Abstract
The Helix Research Institute (HRI) in Japan is releasing 4356 HUman Novel Transcripts and related information in the newly established HUNT database. The institute is a joint research project principally funded by the Japanese Ministry of International Trade and Industry, and the clones were sequenced in the governmental New Energy and Industrial Technology Development Organization (NEDO) Human cDNA Sequencing Project. The HUNT database contains an extensive amount of annotation from advanced analysis and represents an essential bioinformatics contribution towards understanding of the gene function. The HRI human cDNA clones were obtained from full-length enriched cDNA libraries constructed with the oligo-capping method and have resulted in novel full-length cDNA sequences. A large fraction has little similarity to any proteins of known function and to obtain clues about possible function we have developed original analysis procedures. Any putative function deduced here can be validated or refuted by complementary analysis results. The user can also extract information from specific categories like PROSITE patterns, PFAM domains, PSORT localization, transmembrane helices and clones with GENIUS structure assignments. The HUNT database can be accessed at http://www.hri.co.jp/HUNT.
INTRODUCTION
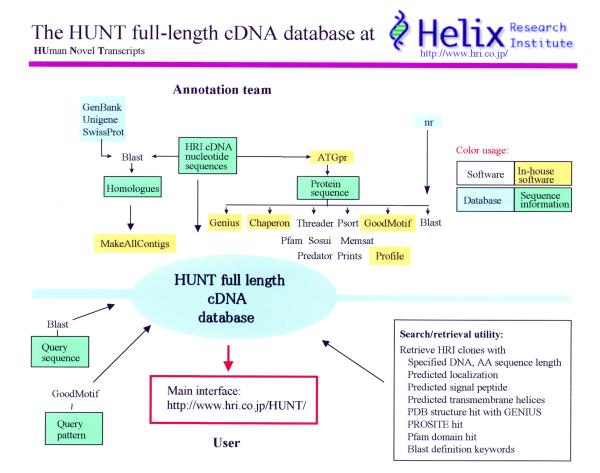
The HUNT database aims to make publicly available novel full-length clones and related analysis from the Helix Research Institute (HRI), Japan. A characteristic of the present sequencing project is that we emphasize full-length clones because of the many advantages of and suitability for high throughput functional analysis (1). The HRI human cDNA clones have been obtained from full-length enriched cDNA libraries constructed with the oligo-capping method (2), and the selection procedure was based upon BLAST analysis of the 5′- and 3′- one-pass sequences and start codon prediction by the ATGpr software (3). This has resulted in novel full-length cDNA sequences now released via the DNA Data Bank of Japan (DDBJ). We took advantage of excellent software from the scientific community to analyze these for structure and function annotation (Fig. 1). All the diverse sets of information thus obtained are ordered and made publicly accessible via the HUNT database, which has been specifically developed for this purpose. The user can efficiently extract data either by browsing the database or by using one of a set of specific information extracting tools.
Figure 1.
Overview of the HUNT full-length cDNA database. Programs and databases used to annotate HUNT are shown together with utilities available in the user interface.
THE HUNT FULL-LENGTH HUMAN cDNA DATABASE
Similarity search with nucleotide sequence
This inaugural release of the HUNT database contains 4356 full-length cDNA clones each with an individual web page displaying the main annotation (Fig. 2). The first section lists the clone name and a link to the corresponding GenBank entry followed by a link to the actual nucleotide sequence. We note here that all sequences and analysis data are publicly available from the web site or by following the direct link to the corresponding GenBank entries. The sequence was used as the query for similarity searches and results here are displayed in an easily comprehensible form for each of the GenBank, UniGene and SwissProt databases. The user can quickly grasp the presence of interesting genes as a header explaining possible functions of the hit sequences is displayed together with search characteristics of the BLAST alignment. Assembly with similar human cDNAs is provided using MakeAllContigs (4), which produces all possible contigs from a set of sequences taking the length of non-aligned regions, alignment lengths and identities into account. Library and tissue information, strand, 5′-end position together with links to the corresponding GenBank entry are summarized in a comprehensible table form. The clone selection procedures were designed so that redundant sequences would be excluded as much as possible. We found, however, in an all-against-all comparison of the 4356 sequences on the nucleotide level using thresholds of 95% identity and 300 aligned base pairs or 98% identity and 100 aligned base pairs, 3635 clusters including 3085 singles and 413 doublets with a rapidly decreasing number of triplets, quartets, etc. Here, alternative splice-forms were observed, but these findings have not been quantified.
