five subtypes was characterized by over-expression of ERBB2 and poor prognosis. A second tumor subtype, lacking expression of estrogen receptor α (ER) and also with a poor clinical prognosis, has been termed “basal,” as it resembles the pattern found in basal epithelial cells of the normal mammary gland. This basal tumor type differs from two other subtypes, luminal A and luminal B subtypes, both of which are ER positive and resemble cells that line the duct and give rise to the majority of breast cancers.2,3 Additionally, much work has been done on the hormonal status of breast cancers; DNA microarray and SAGE (Serial Analysis of Gene Expression) studies have focused on the ability of these gene profiling techniques to accurately discriminate ERα(+) from ERα(−) phenotypes.4–7 Furthermore, various laboratories have identified gene expression signatures that correlate with prognosis and can be used to predict the risk of disease recurrence and outcome in breast cancer patients.8,9
According to Sotiriou and Pusztai,10 global gene expression profiling has employed three different strategies to develop genomic signatures that may provide better prediction of clinical outcome. First, in the ‘top-down’ approach, gene expression data from tumors (or cell line models) are correlated with the clinical outcome of patients to identify prognostic gene signatures (eg, 70- and 76-gene poor-prognosis signatures). Second, in the ‘bottom-up’ approach the prognostic predictor is derived from a gene expression signature related to a biological pathway or process (eg, wound-response, invasiveness and stromal related poor-prognosis signatures). Third, in the candidate-gene list approach a set of biomarkers are prospectively selected on the basis of previous biological knowledge (eg, recurrence score signature)10.
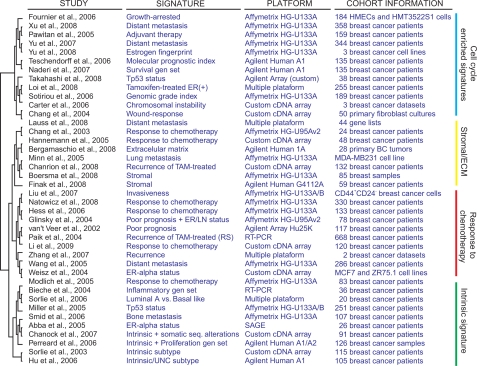
Among the myriad of prognostic or predictive gene expression signatures generated, only four genetic assays have been currently licensed for commercial use: the 70-gene ‘poor-prognosis’ signature (MammaPrint, Agendia BV, Amsterdam), the 21-gene ‘recurrence score’ signature (Oncotype DX, Genomic Health, Redwood City, California), the 97-gene ‘genomic grade index’ signature (MapQuant Dx, Ipsogen, Marseille, France), and the 2-gene ratio signature (Theros, Biotheranostics, San Diego, California). Some of these signatures have been previously compared. Fan et al demonstrated that 5 gene expression signatures, the intrinsic subtypes, the 70-gene, the 2-gene expression ratio, the 21-gene, and the wound response signature, had similar performance in predicting outcome.11 However, comparisons of the gene lists derived from these studies have shown a limited or zero overlap between signatures. The reasons for this disparity have been attributed to differences in the group of patient analyzed (ER status, tumor grade, stage, etc), in sample preparation (bulk, micro-dissected, etc.), in microarray platforms (high or low coverage of the human genome) and the statistical methods used (supervised or unsupervised methods, gene selection, construction of the classifiers, etc.). In this sense, Ein-Dor et al demonstrated that many equally prognostic or predictive gene sets can be obtained from the same study.12 These data showed that each gene signature identify different molecular features, which are predictive of the clinical outcome by looking a partial picture of breast cancer biology. More importantly, these data suggest that combining multiples gene expression signatures may provide an integrated view that would be useful to define the most relevant breast cancer biomarkers.
In the present review, we provide a comprehensive integration of 42 breast cancer gene expression signatures demonstrating that the overlap between gene expression signatures is greater than previously estimated by the comparison of a reduced set of gene lists.11 In addition, we demonstrate that the gene expression meta-signature is a powerful predictor of clinical outcome in patients with early-stage breast cancers. We also discuss the most relevant set of genes recurrently identified in these signatures re-analysis.
Materials and Methods
Identification of common gene expression features among breast cancer signatures
We employed the GeneSigDB (release 2.0) online resource (http://compbio.dfci.harvard.edu/genesigdb) for the detection of gene overlapping among breast cancer gene expression signatures available in this database.13 GeneSigDB is a manually curated and standardized (EnsEMBL gene identifiers) database of gene expression signatures (n = 957), which focuses on cancer and stem cell studies. We selected the most relevant gene signatures derived from 42 breast cancer gene expression profiling studies (from 2002 to 2009) (see additional file 1). For the selected signatures, the GeneSigDB web application provide one gene per signature heatmap-style plot colored in red or grey according to presence or absence of gene overlap, respectively.
Data extraction and hierarchical clustering
GeneSigDB data management was performed using a customizable HEM2TEM (for HeatMap to TExtMatrix) java tool developed by us for extracting a plain text matrix from the XML/HTML heatmap previously described. To enable unsupervised classification and illustration of the commonly overlapped genes between the 42 breast cancer gene expression signatures, we used the Multi Experiment Viewer (MeV 4.5) software (http://www.tm4.org/mev/).14 Two-way (by gene and by signature) hierarchical clustering was used to examine the relationships among the 42 breast cancer gene expression signatures. Hierarchical clustering was based on Spearman’s rank correlation distance metric and the complete linkage clustering method. Furthermore, we tested whether semantic terms (signature name, platform name or biological process) differed across clusters using the Fisher’s exact test. All P values were two sided, and P < 0.05 was considered significant. Subsequently, we selected the most frequently overlapped genes by applying a cutoff of 5 gene signatures (12% of 42 signatures) to generate the gene expression meta-signature for further analysis.
Data mining analysis
For automated functional annotation and classification of genes of interest based on Gene Ontology (GO) terms, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/)15.
In order to identify the molecular pathways that were mainly affected by the meta-signature, we look for protein/gene interaction networks in the common core of overlapped genes. The protein-protein interaction network was generated using the STRING database (‘Search Tool for the Retrieval of Interacting Genes/Proteins’) (http://string.embl.de/).16 This bioinformatic tool was used with the aims to collect, predict and unify most types of protein-protein associations, including direct and indirect associations. STRING runs a set of prediction algorithms and transfers known interactions from model organisms to other species based on predicted orthology of the respective proteins.16 In order to identify each gene in the database, we used both gene names and EnsEMBL gene identifiers in the ‘protein-mode’ application. The analysis input options were ‘co-occurrence’, ‘co-expression’, ‘experiments’, ‘databases’, and ‘text mining’ data at high confidence level of predicted human orthology groups. All of the raw data reported as additional files in this article are publicly available at the journal web site.
