Abstract
The title compound, C12H14N4, has a crystallographically imposed centre of symmetry. Intermolecular N—H⋯N hydrogen bonds between amino groups link adjacent molecules into a three-dimensional network where ten-membered hydrogen-bonded rings are observed.
Related literature
For a related compound, see: Dobrzycki & Wozniak (2007 ▶).
Experimental
Crystal data
C12H14N4
M r = 214.27
Monoclinic,
a = 9.646 (4) Å
b = 7.476 (3) Å
c = 7.751 (3) Å
β = 95.773 (5)°
V = 556.1 (4) Å3
Z = 2
Mo Kα radiation
μ = 0.08 mm−1
T = 291 K
0.14 × 0.12 × 0.10 mm
Data collection
Bruker SMART 1K CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2000 ▶) T min = 0.989, T max = 0.992
2698 measured reflections
979 independent reflections
724 reflections with I > 2σ(I)
R int = 0.075
Refinement
R[F 2 > 2σ(F 2)] = 0.051
wR(F 2) = 0.156
S = 1.09
979 reflections
73 parameters
H-atom parameters constrained
Δρmax = 0.18 e Å−3
Δρmin = −0.30 e Å−3
Data collection: SMART (Bruker, 2000 ▶); cell refinement: SMART; data reduction: SAINT (Bruker, 2000 ▶); program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810012511/bv2140sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810012511/bv2140Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1A⋯N2i | 0.90 | 2.39 | 3.224 (2) | 154 |
| N2—H2A⋯N1ii | 0.90 | 2.35 | 3.124 (2) | 145 |
Symmetry codes: (i) 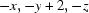 ; (ii)
; (ii) 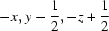 .
.
Acknowledgments
WH acknowledges the National Natural Science Foundation of China (grant No. 20871065) and the Jiangsu Province Department of Science and Technology (grant No. BK2009226) for financial aid.
supplementary crystallographic information
Comment
The crystal structure of 3,3',4,4'-tetrammoniobiphenyl tetrachloride dihydrate (Dobrzycki & Wozniak, 2007) has been reported in literature. In this paper, we report the X-ray single-crystal structure of 3,3',4,4'-tetrammoniobiphenyl (I).
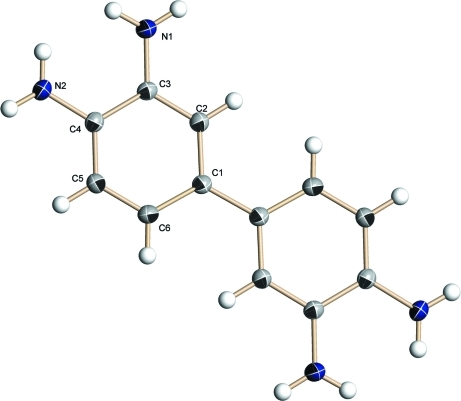
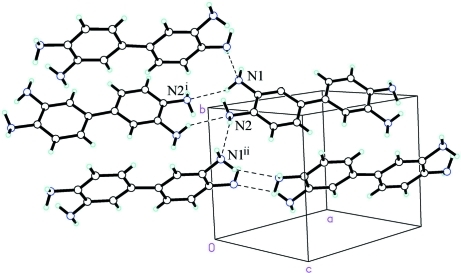
The molecular structure of (I) is illustrated in Fig. 1. Two amino groups in the 3-position lie in the opposite sides of the molecular plane. The dihedral angle between phenyl rings of adjacent molecules is 86.3 (2)°. Intermolecular N—H···N hydrogen bonds between amino groups link adjacent molecules into a three-dimensional network, where ten-membered hydrogen-bonded rings are observed (Fig. 2).
Experimental
The title compound was purchased directly from TCI. Single crystals suitable for X-ray diffraction were grown from a methanol solution by slow evaporation in air at room temperature for one week.
