Abstract
Recent increases in the production of genomic data are yielding new opportunities and challenges for biologists. Among the chief problems posed by next-generation sequencing are assembly and analyses of these large data sets. Here we present an online server, http://EvoPipes.net, that provides access to a wide range of tools for bioinformatic analyses of genomic data oriented for ecological and evolutionary biologists. The EvoPipes.net server includes a basic tool kit for analyses of genomic data including a next-generation sequence cleaning pipeline (SnoWhite), scaffolded assembly software (SCARF), a reciprocal best-blast hit ortholog pipeline (RBH Orthologs), a pipeline for reference protein-based translation and identification of reading frame in transcriptome and genomic DNA (TransPipe), a pipeline to identify gene families and summarize the history of gene duplications (DupPipe), and a tool for developing SSRs or microsatellites from a transcriptome or genomic coding sequence collection (findSSR). EvoPipes.net also provides links to other software developed for evolutionary and ecological genomics, including chromEvol and NU-IN, as well as a forum for discussions of issues relating to genomic analyses and interpretation of results. Overall, these applications provide a basic bioinformatic tool kit that will enable ecologists and evolutionary biologists with relatively little experience and computational resources to take advantage of the opportunities provided by next-generation sequencing in their systems.
Keywords: bioinformatics, next-generation sequencing, genomic analyses, evolutionary genomics, ecological genomics
Introduction
Next-generation sequencing approaches are providing exciting new opportunities for biologists to explore new and old questions with greater detail than previously imagined. Ecology and evolutionary biology are not exceptions, and the frontiers of these fields are being pushed by the growing genomic data available on public databases and the ability of a single lab to quickly sequence multiple transcriptomes or genomes. Although these data have created outstanding research opportunities, the introduction of genomic approaches into more traditional ecological and evolutionary biology settings poses unique challenges because many labs currently lack the bioinformatic skills and tools needed to analyze data at such a large scale. Traditionally, the development of custom in-house bioinformatic tools was the solution for these problems, and such custom tools will always play a vital role in bioinformatics research. However, many questions studied by ecology and evolutionary biology research programs use similar approaches and have shared bioinformatic needs. Thus, a basic bioinformatic tool kit would be a useful resource for disparate labs across ecological and evolutionary biology.
To address this growing issue, we announce the release of a public server of bioinformatic pipelines designed especially for ecologists and evolutionary biologists employing genomic approaches. The server, http://EvoPipes.net, currently provides pipelines and downloadable tools for basic processing of genomic data, such as SnoWhite for cleaning raw next-generation sequence data and SCARF1 for scaffolding assemblies against reference data sets. A suite of tools are also available for data analyses including pipelines to construct gene family phylogeny and summarize duplications (DupPipe), identify orthologs (RBH Orthologs), find reading frame and translate transcriptome or genomic data to protein sequences (TransPipe), and identify simple sequence repeats (SSRs) or microsatellites in a collection of genomic data (findSSRs). Combined with other tools available for download for simulating data sets (eg, NU-IN2), EvoPipes.net provides a starting point and basic tool kit for ecologists and evolutionary biologists wishing to take advantage of genomic approaches (Fig. 1). Below, we provide further details of the server and the currently available pipelines.
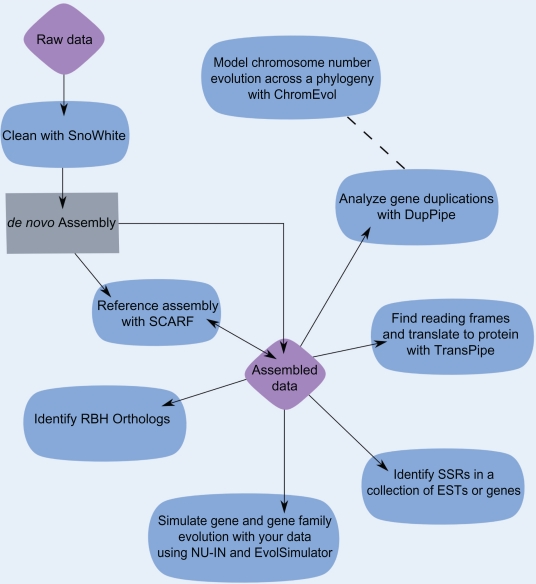
Figure 1.