Gene expression meta-signature and survival analysis
To further investigate the prognostic value of the gene expression meta-signature, we did survival analyses in a publicly available breast cancer microarray study. We selected van de Vijver data set due to the biological diversity of breast tumors included in this study.17 Briefly, van de Vijver’s data set included 295 early-stage breast cancer samples (226 ER-positive and 69 ER-negative), some of whom were lymph-node-negative (n = 151) and the others were lymph-node-positive (n = 144). The patients had all been treated by radical mastectomy or breast-conserving surgery, followed in some cases by radiotherapy; and a fraction of patients had received adjuvant treatment. Data on relapse-free survival (defined as the time to a first event) and overall survival were available for all patients. The gene expression profile was derived by researchers from the Netherlands Cancer Institute and Rosetta Inpharmatics—Merck using Agilent Hu25K oligonucleotide (60mer) microarray (Agilent Technologies, Palo Alto, CA—USA). The gene expression matrix and the associated clinical data were obtained from the Rosetta Inpharmatics website17 (http://www.rii.com/publications/2002/nejm.html).
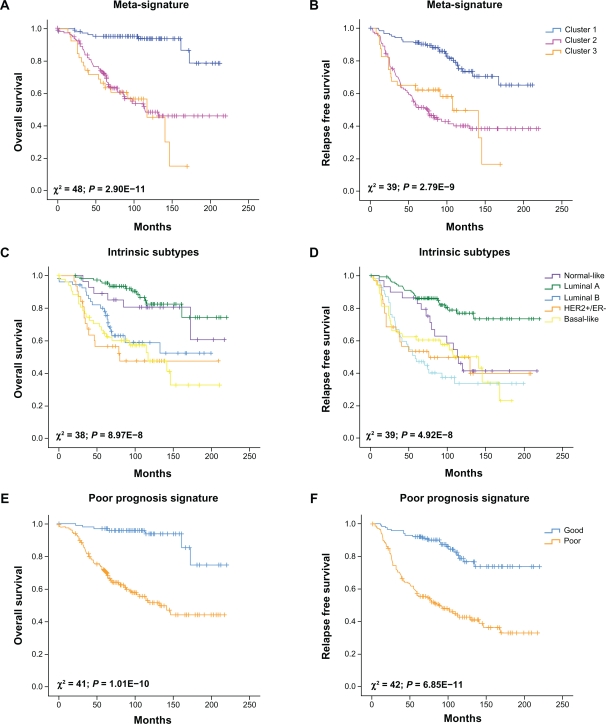
In an unsupervised analysis, 295 tumor samples were grouped by similarity of the 117 gene list meta-signature by complete linkage clustering by using the Multi Experiment Viewer software. The samples were segregated into three classes (from Cluster 1 to Cluster 3) based on the second bifurcation of the clustering dendrogram. In addition, we integrated the gene expression meta-signature with four prognostic or predictive gene signatures (Intrinsic subtype, Poor-prognosis, Recurrence Score and Wound Response signatures) to evaluate the data set. Tumor classification according to the four prognostic or predictive gene signatures were stablished based on data provided by Fan et al 2006.11 Kaplan–Meier survival curves and, log-rank statistics and the Cox proportional hazard method were performed by using the SPSS® statistic software package (SPSS Inc., Chicago). The multivariate Cox proportional-hazard model included: estrogen receptor status (ER-positive vs. ER-negative), tumor grade (grade 1 vs. 2 and grade 1 vs. 3), lymph node status (LN-negative vs. 1–3 LN-positives and LN-negative vs. > 3 LN-positives), age (as a continuous variable), tumor size (diameter ≤ 2 cm vs. diameter > 2 cm), treatment received (no adjuvant therapy vs. chemotherapy/hormonal therapy), and gene expression meta-signature predictive clusters (cluster 1 vs. cluster 2/3). Overall survival and relapse-free survival were the end points.
Results and Discussion
Based on a novel gene list meta-analysis approach, a systematic review of 42 gene signatures of breast cancer was performed in order to identify and compare the most relevant breast cancer biomarkers. The study approach underwent four phases: (a) detection of overlapping genes among the different signatures, (b) examination of the relationship between gene expression signatures by a two-way unsupervised analysis, (c) identification of the molecular pathways that are mainly affected by the gene expression meta-signature followed by (d) validation of the gene expression meta-signature’s prognostic value in a set of 295 patients with early-stage breast cancers obtained from van de Vijver et al study17.
Identification of the gene expression meta-signature and data mining analysis
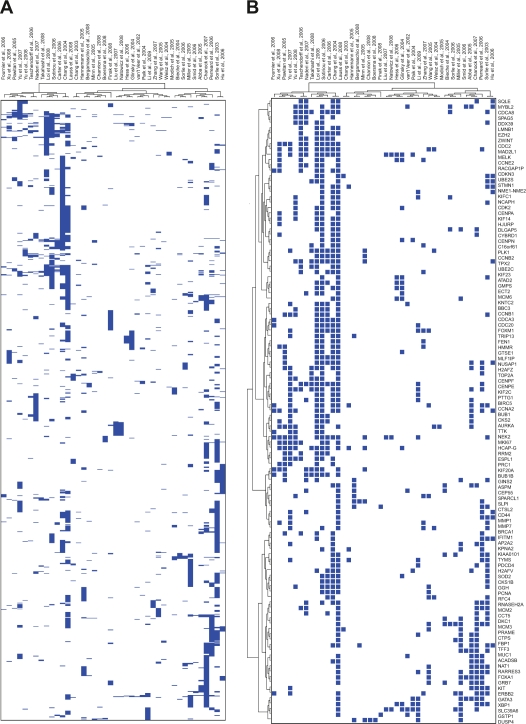
Among the 42 gene expression signatures (see additional file 1), a total of 946 transcripts were identified as overlapping in more than one study (Fig. 1A, Additional file 2). Of the 946 transcripts, 117 genes were identified in more than four studies, representing a set of the most frequents breast cancer biomarkers in this analysis (Fig. 1B). Additional file 2 shows the most common overlapping genes between breast cancer signatures.
Figure 1.
Overlap beween gene identifiers across 42 breast cancer gene expression signatures. A) Heatmap representation of 946 genes overlapping in more than one gene expression signature. B) Heatmap representation of 117 genes overlapping in at least 5 out of 42 gene expression signatures analyzed. Easch row is a gene and each column is a breast cancer gene expression signature. Presence of a gene is indicated by a blue box, and absence is white.
Hierarchical clustering analysis of the 42 gene expression studies classified the signatures in four groups: the intrinsic subtype signatures, the response to chemotherapy related signatures, the stromal/extracellular matrix (ECM) related signatures and the signatures enriched in cell cycle genes (Fig. 2). It can be clearly seen that related signatures such us intrinsic subtypes and ER-alpha status on the one hand, or stromal and extracellular matrix signatures on the other hand, have a large overlap relative to other gene expression signatures. Furthermore, it is interesting to note that the most common signatures cluster found was associated with the enrichment of cell cycle genes (Fig. 2). Non-statistically significant associations were detected between signatures clusters and the microarray platforms employed for gene expression profiling (P > 0.05).
Figure 2.
Hierarchical clustering analysis of the 42 breast cancer gene expression studies, classified them in four groups: the intrinsic subtypes, response to chemotherapy, stromal/extracellular matrix (ECM) and signatures enriched in cell cycle genes. It can clearly be seen that related signatures such us intrinsic subtype and ER-alpha status on the one hand, or stromal and extracellular matrix signatures on the other hand, have a large overlap relative to other gene expression signatures.