Refinement
H atoms were placed in geometrically idealized positions and refined as riding, with C—H = 0.93 Å and N—H = 0.86–0.90 Å, and with Uiso(H) = 1.2Ueq(C,N).
Figures
Fig. 1.
The molecular structure of the title compound with the atom-numbering scheme. Displacement ellipsoids are drawn at the 30% probability level.
Fig. 2.
Perspective view of the hydrogen bonding interactions in the crystal packing of (I), where the hydrogen bonds are shown as dashed lines. [Symmetry codes: (i) -x, -y + 2, -z; (ii) -x, y - 1/2, -z + 1/2.]
Crystal data
| C12H14N4 | F(000) = 228 |
| Mr = 214.27 | Dx = 1.280 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 931 reflections |
| a = 9.646 (4) Å | θ = 2.5–27.0° |
| b = 7.476 (3) Å | µ = 0.08 mm−1 |
| c = 7.751 (3) Å | T = 291 K |
| β = 95.773 (5)° | Block, colourless |
| V = 556.1 (4) Å3 | 0.14 × 0.12 × 0.10 mm |
| Z = 2 |
Data collection
| Bruker SMART 1K CCD area-detector diffractometer | 979 independent reflections |
| Radiation source: fine-focus sealed tube | 724 reflections with I > 2σ(I) |
| graphite | Rint = 0.075 |
| ω scans | θmax = 25.0°, θmin = 2.1° |
| Absorption correction: multi-scan (SADABS; Bruker, 2000) | h = −9→11 |
| Tmin = 0.989, Tmax = 0.992 | k = −6→8 |
| 2698 measured reflections | l = −8→9 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.051 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.156 | H-atom parameters constrained |
| S = 1.09 | w = 1/[σ2(Fo2) + (0.0926P)2 + 0.0016P] where P = (Fo2 + 2Fc2)/3 |
| 979 reflections | (Δ/σ)max < 0.001 |
| 73 parameters | Δρmax = 0.18 e Å−3 |
| 0 restraints | Δρmin = −0.30 e Å−3 |
Special details
| Experimental. The structure was solved by direct methods (Bruker, 2000) and successive difference Fourier syntheses. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.42719 (17) | 0.9895 (2) | 0.4590 (2) | 0.0335 (5) | |
| C2 | 0.37707 (18) | 1.0872 (2) | 0.3125 (2) | 0.0356 (5) | |
| H2 | 0.4378 | 1.1639 | 0.2629 | 0.043* | |
| C3 | 0.24074 (18) | 1.0749 (2) | 0.2378 (2) | 0.0336 (5) | |
| C4 | 0.14684 (18) | 0.9615 (2) | 0.3120 (2) | 0.0341 (5) | |
| C5 | 0.1965 (2) | 0.8586 (2) | 0.4523 (2) | 0.0391 (6) | |
| H5 | 0.1367 | 0.7785 | 0.4991 | 0.047* | |
| C6 | 0.3330 (2) | 0.8714 (3) | 0.5255 (2) | 0.0421 (6) | |
| H6 | 0.3629 | 0.8003 | 0.6205 | 0.051* | |
| N1 | 0.18955 (16) | 1.1838 (2) | 0.0986 (2) | 0.0442 (5) | |
| H1A | 0.1515 | 1.1130 | 0.0127 | 0.053* | |
| H1B | 0.2437 | 1.2600 | 0.0562 | 0.053* | |
| N2 | 0.00747 (15) | 0.9522 (2) | 0.23637 (19) | 0.0418 (5) | |
| H2A | −0.0484 | 0.9167 | 0.3161 | 0.050* | |