An outline of data analyses possible with tools available on the http://EvoPipes.net server. Purple diamonds represent sequence data provided by the user, the blue ovals indicate analyses that may be conducted with EvoPipes.net tools, and the gray rectangle represents steps requiring software from other sources. Note that ChromEvol requires chromosome number and phylogenetic information as input rather than sequence data. However, ChromEvol results can be informed by genomic analyses with DupPipe.
Server and Pipeline Descriptions
Server interface
EvoPipes.net is organized with simplicity in mind and possesses a relatively clean interface to increase its accessibility to a broad range of researchers. A straightforward system for uploading data and e-mail retrieval of results makes the server accessible to anyone familiar with web browsing (Fig. 2). After selecting a data analysis pipeline from the homepage, the user is taken to a simple upload page. A set of custom perl scripts collects uploaded data and queues them for the appropriate pipeline run. Once the pipeline run is finished, they will receive an e-mail from EvoPipes.net with a link to retrieve the results. Links to documentation describing each pipeline are located on their respective data upload pages. A forum is also maintained on the server for discussion of ecological and evolutionary genomic analyses, available pipelines, and interpretation of results.
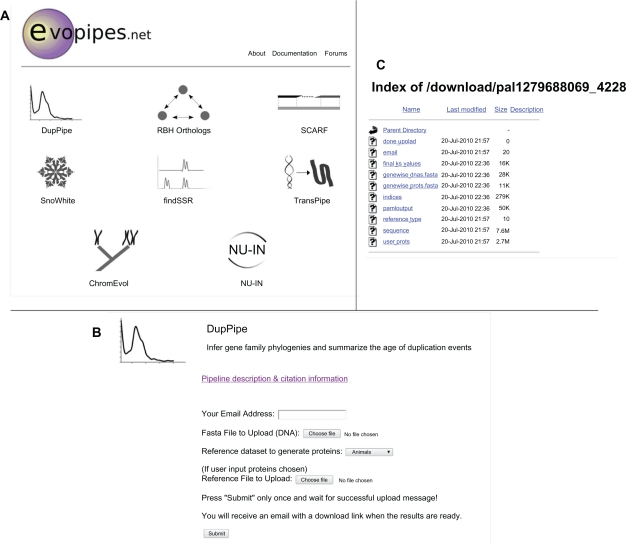
Figure 2.
A) The EvoPipes.net home page with icons representing each bioinformatic analysis available on the server. B) Example upload page for the DupPipe. C) Example results download page for the DupPipe.
Pipelines executable on the server
DupPipe
The DupPipe is useful for a range of studies of gene family evolution. The original DupPipe was developed to identify ancient genome duplication events in plants,3–5 but the pipeline can be used for any application that requires duplicate genes and gene families to be identified within a genome, such as examining paralog resolution to evaluate assembly quality. The pipeline uses a set of custom perl scripts to identify duplicate gene pairs and their divergence, in terms of substitutions per synonymous site (Ks). Duplicate pairs are identified as sequences that demonstrate 40% sequence similarity over at least 300 base pairs from a discontinguous MegaBLAST search.6,7 Reading frames for duplicate pairs are identified by comparison to a selected database of protein sequences. The user may select to use NCBI’s plant RefSeqs, animal RefSeqs, or upload a custom protein data set. Each duplicated gene is searched against all proteins in the chosen data set using BLASTX.8 Best hit proteins are paired with each gene at a minimum cutoff of 30% sequence similarity over at least 150 sites. Genes that do not have a best hit protein at this level are removed before further analyses. To determine reading frame and generate estimated amino acid sequences, each gene is aligned against its best hit protein by GeneWise 2.2.2.9 GeneWise is an algorithm that uses a combination of hidden Markov models (HMM) to align a DNA sequence to a homologous protein sequence in order to annotate the coding regions of the DNA. Using the highest scoring Gene-Wise DNA-protein alignments to indicate the correct orientation of the read, custom perl scripts are used to remove stop and “N” containing codons and produce estimated amino acid sequences for each gene. Amino acid sequences for each duplicate pair are then aligned using MUSCLE 3.6.10 The aligned amino acids are subsequently used to align their corresponding DNA sequences using RevTrans 1.4.11 Ks values for each duplicate pair are calculated using the maximum likelihood method implemented in codeml of the PAML package12 under the F3 × 4 model.13 Gene families are then constructed by single-linkage clustering, and the node Ks values are calculated. Finally, a tab de-limited file containing each duplicated node Ks value and gene annotation is provided.