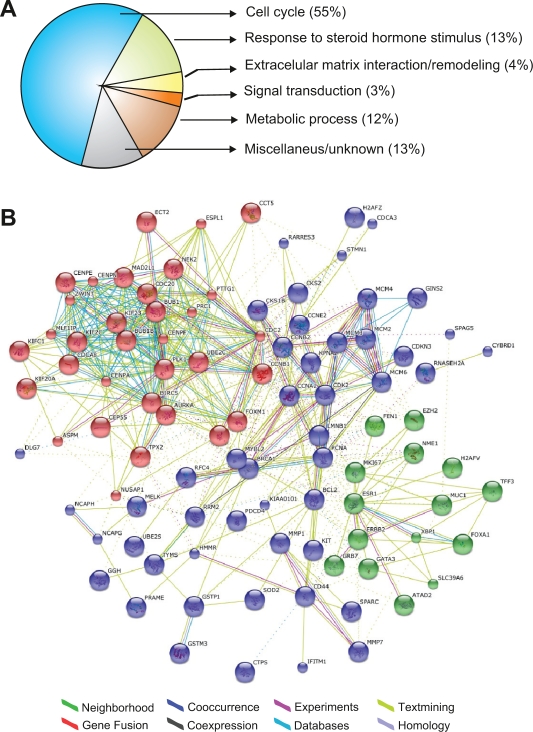
Gene Ontology annotation of the 117 gene meta-signature showed that approximately 55% of the transcripts are involved in cell cycle regulation, 13% are related to response to steroid hormone stimulus, 4% are related to extracellular matrix interaction/remodeling and 3% are related to other signal transduction pathways (Fig. 3A, additional file 2). Additionally, Figure 3B shows a protein-protein interaction network associating the common core of genes across gene expression signatures. The graph was generated employing the STRING on-line resource based on high confidence data. STRING is a comprehensive tool integrating protein association information with the capability to transfer known interactions from model organisms to other species. The generated graph (Fig. 3B) indicates strong interactions among a set of 95 proteins derived from the 117 gene meta-signature (81% of coverage). Furthermore, the network architecture suggests the existence of three functional modules (sets of genes that act in concert to carry out a specific function): a module related with the response to steroid hormone stimulus (green circles in Fig. 3B), and two modules related with the cell cycle signaling pathway (Fig. 3B).
Figure 3.
Data mining analysis of the gene expression meta-signature. A) Gene ontology (GO) classification of the 117 gene list meta-signature with specific gene ontology annotations based on biological processes or molecular function terms. B) Graph of protein-protein interactions among the 117 gene expression metasignature generated using the STRING database. In the network: links between proteins means the various interactions data supporting the network, colored by evidence type.
Gene expression meta-signature analysis and its clinical relevance as prognostic marker
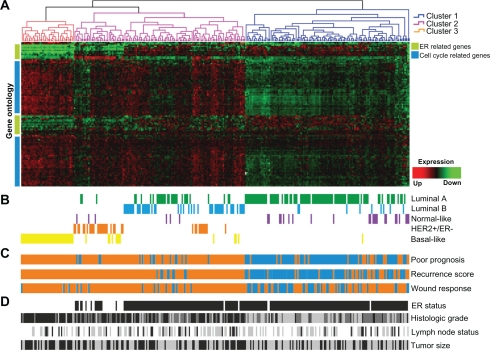
To further explore the prognostic value of gene expression meta-signature, we performed univariate and multivariate analysis of 295 breast cancer patients obtained from a publicly available breast cancer gene expression data set.17 We first used hierarchical clustering (HCL) analysis to separate the patients into groups according the similarity in the gene expression meta-signature, and then determined the overall and relapse-free survival rates for these groups.
The HCL analysis classified the patients into 3 clusters (Fig. 4A). To further elucidate the reasons driving the separation of breast carcinomas in three major groups, we integrated the gene expression meta-signature with four prognostic or predictive gene signatures (Fig. 4B–C). Interestingly, meta-signature cluster 1 was highly associated with normal-like and luminal A breast carcinomas intrinsic subtypes (P < 0.0001), cluster 2 was associated to luminal B and HER2+/ER− subtypes (P < 0.0001), and the meta-signature cluster 3 was mainly composed by basal-like breast carcinomas (P < 0.0001) (Fig. 4B). The meta-signature clusters 2 and 3 were also correlated with breast carcinomas that expressed the 70-gene poor-prognosis signature, the high recurrence score signature and the activated wound-response signature (P < 0.0001) (Fig. 4C). In addition, we identified important clinico-pathological variables that highly correlated with the meta-signature clusters such as: ER status (P < 0.0001), tumor grade (P < 0.0001), and tumor size (P = 0.003) (Fig. 4D).
Figure 4.
Cross-validation of the gene meta-signature with a single data set of 295 breast cancer samples and integration with 4 pronostic or predictive gene expression signatures. A) Meta-signature hierarchical clustering, cluster 1 (blue), cluster 2 (pink), cluster 3 (orange). Gene ontology clustering (left of the graph), the green bar indicates genes related to steroid hormone stimulus, the blue bar indicates cell cycle related genes. B) Intrinsic Subtype signature. C) Poor prognosis signature: good prognosis (blue), poor prognosis (orange); Recurrence score: high (orange); intermediate (blue light), low recurrence score (blue) and Wound response: activated (orange); quiescent (blue). D) Clinicopathological data. Estrogen Receptor (ER) status: positive (black) and negative (white). Histological grade: high (black); moderate (grey) and low grade (light grey). Lymph node (LN) status: negative (white); 1–3 positive (grey); >3 positive (black). Tumor size: ≤2 cm (grey) and >2 cm (black).
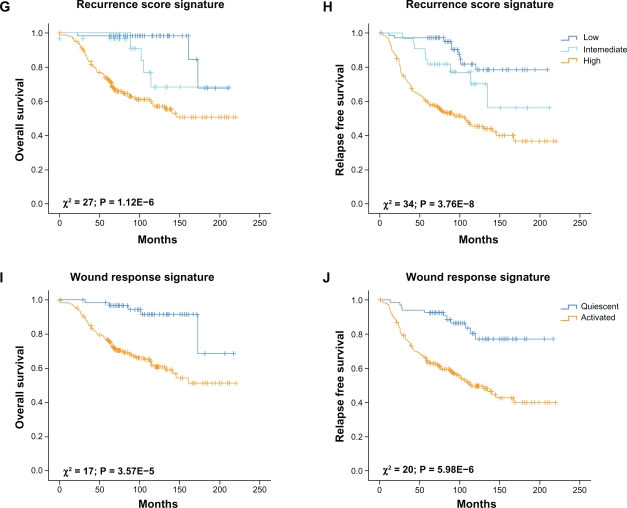
Kaplan–Meier analysis revealed that the meta-signature cluster 2 and 3 were particularly associated with shorter overall survival (P = 2.90E-11; Fig. 5A) and relapse-free survival (P = 2.79E-9; Fig. 5B) comparing with the cluster 1. In addition, the meta-signature and the 70-gene poor prognosis signature were the most predictive models in the comparative analysis of their Kaplan–Meiers survival curves as reflected by their having the lowest nominal P-values (Fig. 5 A–J).
Figure 5.
Kaplan–Meier curves of overall and relapse-free survival among the 295 early-stage breast cancer patients obtained from van de Vijver et al study (2002) according to the meta-signature (A and B), Intrinsic Subtypes (C and D), Poor Prognosis Signature (E and F), Recurrence Score (G and H) and Wound Response (I and J).