Figure 2.
Example of a web page for an individual clone. Underlined words indicate active links to more detailed information.
Protein sequence prediction and similarity search
Much of the analysis data contained in HUNT is based upon the corresponding amino acid sequence, and we bring the protein sequence together with the full output from the ATGpr prediction software (3) developed here at HRI. The coding region predictions from ATGpr are based upon linear discriminant analysis of empirical observations and the resulting putative protein sequences are unique to the HUNT database. Results from a BLASTp search against the current version of the non-redundant nr database for the sequence receiving the highest ATGpr reliability are tabulated together with a link to the actual BLASTp listing. Using a threshold of 10–10 for the E-value from BLASTp we found that 1753 had no hits in the nr database, and therefore that there are homologous proteins to the remaining 2603. Among these, however, 823 were hypothetical proteins and we conclude that there are 2576 truly novel proteins among the first 4356 sequences in the HUNT database, and it becomes necessary to employ other methods to get clues about possible function as described in the following.
LOCALIZATION AND TRANSMEMBRANE SEGMENTS
Sorting signals
We include protein localization in our annotation as subsequent motif, structure and function predictions can be evaluated in principle against this knowledge. Sorting signals usually determine protein localization and we employ the PSORT program (5) for prediction of signal sequences, cleavage sites, transmembrane segments and topology, and protein localization sites in cells. The PSORT procedure is based upon sequence features such as N-terminal positively charged regions and regions of high hydrophobicity that are combined into a subcellular localization prediction. The present version has been trained on a set composed of 1531 yeast sequences from the SwissProt and YPD databases.
Helices of membrane proteins
Transmembrane segments are also predicted with the SOSUI (6) and MEMSAT (7) systems. SOSUI predicts the secondary structure of membrane proteins based on physico-chemical properties of amino acid sequences. The predictions follow three steps: discrimination of membrane proteins from soluble proteins, prediction of the existence of transmembrane helices and determination of transmembrane helical regions. MEMSAT predicts the secondary structure and topology of integral membrane proteins based on recognition of topological models. Well-characterized membrane proteins have been used to create a set of statistical tables and the membrane topology is then predicted using dynamic programming for maximizing an expectation-value. We tabulate the predicted transmembrane regions and the corresponding scores from these three systems, PSORT, SOSUI and MEMSAT as it then becomes particularly easy to compare the predictions and obtain a suggestion about the reliability of the results.
MOTIF SEARCH
Conserved regions, active sites and domains
In the case where there is not sufficient sequence similarity to warrant a family relationship it is often very helpful to search for specific patterns conserved during evolution even if the overall similarity has vanished. PROSITE (8) and PRINTS-S (9) are both databases for motif analysis, and we have developed the GoodMotif procedure, which searches for PROSITE patterns and evaluates the quality of hits as the likelihood of a correct relationship (10). We have also automated the FingerPRINTScan (11) calculations of PRINTS-S fingerprints making updates for the whole HUNT database an easy task. Another very popular approach for automated sequence analysis based upon the domain structure of proteins is the PFAM database (12). This database contains a large collection of protein domain families and corresponding profile hidden Markov models (HMM) for analyzing novel sequences. We display the identifier of the Pfam HMM on the main web page for the individual protein if there is a hit, and a link provides the way to the actual alignment data for that sequence.