| H2B | −0.0130 | 1.0651 | 0.2025 | 0.050* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0327 (11) | 0.0338 (10) | 0.0336 (10) | 0.0017 (8) | 0.0013 (8) | −0.0006 (8) |
| C2 | 0.0326 (11) | 0.0381 (11) | 0.0362 (10) | −0.0008 (8) | 0.0043 (8) | 0.0023 (8) |
| C3 | 0.0355 (11) | 0.0348 (10) | 0.0300 (9) | 0.0026 (8) | 0.0004 (8) | −0.0012 (7) |
| C4 | 0.0327 (11) | 0.0353 (10) | 0.0337 (10) | −0.0013 (8) | 0.0007 (8) | −0.0053 (8) |
| C5 | 0.0376 (12) | 0.0392 (11) | 0.0397 (11) | −0.0082 (8) | −0.0003 (9) | 0.0049 (8) |
| C6 | 0.0422 (12) | 0.0420 (11) | 0.0404 (11) | −0.0036 (9) | −0.0046 (9) | 0.0092 (8) |
| N1 | 0.0434 (11) | 0.0480 (10) | 0.0396 (10) | −0.0045 (7) | −0.0033 (8) | 0.0113 (7) |
| N2 | 0.0324 (10) | 0.0493 (11) | 0.0424 (10) | −0.0036 (7) | −0.0026 (7) | 0.0017 (7) |
Geometric parameters (Å, °)
| C1—C2 | 1.395 (3) | C4—N2 | 1.413 (2) |
| C1—C6 | 1.401 (3) | C5—C6 | 1.384 (3) |
| C1—C1i | 1.491 (3) | C5—H5 | 0.9300 |
| C2—C3 | 1.386 (2) | C6—H6 | 0.9300 |
| C2—H2 | 0.9300 | N1—H1A | 0.8999 |
| C3—N1 | 1.401 (2) | N1—H1B | 0.8600 |
| C3—C4 | 1.405 (2) | N2—H2A | 0.9000 |
| C4—C5 | 1.379 (3) | N2—H2B | 0.9000 |
| C2—C1—C6 | 116.41 (17) | C4—C5—C6 | 121.72 (17) |
| C2—C1—C1i | 121.8 (2) | C4—C5—H5 | 119.1 |
| C6—C1—C1i | 121.8 (2) | C6—C5—H5 | 119.1 |
| C3—C2—C1 | 122.83 (17) | C5—C6—C1 | 121.21 (18) |
| C3—C2—H2 | 118.6 | C5—C6—H6 | 119.4 |
| C1—C2—H2 | 118.6 | C1—C6—H6 | 119.4 |
| C2—C3—N1 | 121.97 (16) | C3—N1—H1A | 108.3 |
| C2—C3—C4 | 119.50 (16) | C3—N1—H1B | 119.9 |
| N1—C3—C4 | 118.29 (16) | H1A—N1—H1B | 108.9 |
| C5—C4—C3 | 118.20 (17) | C4—N2—H2A | 109.9 |
| C5—C4—N2 | 122.70 (16) | C4—N2—H2B | 104.2 |
| C3—C4—N2 | 119.05 (16) | H2A—N2—H2B | 110.4 |
| C6—C1—C2—C3 | 2.1 (3) | N1—C3—C4—N2 | 4.4 (2) |
| C1i—C1—C2—C3 | −177.55 (18) | C3—C4—C5—C6 | 3.2 (3) |
| C1—C2—C3—N1 | 175.14 (17) | N2—C4—C5—C6 | −179.28 (17) |
| C1—C2—C3—C4 | 0.8 (3) | C4—C5—C6—C1 | −0.3 (3) |
| C2—C3—C4—C5 | −3.4 (3) | C2—C1—C6—C5 | −2.3 (3) |
| N1—C3—C4—C5 | −177.99 (15) | C1i—C1—C6—C5 | 177.30 (19) |
| C2—C3—C4—N2 | 178.99 (15) |
Symmetry codes: (i) −x+1, −y+2, −z+1.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···N2ii | 0.90 | 2.39 | 3.224 (2) | 154 |
| N2—H2A···N1iii | 0.90 | 2.35 | 3.124 (2) | 145 |
Symmetry codes: (ii) −x, −y+2, −z; (iii) −x, y−1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BV2140).
References
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810012511/bv2140sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810012511/bv2140Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report