To further evaluate the independent prognostic value of the gene expression meta-signature, we next performed a multivariate Cox proportional-hazard analysis that included the most relevant and traditional prognostic factors such as: ER status, tumor grade, nodal status, tumor size, etc. This analysis demonstrated that the gene expression meta-signature was statistical significant predictor of both overall survival and relapse-free survival (Table 1).
Table 1.
Multivariate Cox proportional hazard analysis of standard clinical prognosis factors with the gene expression meta-signature predictor.
| Variable* |
Overall survival |
Relapse-free survival |
||
|---|---|---|---|---|
| Hazard ratio (95% CI) | P-value | Hazard ratio (95% CI) | P-value | |
| Age, per decade | 0.68 (0.46–1.01) | 0.056 | 0.57 (0.41–0.79) | 0.001 |
| ER status | 0.64 (0.39–1.05) | 0.076 | 0.93 (0.60–1.45) | 0.763 |
| Tumor grage 2 vs. 1 | 3.25 (1.10–9.60) | 0.033 | 1.66 (0.88–3.13) | 0.120 |
| Tumor grade 3 vs. 1 | 2.69 (0.90–8.01) | 0.076 | 1.37 (0.70–2.68) | 0.358 |
| Size | 1.71 (1.05–2.79) | 0.031 | 1.47 (1.01–2.16) | 0.045 |
| Lymph node 1–3 (+) vs. 0 | 0.95 (0.44–2.05) | 0.904 | 1.13 (0.63–2.04) | 0.683 |
| Lymph node > 3 (+) vs. 0 | 1.77 (0.75–4.21) | 0.195 | 2.04 (1.01–4.13) | 0.047 |
| Treatment | 0.83 (0.39–1.76) | 0.622 | 0.55 (0.30–1.00) | 0.049 |
| Meta-signature | 5.18 (2.33–11.5) | 5.5E-5 | 3.18 (1.91–5.31) | 9.5E-6 |
Notes:
Size was a binary variable (0 = diameter of 2 cm or less, 1 = greater than 2 cm.), age was a continuous variable formatted as decade-years. Hazard ratio for meta-signature was calculated comparing the clusters 2 and 3 relative to cluster 1. Variables found to be significant (P < 0.05) in the Cox proportional hazard model are shown in bold.
The results show that the 117-gene meta-signature was highly informative in identifying patients with good and poor prognosis outcome based on the expression profiles obtained from van de Vijver data set.17 In addition, the meta-signature added important prognostic information beyond that provided by the standard clinical predictors. In fact, the meta-signature was the most predictive variable in the analysis as reflected by their having the lowest nominal P-values (see Table 1). We identify the most representative differentially expressed transcripts between meta-signature clusters using a supervised statistical method (ANOVA test). The most statistically significant transcripts up-regulated between clusters are represented in Table 2.
Table 2.
Most highly up-regulated transcripts from meta-siganture gene list in van de Vijver et al 2002 data set.
| Gene name | Entrez ID | F-ratio | Biomarker |
|---|---|---|---|
| Response to steroid hormone stimulus | |||
| FOXA1 (forkhead box A1) | 3169 | 518.77 | Cluster 1 |
| GATA3 (GATA binding protein 3) | 2625 | 167.42 | Cluster 1 |
| ESR1 (estrogen receptor 1) | 2099 | 152.04 | Cluster 1 |
| XBP1 (X-box binding protein 1) | 7494 | 141.47 | Cluster 1 |
| Cell cycle and mitotic spindle | |||
| CENPA (centromere protein A) | 1058 | 243.97 | Cluster 3 |
| BUB1 (budding uninhibited by benzimidazoles 1 homolog) | 699 | 234.41 | Cluster 3 |
| HJURP (Holliday junction recognition protein) | 55355 | 230.73 | Cluster 3 |
| CCNB2 (cyclin B2) | 9133 | 227.29 | Cluster 3 |
| KIF2C (kinesin family member 2C) | 11004 | 213.76 | Cluster 3 |
| KIF20A (kinesin family member 20A) | 10112 | 202.88 | Cluster 3 |
| CDC20 (cell division cycle 20 homolog) | 991 | 187.72 | Cluster 3 |
| PRC1 (protein regulator of cytokinesis 1) | 9055 | 183.64 | Cluster 3 |
| CEP55 (centrosomal protein 55 kDa) | 55165 | 182.11 | Cluster 3 |
| FOXM1 (forkhead box M1) | 2305 | 169.99 | Cluster 3 |
| UBE2C (ubiquitin-conjugating enzyme E2C) | 11065 | 167.93 | Cluster 3 |
| CDCA8 (cell division cycle associated 8) | 55143 | 167.46 | Cluster 3 |
| KIFC1 (kinesin family member C1) | 3833 | 161.72 | Cluster 3 |
| AURKA (aurora kinase A) | 6790 | 155.97 | Cluster 3 |
| TTK (TTK protein kinase) | 7272 | 149.15 | Cluster 3 |
| CENPN (centromere protein N) | 55839 | 147.54 | Cluster 3 |
| MAD2L1 (MAD2 mitotic arrest deficient-like 1) | 4085 | 141.72 | Cluster 3 |
| BIRC5 (baculoviral IAP repeat-containing 5) | 332 | 141.26 | Cluster 3 |
| KIF14 (kinesin family member 14) | 9928 | 136.10 | Cluster 3 |
| CCNA2 (cyclin A2) | 890 | 130.65 | Cluster 3 |
| PTTG1 (pituitary tumor-transforming 1) | 9232 | 126.93 | Cluster 3 |
| Metabolism and Miscellaneous | |||
| MELK (maternal embryonic leucine zipper kinase) | 9833 | 194.99 | Cluster 3 |
| CTPS (CTP synthase) | 1503 | 145.28 | Cluster 3 |
| EZH2 (enhancer of zeste homolog 2) | 2146 | 132.67 | Cluster 3 |
| SOD2 (superoxide dismutase 2, mitochondrial) | 6648 | 127.96 | Cluster 3 |
| TRIP13 (thyroid hormone receptor interactor 13) | 9319 | 126.41 | Cluster 3 |
Gene expression modules associated with the meta-signature
Response to steroid hormone stimulus module
Approximately two-thirds of all breast cancers are ERα(+) at the time of diagnosis and the expression of this receptor is determinant of a tumor phenotype that is associated with hormone-responsiveness. Patients with tumors expressing ERα have a longer disease-free interval and overall survival than patients with tumors that lack ERα expression.18 Several studies have been carried out using cDNA and oligonucleotide microarrays identifying breast cancer subclasses possessing distinct biological and clinical properties.1,19 Among the distinctions made to date, the clearest separation was observed between ERα (+) and ERα (−) tumors. It has been suggested that there are sets of genes expressed in association with ERα that could play an important role in determining the hormone-responsive breast cancer phenotype.20 Functional annotation of the 117 gene meta-signature identified several genes related to the response to steroid hormone stimulus, such us ESR1 (ERα), XBP1, FOXA1, GATA3, MUC1, TFF3, BCL2, etc. The expression of this gene set has been shown to correlate with a specific breast cancer phenotype, defined as luminal type A, carrying an improved disease-free survival and overall survival when is compared with tumors that do not express it. The XBP1 transcription factor is an estrogen-regulated gene that is known to augment ER-mediated transcription itself, thereby initiating a feed-forward pathway.21,22 FOXA1 encodes a transcription factor protein that is known to bind to condensed heterochromatin via its winged helix DNA binding domains, functioning as a major factor to facilitate subsequent association of ER with chromatin of estrogen-target genes (eg, TFF1, XBP1 genes).23 Recently, it was demonstrated that GATA3 is required for estradiol stimulation of cell-cycle progression of breast cancer cells. GATA3 binds to cis-regulatory elements located within the ESR1 promoter, and this is required for transcriptional modulation of the ESR1 gene. Reciprocally, ERα directly stimulates transcription of the GATA3 gene, indicating that these two factors are involved in a positive cross-regulatory loop.24 It has been reported that GATA3 may be involved in growth control and differentiation of breast epithelial cells mediating the transcriptional activation of several genes such as those encoding cytokeratins 5, 6 and 17, and trefoil factors 1 and 3.25 Parikh and colleagues (2005) suggested that GATA3 expression might be associated with responsiveness to hormone therapy in breast cancer patients.26 Moreover, some of the genes in the cluster are ERα/GATA3–regulated genes such as MUC1, TFF3, and FOXA1, thus showing the functional clustering of a transcription factor and some of its direct targets.5 In this sense, we previously demonstrated that GATA3 is a mediator for the transcriptional up-regulation of MUC1 oncogene expression in some breast cancers.27 MUC1 gene encodes a highly glycosylated protein located on the apical surface of mammary epithelia that is aberrantly over-expressed in