STRUCTURE ASSIGNMENT
Profile and secondary structure prediction
It is well known in protein structure prediction studies that a significant improvement in performance can be obtained by incorporating evolutionary information. For this purpose and for the value per se we also list a profile calculated from multiple sequence alignments. The profile is the frequency of occurrence of each of the 20 standard amino acids in each position of the alignment. The profile is used as input to CHAPERON secondary structure predictions, where CHAPERON is our in-house software for protein sequence analysis (13). The HUNT database provides results from two different secondary structure prediction programs. CHAPERON’s neural network is capable of correlated predictions by employing multiple output units and the activities for these units are reported making judgment about the reliability of individual secondary structure elements possible. Predator is also a secondary structure prediction program enjoying a very high reputation and is an approach based on recognition of potentially hydrogen-bonded residues (14).
Intermediate sequence search and fold recognition
If the similarity between a pair of sequences drops below the twilight zone it may be difficult to ascertain a relationship. It is still possible, however, that tertiary structures of proteins related to sequences in the HUNT database have been determined despite a low sequence similarity, and to find these we use the GENIUS program developed at HRI (15,16) and the THREADER software (17). GENIUS makes use of a conventional sequence search but in a manner that includes intermediate sequences which makes it possible to reliably link a sequence to a protein of known structure for a larger fraction of novel sequences. The THREADER program is well documented and it suffices here to say that the HUNT protein sequences are threaded upon a database of structures and domains, and the top scoring candidates are tabulated according to their Z scores.
DATA ACCESS/RETRIEVAL
The HUNT database has a ‘Similarity search’ interface where the user can paste a nucleotide or amino acid sequence as the query in a BLAST search, or a search can be conducted with a user-defined pattern. The database is also equipped with a ‘Content search’ interface, where the user can click his way to a list of all HRI clones exhibiting for example PDB hits from a GENIUS search, or a list of all clones predicted to be localized in the nucleus. There is also a ‘BLAST definition keyword search’ utility where the user can search for all clones having hits to, for example, kinases as found in the BLAST listing from a search with all HRI clones against the nr database. These tools are available from the HUNT homepage.
OUTLOOK
We have designed the HUNT database for public release of annotated full-length cDNA sequences. Various independent programs have been applied to each individual clone, and a novel function prediction may be taken from these data. However, at present, a human expert is required to analyze the results in order to come up with a consensus prediction. Ideally, a weighting scheme or clustering should be applied to the annotations so as to evaluate these data. That such a procedure is possible has recently been demonstrated (18). The HRI will prepare and release novel genes to this database, which is estimated to grow 5-fold within the first half year of its public release. It will therefore become more important to automate the integration of these data for a consensus functional assignment.
The data input and annotation are carried out by individual researchers at the HRI with a master script bringing the data into a coherent picture for each clone. Each of us welcome feedback and we hope that this value enhancement to the full-length cDNA sequences may help worldwide proliferation of the HUNT database (http://www.hri.co.jp/HUNT/).
Acknowledgments
ACKNOWLEDGEMENTS
We would like to acknowledge the NEDO Human cDNA Sequencing Project, the Research Association for Biotechnology having made this research possible together with the following researchers having contributed to the full-length cDNA sequencing: (i) K. Ninomiya, A. Shiratori, H. Sudo, M. Wagatsuma, T. Hosoiri, Y. Kaku, H. Kodaira, H. Kondo, M. Sugawara, M. Takahashi, Y. Chiba, S. Ishida, K. Murakawa, Y. Ono, S. Takiguchi and S. Watanabe; (ii) T. Ishibashi, K. Fujimori, H. Tanai, M. Kimata, M. Watanabe, S. Hiraoka and K. Kanehori; (iii) A. Takahashi-Fujii, H. Hara, T. Tanase, Y. Nomura, S. Togiya, F. Komai, R. Hara, K. Takeuchi, M. Arita, T. Nabekura and A. Oshima; (iv) N. Sasaki, S. Aotsuka, Y. Yoshikawa and H. Matsunawa. We are also very grateful to S. Ishii, Y. Kawai, K. Saito, J. Yamamoto and A. Wakamatsu in the HRI genomics laboratory for cDNA library construction, 5′- and 3′-end one-pass sequencing and clone selection.
