Abstract
In the title compound, C17H13N3OS, the dihedral angle between the ring systems is 2.22 (5)°. The N—H grouping participates in both intra- and intermolecular N—H⋯O hydrogen bonds, the latter leading to dimers related by a twofold rotation axis.
Related literature
For related structures, see: Teo et al. (1993 ▶); Chen (1994 ▶); Sawusch & Schilde (1999 ▶); Chu et al. (2003 ▶); Liu et al. (2004 ▶). For related literature, see: Harnden et al. (1978 ▶); Hatheway et al. (1978 ▶); Londershausen (1996 ▶); Tewari & Mishra (2001 ▶).
Experimental
Crystal data
C17H13N3OS
M r = 307.36
Monoclinic,

a = 27.0144 (8) Å
b = 7.4021 (2) Å
c = 14.0523 (4) Å
β = 97.466 (1)°
V = 2786.12 (14) Å3
Z = 8
Mo Kα radiation
μ = 0.24 mm−1
T = 100 K
0.40 × 0.28 × 0.20 mm
Data collection
Bruker APEXII with a CCD detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2008 ▶) T min = 0.875, T max = 0.956
57380 measured reflections
3359 independent reflections
2928 reflections with I > 2σ(I)
R int = 0.041
Refinement
R[F 2 > 2σ(F 2)] = 0.030
wR(F 2) = 0.082
S = 1.05
3359 reflections
203 parameters
H-atom parameters constrained
Δρmax = 0.37 e Å−3
Δρmin = −0.26 e Å−3
Data collection: APEX2 (Bruker, 2008 ▶); cell refinement: SAINT (Bruker, 2008 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: DIAMOND (Brandenburg, 2006 ▶); software used to prepare material for publication: SHELXTL (Sheldrick, 2008 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810013176/fk2016sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810013176/fk2016Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N3—H3⋯O15 | 0.88 | 2.24 | 2.8483 (14) | 126 |
| N3—H3⋯O15i | 0.88 | 2.22 | 2.9161 (13) | 136 |
Symmetry code: (i)  .
.
Acknowledgments
We thank the Deutsche Forschungsgemeinschaft and the Government of Lower Saxony for their financial support in the acquisition of the diffractometer.
supplementary crystallographic information
Comment
The chemistry of pyrazole derivatives has been the subject of much interest due to their importance for various applications and their widespread potential and proven biological and pharmacological activities such as anti-inflammatory (Tewari et al., 2001), antimicrobial, antiviral (Harnden et al., 1978), anti-tumor (Hatheway et al., 1978), anti-fungal and pesticidal substances (Londershausen et al., 1996).
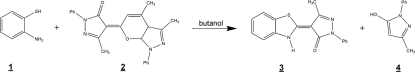
The new pyrazole derivative (E)-4-(benzo[d]thiazol-2-(3H)-ylidene)-3-methyl-1- phenyl-1H-pyrazol-5(4H)-one, 3, was synthesised according to reaction scheme 1. The solid state structure of 3 (Fig. 1) shows the typical structural features of its three subunits: a phenyl rest, a pyrazole derivative and a benzothiazole-like fragment.
In the pyrazol moiety the shortening of the bond between N12 and C13 [1.307 (2) Å] corresponds to a double bond in accordance with the formulation of the double bond in scheme 1.
All of the three subunits are for themselves planar, deviations from the least-square planes are small. Within the benzo[d]thiazole the greatest deviations result from the fact that the phenyl ring is planar whereas the remaining atoms of the thiazole ring are out of this plane with a maximum at C2 [-0.055 (2) Å]. Moreover, the complete molecule adopts are slightly curved conformation (Fig. 2). In the case of the 1,3-benzothiazole-pyrazole bond planarity should be result from the double bond [d(C—C) = 1.388 (2) Å] between the two subunits. Nevertheless, the interplanar dihedral angle between both ring systems is 2.22 (5)°. Between the pyrazole and the phenyl fragment the carbon-carbon single bond is shortened [d(C—C) = 1.416 (2) Å] but even longer than a double bond. In this case, the interplanar dihedral angle between both fragments is 7.16 (6)°. Similar values between these two subunits were obsereved earlier (Liu et al. 2004).
In the solid state, 3 forms dimers (Fig. 3) by bifurcated hydrogen bonds between the NH-group [N3] of the 1,3-benzothiazole-rest and O15 of a neighbouring molecule, both related by a twofold rotation axis. The second part of the bifurcated hydrogen bond system combines the NH-group with O15 within the same molecule.
Experimental
1.98 g (0.1 mol) (E)-3-methyl-4-(4-methyl-1-phenylpyrano[2,3-c]pyrazol-6(1H)-ylidene)-1-phenyl-1H-pyrazol-5(4H)-one, 2, were refluxed for 72 h with 2.136 ml (0.02 mol) 2-aminobenzenethiol, 1, in 50 ml n-butanol. On cooling the product forms transparent colourless crystals, which were filtered off in a yield of 50%. A suitable single crystal was selected under a polarization microsope and mounted on a 50 µm MicroMesh MiTeGen MicromountTM using FROMBLIN Y perfluoropolyether (LVAC 16/6, Aldrich).