approximately 90% of human breast cancers.28,29 MUC1 protein over-expression has been associated with cell adhesion inhibition as well as increased metastatic and invasive potential of tumor cells. This over-expression allows MUC1 to interact with members of the ERBB family of receptor tyrosine kinases30 In addition, the MUC1 cytoplasmic domain, which comprises the last 72-aa, also interacts with diverse effectors that have been linked to transformation, such as c-Src, β-catenin, and IKβ/NF-KB.30–32 Interestingly, MUC1stimulates ERα-mediated transcription by direct binding to the ERα DNA binding domain and contributes to E2-mediated growth and survival of breast cancer cells.33 It has also been shown that MUC1 levels can be regulated by estrogen since ERα can bind to putative binding sites derived from the MUC1 promoter in-vitro.34 The identified module across gene expression signatures may be of value as breast cancer prognostic or predictive indicators analyzed as a group, playing an important role in controlling ER-E2-mediated effects in breast cancer cells. It is also likely that groups of co-regulated genes in ERα (+) breast cancers may be associated to the hormonal control of mammary epithelial cells growth and differentiation. In addition, a better understanding of the signaling networks controlled or associated with the estrogen response may lead to the identification of novel breast cancer therapeutic targets.
Cell cycle module and the mitotic spindle related genes
A common observation in cancer gene expression profiling is the systematic up-regulation of proliferation/cell cycle related genes among human cancer cells. The up-regulation of these genes is consistent with the fact that cancer is a disease that disrupts normal cell cycle control. Moreover, both in interphase and during mitosis, surveillance mechanisms (checkpoints) ensure that cell cycle events occur in the correct order by delaying crucial transitions until previous processes have been completed. Lesions in the processes and checkpoints mentioned above inevitably lead to genetic imbalances, a hallmark of cells in most solid tumors.
As was previously described, functional annotation of the 117 gene meta-signature identified 64 genes related to the cell cycle process. In addition, according to the gene/protein network analysis the 64 genes were divided in two modules: 32 genes (50%) related to the mitotic spindle biology and 32 genes (50%) related with cell cycle progression per se (red circles and part of blue circles in Figure 3, respectively). More importantly, the mitotic spindle module consists of 32 genes of which many have been associated with gene over-expression and poor prognosis in breast cancer such as PTTG1, ESPL1, TOP2A, NEK2, AURKA, TPX2, PLK1, etc.
PTTG1 also called securin gene encodes an anaphase-pomoting complex (APC) substrate that associates with a separin (ESPL1) until activation of the APC. In human tumours, high securin expression has been related to increased cell proliferation and angiogenic phenotype.35,36 Although the role of securin in breast carcinoma is not thoroughly studied, Solbach et al (2004)37 published an initial observation on securin mRNA over-expression in association with lymph node involvement and tumor recurrence. According to this study, the most significantly deregulated proliferation-associated genes were securin and topoisomerase DNA II alpha (TOP2A), other of the cell cycle module genes. TOP2A is located close to ERBB2 on chromosome 17q12 and copy number changes of TOP2A have frequently been linked to ERBB2 amplified breast cancers.38
Interestingly, in another study it has been demonstrated that BRCA1 regulates transcriptional expression of multiple cell cycle genes, including the genes mentioned above PTTG1 and ESPL1 as well as NEK2, BUB1, PLK1 and the progression genes CDC2 and CDC20. In this sense, it was demonstrated that NEK2 plays a critical role in carcinogenesis, tumor invasion, and tumorigenic growth of breast carcinoma, and that inhibition of NEK2 expression with siRNA causes suppression of cancer growth and invasion in both ER(+) and ER(−) cells.39 Another mitotic spindle related gene that has gained interest recently is AURKA (Aurora Kinase A). AURKA has well-established but perhaps not yet fully understood roles in centrosome function and duplication, mitotic entry, and bipolar spindle assembly. By the G2 phase of the cell cycle through anaphase, it can be detected in the pericentriolar material. Additionally, it spreads to mitotic spindle poles and midzone microtubules during metaphase.40 In a wide range of tumor types compared with essentially non-proliferating matched normal tissue, AURKA is strongly expressed at high frequency. This high level of expression is often associated with amplification of the region of chromosome 20 encoding AURKA.41 A number of recent findings have considerably advanced our understanding of the regulation of AURKA. The first insight came when a search for proteins interacting with AURKA revealed TPX2 as a prominent interaction partner of this kinase in mitotic human cells.42 TPX2 is not only a prominent component of the mitotic spindle,43 but also a key player in a spindle assembly process that is regulated by the small GTPase Ran.44 After the breakdown of the nuclear envelope, inactive cytoplasmic AURKA is transported to the proximal ends of the microtubules and activated by the spindle protein TPX2, where it plays an as yet not fully defined role in the Ran spindle assembly process.40,45 AURKA is also linked to the process of G2-M transition, with suppression of expression leading to G2-M arrest and apoptosis and ectopic expression leading to bypass of the G2-M DNA damage-activated checkpoint in model systems.46,47 In this sense, AURKA also regulates the activity of the PLK1 enzyme. One of PLK1’s important early mitotic functions is to activate CDK1.48 Recent work in mammalian cells revealed that phosphorylation of PLK1 by AURKA leads to the burst of PLK1 activity at the G2-M transition and efficient entry into mitosis and ensures timely entry into mitosis.49 Moreover, the adaptation and recovery functions of PLK1 take place at the G2-M transition, when PLK1 activity starts to increase.48 Thus, successful resumption of cell cycle progression at G2-M and mitotic entry relies on the activation of PLK1 by AURKA mediated phosphorylation within the activation loop of PLK1.49 Also, PLK1 is overexpressed in human tumors and has prognostic potential in cancer, indicating its involvement in carcinogenesis and its potential as a therapeutic target. In breast cancers, PLK1 has been found to be highly expressed in preinvasive in situ carcinomas.50 Several PLK1 inhibitors are in different phases of clinical development for anticancer therapy.51 As we have mentioned before, PLK1 activates CDK1, which has been strongly associated with breast cancer clinical outcome especially for node negative cancer patients.52 Following with the mitotic spindle module genes, PLK1 can enhance the transcription of multiple proteins necessary for mitotic progression via its effect on FOXM1.53–55 As the genes mentioned before, FOXM1 transcription factor is involved in the G2-M phase of the cell cycle. Consistent with a role in proliferation, elevated expression of FOXM1 has been reported in basal cell carcinoma.56 Furthermore, analysis of microarray data from primary breast cancers revealed that FOXM1 expression is increased in infiltrating ductal carcinoma.55 Microarray data from cells treated with FOXM1 siRNA identified several genes that are regulated by FOXM1, including CENPA, NEK2, and KIF20A, which also belong to the identified mitotic spindle module genes.57 FOXM1 also plays a role in regulating G2-M by inducing expression of cyclin A and CDC25B. Cyclin A binding to CDK1 promotes entry into mitosis, whereas CDC25B dephosphorylates CDK1, thereby promoting CDK1 activity58.