findSSR
Short Sequence Repeats (SSRs) or microsatellites are variable regions of eukaryotic genomes characterized by repeated sequence motifs. Varying in the number of these repeat units and amenable to PCR amplification, SSRs have long been a valuable source of variation for population genetic studies.14,15 findSSR is a pipeline that identifies all genes containing SSRs or microsatellites, including di-, tri-, tetra- and penta-nucleotide repeats of at least five repeats in a fasta formatted sequence collection. Similar in scope to previous approaches,16 findSSRs was designed to pull out SSRs that would be useful for genotyping. For this purpose, unlike other programs, findSSRs only reports repeats found greater than 20 nucleotides from the ends of the sequence, leaving room for primers on either side. Output includes a list of each sequence name for genes containing an SSR, the location of the SSR, repeated motif, number of repeats, and the total length of the sequence examined. The program has been used to identify microsatellites that have been developed into markers used in several publications.17–19
RBH Orthologs
The RBH Ortholog pipeline identifies reciprocal-best-BLAST-hit orthologs for a set of uploaded fasta formatted data files. The identification of orthologs is useful in a variety of studies, from identifying accelerated amino acid evolution and genes putatively experiencing positive selection to deducing phylogenetic relationships among taxa. The user uploads up to five separate fasta formatted DNA sequence files and provides a three letter code to uniquely identify each file. The RBH Ortholog pipeline uses a set of custom perl scripts to iteratively MegaBLAST6,7 all combinations of the files against each other, including all reciprocal searches. These results are parsed to identify sequences with greater than 70% sequence similarity over at least 100 bp. From these matches, a final list of orthologs that are reciprocal best hits of each other is parsed and provided to the user along with an index to translate the coded names back to the original fasta headers if needed. Using this pipeline, orthologs have been identified in a number of previous studies to compare the divergence of species17 and construct nuclear ortholog phylogenies.3–5
SCARF
SCARF1 is a next-generation sequence assembly tool for evolutionary genomics that was designed especially for assembling 454 EST sequences against high quality reference sequences from related species. Using a reference sequence library to orient contigs, SCARF knits together low-coverage contigs that do not assemble during traditional de novo assembly. SCARF is especially well suited for non-contiguous or low depth data sets such as EST (expressed sequence tag) libraries, and can also be used to sort and assemble a pool of next-generation sequence data according to a set of reference sequences (eg, for metagenomics). Significant increases in contig length and reduction of unigene number may be obtained with SCARF reference assemblies. For example, SCARF nearly doubled the number of contigs greater than 1 kb by scaffolding a MIRA de novo assembly of 100,000 454 FLX reads for Centaurea solstitialis against a more complete Sanger reference data set of the same species.1 Overall, SCARF yielded an average increase of 6.04% (excluding gaps) and a 9.4% decrease in unigene number for four different individual de novo assemblies of C. solstitialis. Similar to the previous pipelines, the user uploads their fasta formatted assembly and reference sequence set. Although the stand-alone version of SCARF has numerous options, EvoPipes. net uses the default settings and only allows the user to select if repeat correction should be enabled. Once a SCARF run is finished on the server, the user receives an e-mail with links to the SCARFed assembly.
TransPipe
TransPipe provides bulk translation and reading frame identification for a set of fasta formatted sequences. The identification of reading frames and translated protein sequences are crucial steps for codon based analyses, such as tests for evidence of accelerated amino acid evolution, calculation of Ka/Ks ratios, or protein guided DNA alignments. Using GeneWise’s HMM algorithm, the TransPipe identifies only the regions of genes or EST reads that align to the protein sequence and starts them in-frame. An added benefit of this approach is that any missed vectors and adapters are also removed, as well as all non-coding DNA from the data set, such as introns and UTRs. This simplifies the comparison of transcriptome, whole genome shotgun, and genespace data.
The reading frame and protein translation for each sequence are identified by comparison to available protein sequences provided by the user or available on GenBank.20 Using BlastX,8 best hit proteins are paired with each gene at a minimum cutoff of 30% sequence similarity over at least 150 sites. Genes that do not have a best hit protein at this level are removed. To determine reading frame and generate estimated amino acid sequences, each gene is aligned against its best hit protein by GeneWise 2.2.2.9 Using the highest scoring GeneWise DNA-protein alignments, custom Perl scripts are used to remove stop and ‘N’ containing codons and produce estimated amino acid sequences for each gene. Output includes paired DNA and protein sequences with the DNA sequence’s reading frame corresponding to the protein sequence.