Refinement
Hydrogen atoms were clearly identified in difference Fourier syntheses, idealized and refined at calculated positions riding on the carbon atoms with C—H = 0.98 Å for methyl H atoms, 0.95 Å for aromatic H atoms and N—H = 0.88 Å. Methyl groups were allowed to rotate around the C—C–bond. Three common isotropic displacement parameters for the H–atoms of the three different subunits were refined.
Figures
Fig. 1.
Ball-and-stick model of 3 with the atomic numbering scheme used; with exception of the hydrogen atoms, which are shown as spheres with common isotropic radius, all other atoms are represented as thermal displacement ellipsoids showing the 50% probability level of the corresponding atom.
Fig. 2.
Side view of the ball-and-stick model of 3 showing slightly curved conformation of the different ring systems.
Fig. 3.
Hydrogen bonded dimers of 3 with the intra- and intermolecular bifurcated hydrogen bonds indicated as broken lines; symmetry code 1) -x+2, y, -z+3/2.
Fig. 4.
The formation of the title compound.
Crystal data
| C17H13N3OS | F(000) = 1280 |
| Mr = 307.36 | Dx = 1.466 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -C 2yc | Cell parameters from 9789 reflections |
| a = 27.0144 (8) Å | θ = 2.9–30.6° |
| b = 7.4021 (2) Å | µ = 0.24 mm−1 |
| c = 14.0523 (4) Å | T = 100 K |
| β = 97.466 (1)° | Prism, red |
| V = 2786.12 (14) Å3 | 0.40 × 0.28 × 0.20 mm |
| Z = 8 |
Data collection
| Bruker APEXII with a CCD detector diffractometer | 3359 independent reflections |
| Radiation source: fine-focus sealed tube | 2928 reflections with I > 2σ(I) |
| graphite | Rint = 0.041 |
| φ and ω scans | θmax = 28.0°, θmin = 2.9° |
| Absorption correction: multi-scan (SADABS; Bruker, 2008) | h = −35→35 |
| Tmin = 0.875, Tmax = 0.956 | k = −9→9 |
| 57380 measured reflections | l = −18→18 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.030 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.082 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0331P)2 + 3.8168P] where P = (Fo2 + 2Fc2)/3 |
| 3359 reflections | (Δ/σ)max = 0.001 |
| 203 parameters | Δρmax = 0.37 e Å−3 |
| 0 restraints | Δρmin = −0.26 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 1.059848 (12) | 0.17433 (4) | 1.08323 (2) | 0.01678 (9) | |
| C2 | 1.02064 (5) | 0.21310 (17) | 0.97688 (9) | 0.0151 (2) | |
| N3 | 1.04276 (4) | 0.16259 (14) | 0.90040 (7) | 0.0160 (2) | |
| H3 | 1.0281 | 0.1754 | 0.8411 | 0.0247 (19)* | |
| C4 | 1.12040 (5) | 0.02162 (18) | 0.85676 (10) | 0.0198 (3) | |
| H4 | 1.1098 | 0.0244 | 0.7896 | 0.0247 (19)* | |
| C5 | 1.16636 (5) | −0.04948 (19) | 0.89339 (11) | 0.0235 (3) | |
| H5 | 1.1876 | −0.0965 | 0.8506 | 0.0247 (19)* | |
| C6 | 1.18205 (5) | −0.05341 (19) | 0.99209 (11) | 0.0250 (3) | |
| H6 | 1.2138 | −0.1025 | 1.0152 | 0.0247 (19)* | |
| C7 | 1.15201 (5) | 0.01318 (19) | 1.05696 (10) | 0.0222 (3) | |
| H7 | 1.1626 | 0.0104 | 1.1241 | 0.0247 (19)* | |
| C8 | 1.10583 (5) | 0.08405 (17) | 1.02029 (9) | 0.0174 (2) | |
| C9 | 1.09021 (5) | 0.08893 (17) | 0.92167 (9) | 0.0167 (2) | |