Interestingly, the centromere associated protein family members, the mentioned CENPA, CENPN, CENPE and CENPF are all linked in the spindle module gene. CENPA is essential for the recruitment to the centromere of most other proteins required for kinetochore function,59 as indicated by the observation that RNAi of CENPA causes a failure of chromosome alignment at the metaphase plate.57 Although there is no enough information about gene expression and prognosis of CENPA, CENPN and CENPE in breast cancer, CENPF expression has been associated with poor prognosis and chromosomal instability in patients with primary breast cancer. Little is known about the function of CENPF in cancer, but it has been examined its association with other known tumor parameters.60 It is known that normal kinetochore accumulation of CENPF follows the recruitment of BUB1 that first localizes to outer and inner kinetochore plates in a BUB3 dependent manner.61,62 This is followed by kinetochore accumulation of BUBR1, CENPE, and MAD2.63,62 Systematic silencing of kinetochore components with RNAi has been used to examine the interdependencies in the kinetochore assembly pathway. It has been noted that the order of assembly reflects the requirement of interaction between early and late associating proteins.64 Depletion of CENPF has been reported to decrease the amount of CENPE,64,65 BUBR1, and MAD1 at the kinetochores, suggesting that CENPF may modulate kinetochore maturation and function.53 Moreover, the Forkhead transcription factor FOXM1 as well as the other mentioned G2-specific genes NEK2, KIF20 and CENPA, regulates expression of CENPF. The interdependency between CENPF and BUBR1 is further supported by the observation that depletion of ZWINT, a structural component of the kinetochore, reduces the amount of kinetochore-bound CENPF and BUBR1.66 CENPF also associates with CENPE, a known activator of the kinetochore bound BUBR1.67 The mentioned CENPF associated genes, BUBR1 (BUB1), MAD2 (MAD2L1) and ZWINT are also members of the spindle module genes. Except ZWINT, whose role has not been well characterized, these genes along with other spindle checkpoint genes have shown increased expression in breast carcinomas, which was associated with genetic instability.68,69 Finally, the other members of the mitotic spindle gene cluster are also closely related; the kinesin KIF20, for instance, is a target for PLK170 CEP55, a protein associated with the centrosome directly interacts with KIF23,71 PRC1 a protein involved in cytokinesis, which is at high level during S and G2-M interacts with KIF2C suggesting that PRC1 might play critical roles in tumor cell growth and be a promising target for the development of anticancer drugs to breast cancer.72
In view of this information, it is interesting to note that most genes of the mitotic spindle cluster are involved in the G2-M phase of the cell cycle in which they are more active. Since these genes arose from a breast cancer gene signature meta-analysis of 42 studies, it is possible to believe that these genes, involved in “opening the door to proliferation”, could represent potential targets for breast cancer therapy. Although many of them have been extensively studied in breast carcinoma, there are new ones that might constitute the “key to close the door”.
The other cluster of cell cycle genes is a more heterogeneous group, which mainly includes cyclins, cyclin dependent kinases, cyclin dependent kinases inhibitors and members of the minichromosome maintenance complex (MCM). Several studies have focused on the behavior and localization of different cyclins during tumor progression. Of cyclins that emerged from our analysis, cyclins A2, B1, B2 and E2 are all well characterized; however there is no enough information about their expression in breast cancer. Cyclin A2 is associated with cellular proliferation and can be used for molecular diagnostic as a proliferation marker. It has been demonstrated that this gene is an estrogen-mediated down-regulated.73 A recent study, suggested that an oncogenic role of overexpressed cyclin B1 is mediated in nuclei of breast carcinoma cells, and the nuclear translocation is regulated by PLK1.74 Cyclin E2 has been shown to be overexpressed in breast cancer although the potential role as a diagnostic or prognostic marker is unknown.75 Similarly, little is known of MCM genes in breast cancer. Ha et al postulated that MCM3 is involved in multiple types of human carcinogenesis.76 Recently, MCM2 has been proposed as a useful proliferative marker in breast cancer.77
Conclusions
In summary, microarray technology has allowed the discovery of relevant signatures and consequently the identification of novel genes that may have an impact as breast cancer biomarkers. Our comprehensive comparison of overlapping genes across 42 breast cancer gene expression signatures provides an integrated view of a significant number of transcripts identified as highly modulated in breast tumors. The identification of individual proteins is of high relevance not only for the potential value as prognostic biomarkers but also because may provide insight into mechanisms and pathways of relevance in breast cancer progression. More importantly, this analysis identified the most promising biomarkers for further evaluation in breast cancer such as the cell cycle and mitotic spindle related genes.
Supplementary Data
42 gene expression signatures selected for analysis and their corresponding list of genes.
List of 946 transcripts that were identified as overlapping in more than one of the 42 gene expression signatures analyzed.
Acknowledgments
This work was supported by FONCYT (PICT N°32702, BID 1728 OC/AR), CONICET (PIP N°2131) grants.