Tools presently available for download only
ChromEvol
ChromEvol21 implements a series of likelihood models to study the evolution of chromosome numbers across phylogenies. By comparing the fit of the different models to the phylogenetic data, it may be possible to gain insight regarding the pathways by which the evolution of chromosome number proceeds. For each model, the program infers the set of ancestral chromosome numbers and estimates the location along the phylogeny for which polyploidy and other chromosome number changes occurred. Currently, ChromEvol is only available for download from EvoPipes.net.
NU-IN
NU-IN2 is an adaptation and expansion of the EvolSimulator 2.1.0 genome evolution simulation program.22 NU-IN was designed to expand EvolSimulator in two fundamental ways: 1) Allow synonymous and non-synonymous nucleotide evolution and 2) Permit input of genomes, gene family membership, and gene ‘usefulness’ (the selective retention of particular loci in particular environments). With these changes, the user has the ability to use real genomic (coding) sequence data to initiate a simulation of one or more lineages, generate mutations through SNPs and copy number variation (as well as horizontal gene transfer), evolve genomes by drift and selection, and use output of previous simulations as starting points for further evolution. Currently, NU-IN is only available for download from EvoPipes.net.
SnoWhite
SnoWhite is a pipeline designed to flexibly and aggressively clean sequence reads (gDNA or cDNA) prior to assembly, and is designed to deal with issues encountered in next-generation sequence data. The pipeline takes in and returns fasta formatted sequence and (optionally) quality files from any source that can be converted to fasta format (including Sanger, Roche 454, Illumina, and SOLiD). Several cleaning steps are employed by the SnoWhite pipeline:
To eliminate consistent terminal adapter sequences, SnoWhite can initially remove a user-specified number of bases from the 5’ and/or 3’ end of each read or clip through a user-provided 5’ and/or 3’ sequence tag.
Contaminant removal is further accomplished by passing files to TGI’s Seqclean (http://compbio.dfci.harvard.edu/tgi/software/), a relatively old but still excellent pipeline for sequence trimming.23 Seqclean removes terminal N’s and polyA/T for all reads, and optionally searches for BLAST hits to user-provided contaminant lists at terminal positions (minimum 12 bp match at 92% identity using blastn) and internal positions (minimum 60 bp match at 94% identity using megablast). SnoWhite provides options for executing Seqclean with or without contaminant searches, of just terminal or both terminal and internal types. If contaminants are identified, reads are trimmed through these areas, and the remaining sequences are deleted entirely if they are too short (given a user-defined minimum length) or if the read is entirely low complexity sequence (‘dust’ as determined by BLAST’s -F mD filter).
Following Seqclean, SnoWhite executes additional polyA/T trimming governed by many tunable parameters, including the option to search for and remove sequences with internal polyA/T. Users can define the minimum polyA/T length for trimming, whether to tolerate N’s or other single base interruptions in a polyA/T, and which end(s) to search for A and T repeats. These options can make SnoWhite much more aggressive than Seqclean in trimming polyA/T regions that include some sequence error, which are frequently encountered in error-prone next-generation cDNA datasets.
After SnoWhite’s polyA/T trimming, it is often possible to identify additional contaminants and/or reads containing only low-complexity sequence. SnoWhite automatically passes the data to Seqclean again for a second cleaning, identical to the first.
Finally, SnoWhite optionally implements TagDust,24 a program designed to find sequences that are composed almost entirely of primer/adapter fragments. TagDust computes probabilities that each read is composed predominantly of user-provided contaminant sequences, and reads are eliminated to achieve a dataset-wide false discovery rate set by the user. These primer ‘multimers’ or ‘concatmers’ are a persistent low-abundance feature of many datasets, and are extremely difficult to remove using traditional contaminant searches.
SnoWhite compiles output files detailing the trimming or elimination of each sequence at each of these steps, and creates a log file summarizing the number of sequences trimmed or removed by each filter. The final fasta formatted sequence and quality files are suitable for assembly and analysis by any downstream software. Currently, SnoWhite is available for download only from EvoPipes.net.
Conclusions
Ecologists and evolutionary biologists are increasingly analyzing large-scale genomic data, and resources such as EvoPipes.net will provide a strong starting point for many researchers’ entry into bioinformatics. By providing a unique collection of bioinformatic tools for large-scale data analyses, we hope that the resources and forums at EvoPipes.net will provide a helpful and useful resource for beginning their genomic investigations and exploring potential questions with their data. Future updates to EvoPipes. net will occur as new tools are developed and contributed with the goal of making the server a valuable community resource.