References
- 1.Ota T., Nishikawa,T., Suzuki,Y., Maruyama,K., Sugano,S. and Isogai,T. (1997) Full-length cDNA project toward a high throughput functional analysis. Microb. Comp. Genomics, 2, 204. [Google Scholar]
- 2.Maruyama K. and Sugano,S. (1994) Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with Oligoribonucleotides. Gene, 138, 171–174. [DOI] [PubMed] [Google Scholar]
- 3.Salamov A.A., Nishikawa,T. and Swindells,M.B. (1998) Assessing protein coding region integrity in cDNA sequencing projects. Bioinformatics, 14, 384–390. [DOI] [PubMed] [Google Scholar]
- 4.Irie R., Masuho,Y. and Nagai,K. (2000) An Approach to Alternative Gene Transcripts Discovery through Assembly of Fragmental cDNA Sequences. In Miyano,S., Shamir,R. and Takagi,T. (eds), Currents in Computational Molecular Biology, Frontiers Science Series. Universal Academic Press, Inc., Tokyo, Vol. 30, pp. 109–110.
- 5.Nakai K. and Horton,P. (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci., 24, 34–36. [DOI] [PubMed] [Google Scholar]
- 6.Hirokawa T., Boon-Chieng,S. and Mitaku,S. (1998) SOSUI: Classification and Secondary Structure Prediction System for Membrane Proteins. Bioinformatics, 14, 378–379. [DOI] [PubMed] [Google Scholar]
- 7.Jones D.T., Taylor,W.R. and Thornton,J.M. (1994) A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry, 33, 3038–3049. [DOI] [PubMed] [Google Scholar]
- 8.Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) The PROSITE database, its status in 1999. Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Attwood T.K., Croning,M.D., Flower,D.R., Lewis,A.P., Mabey,J.E., Scordis,P., Selley,J.N. and Wright,W. (2000) PRINTS-S: the database formerly known as PRINTS. Nucleic Acids Res., 28, 225–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suwa M., Salamov,A.A., Nishikawa,T. and Swindells,M.B. (1997) GoodMotif: The system for finding high quality motifs that can discriminate an unique family. In Miyano,S. and Takagi,T. (eds), Genome Informatics Series. Universal Academy Press, Inc., Tokyo, Vol. 8, pp. 342–343.
- 11.Scordis P., Flower,D.R. and Attwood,T.K. (1999) FingerPRINTScan: intelligent searching of the PRINTS motif database. Bioinformatics, 15, 799–806. [DOI] [PubMed] [Google Scholar]
- 12.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L.L. (2000) The Pfam protein families database. Nucleic Acids Res., 28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yudate H.T., Suwa,M. and Masuho,Y. (1999) Chaperon: Protein binary distance matrix prediction and fold recognition. Protein Sci., 8, Suppl. 2, 44. [Google Scholar]
- 14.Frishman D. and Argos,P. (1997) 75% accuracy in protein secondary structure prediction. Proteins, 27, 329–335. [DOI] [PubMed] [Google Scholar]
- 15.Salamov A.A., Suwa,M., Orengo,C.A. and Swindells,M.B. (1999) Combining sensitive database searches with multiple intermediates to detect distant homologues. Protein Eng., 12, 95–100. [DOI] [PubMed] [Google Scholar]
- 16.Salamov A.A., Suwa,M., Orengo,C.A. and Swindells,M.B. (1999) Genome analysis: Assigning protein coding regions to three-dimensional structures. Protein Sci., 8, 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones D.T., Taylor,W.R. and Thornton,J.M. (1992) A new approach to protein fold recognition. Nature, 358, 86–89. [DOI] [PubMed] [Google Scholar]
- 18.Renner A., Lapp,H. and Aszodi,A. (2000) High-throughput function assignment for novel gene products using annotation clustering. In Miyano,S., Shamir,R. and Takagi,T. (eds), Currents in Computational Molecular Biology, Frontiers Science Series. Universal Academic Press, Inc., Tokyo, Vol. 30, pp. 40–41.