Footnotes
Disclosures
This manuscript has been read and approved by all authors. This paper is unique and not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weisz A, Basile W, Scafoglio C, et al. Molecular identification of ERalpha-positive breast cancer cells by the expression profile of an intrinsic set of estrogen regulated genes. J Cell Physiol. 2004;200(3):440–50. doi: 10.1002/jcp.20039. [DOI] [PubMed] [Google Scholar]
- 5.Abba MC, Hu Y, Sun H, et al. Gene expression signature of estrogen receptor alpha status in breast cancer. BMC Genomics. 2005;6(1):37. doi: 10.1186/1471-2164-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sørlie T, Perou CM, Fan C, et al. Gene expression profiles do not consistently predict the clinical treatment response in locally advanced breast cancer. Mol Cancer Ther. 2006;5(11):2914–8. doi: 10.1158/1535-7163.MCT-06-0126. [DOI] [PubMed] [Google Scholar]
- 7.Yu J, Yu J, Cordero KE, et al. A transcriptional fingerprint of estrogen in human breast cancer predicts patient survival. Neoplasia. 2008;10(1):79–88. doi: 10.1593/neo.07859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 9.Glinsky GV, Higashiyama T, Glinskii AB. Classification of human breast cancer using gene expression profiling as a component of the survival predictor algorithm. Clin Cancer Res. 2004;10(7):2272–83. doi: 10.1158/1078-0432.ccr-03-0522. [DOI] [PubMed] [Google Scholar]
- 10.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360(8):790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 11.Fan C, Oh DS, Wessels L, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355(6):560–9. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 12.Ein-Dor L, Kela I, Getz G, Givol D, Domany E. Outcome signature genes in breast cancer: is there a unique set? Bioinformatics. 2005;21(2):171–8. doi: 10.1093/bioinformatics/bth469. [DOI] [PubMed] [Google Scholar]
- 13.Culhane AC, Schwarzl T, Sultana R, et al. GeneSigDB—a curated database of gene expression signatures. Nucleic Acids Res. 2010;38(Database issue):D716–25. doi: 10.1093/nar/gkp1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. Bio Techniques. 2003;34(2):374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 15.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 16.Jensen LJ, Kuhn M, Stark M. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–6. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 18.Shek LL, Godolphin W. Survival with breast cancer: the importance of estrogen receptor quantity. Eur J Cancer Clin Oncol. 1989;25(2):243–50. doi: 10.1016/0277-5379(89)90015-1. [DOI] [PubMed] [Google Scholar]
- 19.Sotiriou C, Neo S, McShane LM, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003;100(18):10393–8. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu ET, Sotiriou C. Defining the galaxy of gene expression in breast cancer. Breast Cancer Res. 2002;4(4):141–4. doi: 10.1186/bcr438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding L, Yan J, Zhu J, et al. Ligand-independent activation of estrogen receptor alpha by XBP-1. Nucleic Acids Res. 2003;31(18):5266–74. doi: 10.1093/nar/gkg731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang MM, Traystman RJ, Hurn PD, Liu T. Non-classical regulation of estrogen receptor-alpha by ICI182,780. J Steroid Biochem Mol Biol. 2004;92(1–2):51–62. doi: 10.1016/j.jsbmb.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Carroll JS, Liu XS, Brodsky AS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122(1):33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Eeckhoute J, Keeton EK, Lupien M, et al. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 2007;67(13):6477–83. doi: 10.1158/0008-5472.CAN-07-0746. [DOI] [PubMed] [Google Scholar]
- 25.Usary J, Llaca V, Karaca G, et al. Mutation of GATA3 in human breast tumors. Oncogene. 2004;23(46):7669–78. doi: 10.1038/sj.onc.1207966. [DOI] [PubMed] [Google Scholar]
- 26.Parikh P, Palazzo JP, Rose LJ, Daskalakis C, Weigel RJ. GATA-3 expression as a predictor of hormone response in breast cancer. J Am Coll Surg. 2005;200(5):705–10. doi: 10.1016/j.jamcollsurg.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 27.Abba MC, Nunez MI, Colussi AG, et al. GATA3 protein as a MUC1 transcriptional regulator in breast cancer cells. Breast Cancer Res. 2006;8(6):R64. doi: 10.1186/bcr1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gendler SJ. MUC1, the renaissance molecule. J Mammary Gland Biol Neoplasia. 2001;6(3):339–53. doi: 10.1023/a:1011379725811. [DOI] [PubMed] [Google Scholar]
- 29.Raina D, Ahmad R, Joshi MD, et al. Direct targeting of the mucin 1 oncoprotein blocks survival and tumorigenicity of human breast carcinoma cells. Cancer Res. 2009;69(12):5133–41. doi: 10.1158/0008-5472.CAN-09-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Liu D, Chen D, Kharbanda S, Kufe D. Human DF3/MUC1 carcinoma-associated protein functions as an oncogene. Oncogene. 2003;22(38):6107–10. doi: 10.1038/sj.onc.1206732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang L, Chen D, Liu D, et al. MUC1 oncoprotein blocks glycogen synthase kinase 3beta-mediated phosphorylation and degradation of beta-catenin. Cancer Res. 2005;65(22):10413–22. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 32.Ahmad R, Raina D, Trivedi V, et al. MUC1 oncoprotein activates the IkappaB kinase beta complex and constitutive NF-kappaB signalling. Nat Cell Biol. 2007;9(12):1419–27. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei X, Xu H, Kufe D. MUC1 oncoprotein stabilizes and activates estrogen receptor alpha. Mol Cell. 2006;21(2):295–305. doi: 10.1016/j.molcel.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 34.Zaretsky JZ, Barnea I, Aylon Y, et al. MUC1 gene overexpressed in breast cancer: structure and transcriptional activity of the MUC1 promoter and role of estrogen receptor alpha (ERalpha) in regulation of the MUC1 gene expression. Mol Cancer. 2006;5:57. doi: 10.1186/1476-4598-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kakar SS. Assignment of the human tumor transforming gene TUTR1 to chromosome band 5q35.1 by fluorescence in situ hybridization. Cytogenet Cell Genet. 1998;83(1–2):93–5. doi: 10.1159/000015139. [DOI] [PubMed] [Google Scholar]
- 36.Ishikawa H, Heaney AP, Yu R, Horwitz GA, Melmed S. Human pituitary tumor-transforming gene induces angiogenesis. J Clin Endocrinol Metab. 2001;86(2):867–74. doi: 10.1210/jcem.86.2.7184. [DOI] [PubMed] [Google Scholar]
- 37.Solbach C, Roller M, Fellbaum C, Nicoletti M, Kaufmann M. PTTG mRNA expression in primary breast cancer: a prognostic marker for lymph node invasion and tumor recurrence. Breast. 2004;13(1):80–1. doi: 10.1016/j.breast.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Jacobson KK, Morrison LE, Henderson BT, et al. Gene copy mapping of the ERBB2/TOP2 A region in breast cancer. Genes Chromosomes Cancer. 2004;40(1):19–31. doi: 10.1002/gcc.20019. [DOI] [PubMed] [Google Scholar]
- 39.Tsunoda N, Kokuryo T, Oda K, et al. Nek2 as a novel molecular target for the treatment of breast carcinoma. Cancer Sci. 2009;100(1):111–6. doi: 10.1111/j.1349-7006.2008.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu J, Bian M, Jiang Q, Zhang C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res. 2007;5(1):1–10. doi: 10.1158/1541-7786.MCR-06-0208. [DOI] [PubMed] [Google Scholar]
- 41.Gautschi O, Heighway J, Mack PC, et al. Aurora kinases as anticancer drug targets. Clin Cancer Res. 2008;14(6):1639–48. doi: 10.1158/1078-0432.CCR-07-2179. [DOI] [PubMed] [Google Scholar]