Acknowledgments
Funding for EvoPipes.net was provided by National Science Foundation Plant Genome Awards 0421630 and 0820451 and Natural Sciences and Engineering Research Council of Canada Awards 327475 and 353026. MSB was supported by an NSERC-BRITE postdoctoral fellowship.
Footnotes
Disclosure
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Barker MS, Dlugosch KM, Reddy ACC, Amyotte SN, Rieseberg LH. SCARF: maximizing next-generation EST assemblies for evolutionary and population genomic analyses. Bioinformatics. 2009;25:535–6. doi: 10.1093/bioinformatics/btp011. [DOI] [PubMed] [Google Scholar]
- 2.Dlugosch KM, Barker MS, Rieseberg LH. NU-IN: Nucleotide evolution and input module for the EvolSimulator genome simulation platform. BMC Research Notes. 2010;3:217. doi: 10.1186/1756-0500-3-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker MS, Kane NC, Matvienko M, et al. Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Molecular Biology and Evolution. 2008;25:2445–55. doi: 10.1093/molbev/msn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker MS, Vogel H, Schranz ME. Paleopolyploidy in the Brassicales: analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biology and Evolution. 2009;1:391–9. doi: 10.1093/gbe/evp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi T, Huang H, Barker MS. Ancient genome duplications during the evolution of kiwifruit (Actinidia) and related Ericales. Annals of Botany. 2010;106:497–504. doi: 10.1093/aob/mcq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. Journal of Computational Biology. 2000;7:203–14. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 7.Ma B, Tromp J, Li M. PatternHunter: faster and more sensitive homology search. Bioinformatics. 2002;18:440–5. doi: 10.1093/bioinformatics/18.3.440. [DOI] [PubMed] [Google Scholar]
- 8.Altschul S, Madden T, Schaffer A, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Research. 2004;14:988–95. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgar R. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wernersson R, Pedersen AG. RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Research. 2003;31:3537–9. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. CABIOS. 1997;13:555–6. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 13.Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Molecular Biology and Evolution. 1994;11:725–36. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 14.Queller DC, Strassmann JE, Hughes CR. Microsatellites and kinship. Trends in Ecology and Evolution. 1993;8:285–8. doi: 10.1016/0169-5347(93)90256-O. [DOI] [PubMed] [Google Scholar]
- 15.Jarne P, Lagoda PJL. Microsatellites, from molecules to populations and back. Trends in Ecology and Evolution. 1996;11:424–9. doi: 10.1016/0169-5347(96)10049-5. [DOI] [PubMed] [Google Scholar]
- 16.Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Research. 2001;11:1441–52. doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kane NC, King MG, Barker MS, et al. Comparative genomic and population genetic analyses indicate highly porous genomes and high levels of gene flow between divergent Helianthus species. Evolution. 2009;63:2061–75. doi: 10.1111/j.1558-5646.2009.00703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kane NC, Rieseberg LH. Selective sweeps reveal candidate genes for adaptation to drought and salt tolerance in common sunflower, Helianthus annuus. Genetics. 2007;175:1823–4. doi: 10.1534/genetics.106.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kane NC, Rieseberg LH. Genetics and evolution of weedy Helianthus annuus populations: adaptation of an agricultural weed. Molecular Ecology. 2008;17:384–94. doi: 10.1111/j.1365-294X.2007.03467.x. [DOI] [PubMed] [Google Scholar]
- 20.Sayers EW, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 2010;38:D5–16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayrose I, Barker MS, Otto SP. Probabilistic models of chromosome number evolution and the inference of polyploidy. Systematic Biology. 2010;59:132–44. doi: 10.1093/sysbio/syp083. [DOI] [PubMed] [Google Scholar]
- 22.Beiko RG, Charlebois RL. A simulation test bed for hypotheses of genome evolution. Bioinformatics. 2007;23:825–31. doi: 10.1093/bioinformatics/btm024. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y-A, Lin C-C, Wang C-O, Wu H-B, Hwang P-I. An optimized procedure greatly improves EST vector contamination removal. BMC Genomics. 2007;8:416. doi: 10.1186/1471-2164-8-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lassmann T, Hayashizaki Y, Daub CO. TagDust—a program to eliminate artifacts from next generation sequencing data. Bioinformatics. 2009;25:2839–40. doi: 10.1093/bioinformatics/btp527. [DOI] [PMC free article] [PubMed] [Google Scholar]