- 42.Kufer TA, Silljé HHW, Körner R, et al. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol. 2002;158(4):617–23. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wittmann T, Wilm M, Karsenti E, Vernos I. TPX2, A novel xenopus MAP involved in spindle pole organization. J Cell Biol. 2000;149(7):1405–18. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gruss OJ, Carazo-Salas RE, Schatz CA, et al. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104(1):83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 45.Andrews PD. Aurora kinases: shining lights on the therapeutic horizon? Oncogene. 2005;24(32):5005–15. doi: 10.1038/sj.onc.1208752. [DOI] [PubMed] [Google Scholar]
- 46.Du J, Hannon GJ. Suppression of p160ROCK bypasses cell cycle arrest after Aurora-A/STK15 depletion. Proc Natl Acad Sci U S A. 2004;101(24):8975–80. doi: 10.1073/pnas.0308484101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cazales M, Schmitt E, Montembault E, et al. CDC25B phosphorylation by Aurora-A occurs at the G2/M transition and is inhibited by DNA damage. Cell Cycle. 2005;4(9):1233–8. doi: 10.4161/cc.4.9.1964. [DOI] [PubMed] [Google Scholar]
- 48.Takaki T, Trenz K, Costanzo V, Petronczki M. Polo-like kinase 1 reaches beyond mitosis—cytokinesis, DNA damage response, and development. Curr Opin Cell Biol. 2008;20(6):650–60. doi: 10.1016/j.ceb.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 49.Macůrek L, Lindqvist A, Lim D, et al. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455(7209):119–23. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- 50.Rizki A, Mott JD, Bissell MJ. Polo-like kinase 1 is involved in invasion through extracellular matrix. Cancer Res. 2007;67(23):11106–10. doi: 10.1158/0008-5472.CAN-07-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chopra P, Sethi G, Dastidar SG, Ray A. Polo-like kinase inhibitors: an emerging opportunity for cancer therapeutics. Expert Opin Investig Drugs. 2010;19(1):27–43. doi: 10.1517/13543780903483191. [DOI] [PubMed] [Google Scholar]
- 52.Weichert W, Kristiansen G, Winzer K, et al. Polo-like kinase isoforms in breast cancer: expression patterns and prognostic implications. Virchows Arch. 2005;446(4):442–50. doi: 10.1007/s00428-005-1212-8. [DOI] [PubMed] [Google Scholar]
- 53.Laoukili J, Kooistra MRH, Brás A, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7(2):126–36. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 54.Wang I, Chen Y, Hughes D, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25(24):10875–94. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu Z, Malureanu L, Huang J, et al. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol. 2008;10(9):1076–82. doi: 10.1038/ncb1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teh M, Wong S, Neill GW, et al. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res. 2002;62(16):4773–80. [PubMed] [Google Scholar]
- 57.Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65(12):5181–9. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- 58.Strausfeld UP, Howell M, Descombes P, et al. Both cyclin A and cyclin E have S-phase promoting (SPF) activity in Xenopus egg extracts. J Cell Sci. 1996;109(Pt 6):1555–63. doi: 10.1242/jcs.109.6.1555. [DOI] [PubMed] [Google Scholar]
- 59.Carroll CW, Silva MCC, Godek KM, Jansen LET, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11(7):896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Brien N, O’Donovan N, Foley D, et al. Use of a panel of novel genes for differentiating breast cancer from non-breast tissues. Tumour Biol. 2007;28(6):312–7. doi: 10.1159/000115527. [DOI] [PubMed] [Google Scholar]
- 61.Taylor SS, Ha E, McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J Cell Biol. 1998;142(1):1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor SS, Hussein D, Wang Y, Elderkin S, Morrow CJ. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J Cell Sci. 2001;114(Pt 24):4385–95. doi: 10.1242/jcs.114.24.4385. [DOI] [PubMed] [Google Scholar]
- 63.Jablonski SA, Chan GK, Cooke CA, Earnshaw WC, Yen TJ. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma. 1998;107(6–7):386–96. doi: 10.1007/s004120050322. [DOI] [PubMed] [Google Scholar]
- 64.Johnson VL, Scott MIF, Holt SV, Hussein D, Taylor SS. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell Sci. 2004;117(Pt 8):1577–89. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- 65.Yang Z, Guo J, Chen Q, et al. Silencing mitosin induces misaligned chromosomes, premature chromosome decondensation before anaphase onset, and mitotic cell death. Mol Cell Biol. 2005;25(10):4062–74. doi: 10.1128/MCB.25.10.4062-4074.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H, Hu X, Ding X, et al. Human Zwint-1 specifies localization of Zeste White 10 to kinetochores and is essential for mitotic checkpoint signaling. J Biol Chem. 2004;279(52):54590–8. doi: 10.1074/jbc.M407588200. [DOI] [PubMed] [Google Scholar]
- 67.Weaver BAA, Bonday ZQ, Putkey FR, et al. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162(4):551–63. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuan B, Xu Y, Woo J, et al. Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin Cancer Res. 2006;12(2):405–10. doi: 10.1158/1078-0432.CCR-05-0903. [DOI] [PubMed] [Google Scholar]
- 69.Scintu M, Vitale R, Prencipe M, et al. Genomic instability and increased expression of BUB1B and MAD2L1 genes in ductal breast carcinoma. Cancer Lett. 2007;254(2):298–307. doi: 10.1016/j.canlet.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 70.Neef R, Preisinger C, Sutcliffe J, et al. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J Cell Biol. 2003;162(5):863–75. doi: 10.1083/jcb.200306009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao W, Seki A, Fang G. Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol Biol Cell. 2006;17(9):3881–96. doi: 10.1091/mbc.E06-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shimo A, Nishidate T, Ohta T, et al. Elevated expression of protein regulator of cytokinesis 1, involved in the growth of breast cancer cells. Cancer Sci. 2007;98(2):174–81. doi: 10.1111/j.1349-7006.2006.00381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vendrell JA, Magnino F, Danis E, et al. Estrogen regulation in human breast cancer cells of new downstream gene targets involved in estrogen metabolism, cell proliferation and cell transformation. J Mol Endocrinol. 2004;32(2):397–414. doi: 10.1677/jme.0.0320397. [DOI] [PubMed] [Google Scholar]
- 74.Suzuki T, Urano T, Miki Y, et al. Nuclear cyclin B1 in human breast carcinoma as a potent prognostic factor. Cancer Sci. 2007;98(5):644–51. doi: 10.1111/j.1349-7006.2007.00444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Payton M, Scully S, Chung G, Coats S. Deregulation of cyclin E2 expression and associated kinase activity in primary breast tumors. Oncogene. 2002;21(55):8529–34. doi: 10.1038/sj.onc.1206035. [DOI] [PubMed] [Google Scholar]
- 76.Ha S, Shin SM, Namkoong H, et al. Cancer-associated expression of minichromosome maintenance 3 gene in several human cancers and its involvement in tumorigenesis. Clin Cancer Res. 2004;10(24):8386–95. doi: 10.1158/1078-0432.CCR-04-1029. [DOI] [PubMed] [Google Scholar]
- 77.Reena RMZ, Mastura M, Siti-Aishah MA, et al. Minichromosome maintenance protein 2 is a reliable proliferative marker in breast carcinoma. Ann Diagn Pathol. 2008;12(5):340–3. doi: 10.1016/j.anndiagpath.2008.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
42 gene expression signatures selected for analysis and their corresponding list of genes.
List of 946 transcripts that were identified as overlapping in more than one of the 42 gene expression signatures analyzed.