| N11 | 0.89806 (4) | 0.38442 (15) | 0.90615 (7) | 0.0151 (2) | |
| N12 | 0.90079 (4) | 0.40940 (15) | 1.00569 (7) | 0.0167 (2) | |
| C13 | 0.94462 (5) | 0.34924 (16) | 1.04238 (9) | 0.0157 (2) | |
| C14 | 0.97279 (5) | 0.28461 (17) | 0.96994 (8) | 0.0150 (2) | |
| C15 | 0.94124 (5) | 0.30754 (16) | 0.87995 (9) | 0.0147 (2) | |
| O15 | 0.94948 (3) | 0.26778 (13) | 0.79710 (6) | 0.01790 (19) | |
| C16 | 0.95996 (5) | 0.35330 (18) | 1.14808 (9) | 0.0194 (3) | |
| H16A | 0.9322 | 0.3982 | 1.1800 | 0.030 (3)* | |
| H16B | 0.9888 | 0.4333 | 1.1628 | 0.030 (3)* | |
| H16C | 0.9689 | 0.2310 | 1.1710 | 0.030 (3)* | |
| C21 | 0.85536 (5) | 0.45211 (17) | 0.84799 (9) | 0.0154 (2) | |
| C22 | 0.81913 (5) | 0.54324 (19) | 0.89193 (9) | 0.0201 (3) | |
| H22 | 0.8227 | 0.5545 | 0.9598 | 0.027 (2)* | |
| C23 | 0.77785 (5) | 0.61730 (19) | 0.83640 (10) | 0.0219 (3) | |
| H23 | 0.7534 | 0.6799 | 0.8666 | 0.027 (2)* | |
| C24 | 0.77185 (5) | 0.60084 (19) | 0.73731 (10) | 0.0224 (3) | |
| H24 | 0.7436 | 0.6522 | 0.6995 | 0.027 (2)* | |
| C25 | 0.80761 (5) | 0.50843 (19) | 0.69414 (10) | 0.0225 (3) | |
| H25 | 0.8035 | 0.4960 | 0.6263 | 0.027 (2)* | |
| C26 | 0.84937 (5) | 0.43350 (18) | 0.74838 (9) | 0.0186 (3) | |
| H26 | 0.8736 | 0.3703 | 0.7179 | 0.027 (2)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.01820 (16) | 0.01755 (16) | 0.01406 (15) | 0.00014 (12) | 0.00009 (11) | 0.00048 (11) |
| C2 | 0.0180 (6) | 0.0128 (5) | 0.0142 (5) | −0.0027 (4) | 0.0010 (4) | 0.0002 (4) |
| N3 | 0.0175 (5) | 0.0166 (5) | 0.0138 (5) | 0.0006 (4) | 0.0019 (4) | 0.0002 (4) |
| C4 | 0.0221 (6) | 0.0161 (6) | 0.0222 (6) | −0.0004 (5) | 0.0062 (5) | 0.0012 (5) |
| C5 | 0.0213 (7) | 0.0184 (6) | 0.0322 (7) | 0.0009 (5) | 0.0090 (5) | 0.0006 (5) |
| C6 | 0.0175 (6) | 0.0221 (7) | 0.0348 (8) | 0.0030 (5) | 0.0009 (5) | 0.0030 (6) |
| C7 | 0.0211 (6) | 0.0196 (6) | 0.0248 (7) | −0.0003 (5) | −0.0013 (5) | 0.0025 (5) |
| C8 | 0.0175 (6) | 0.0142 (6) | 0.0206 (6) | −0.0015 (5) | 0.0026 (5) | 0.0007 (5) |
| C9 | 0.0168 (6) | 0.0133 (6) | 0.0200 (6) | −0.0014 (5) | 0.0024 (5) | 0.0019 (5) |
| N11 | 0.0169 (5) | 0.0162 (5) | 0.0123 (5) | 0.0007 (4) | 0.0025 (4) | −0.0001 (4) |
| N12 | 0.0204 (5) | 0.0177 (5) | 0.0122 (5) | 0.0000 (4) | 0.0026 (4) | −0.0010 (4) |
| C13 | 0.0192 (6) | 0.0132 (6) | 0.0150 (6) | −0.0018 (5) | 0.0031 (5) | −0.0006 (4) |
| C14 | 0.0177 (6) | 0.0138 (6) | 0.0135 (5) | −0.0016 (5) | 0.0016 (4) | −0.0003 (4) |
| C15 | 0.0157 (6) | 0.0129 (5) | 0.0157 (6) | −0.0021 (4) | 0.0024 (4) | 0.0004 (4) |
| O15 | 0.0188 (4) | 0.0220 (5) | 0.0132 (4) | 0.0011 (4) | 0.0034 (3) | −0.0014 (3) |
| C16 | 0.0227 (6) | 0.0208 (6) | 0.0146 (6) | −0.0001 (5) | 0.0017 (5) | −0.0014 (5) |
| C21 | 0.0153 (6) | 0.0131 (5) | 0.0176 (6) | −0.0027 (5) | 0.0015 (4) | 0.0014 (4) |
| C22 | 0.0197 (6) | 0.0226 (7) | 0.0182 (6) | 0.0011 (5) | 0.0037 (5) | −0.0001 (5) |
| C23 | 0.0175 (6) | 0.0212 (6) | 0.0275 (7) | 0.0011 (5) | 0.0050 (5) | 0.0003 (5) |
| C24 | 0.0177 (6) | 0.0209 (7) | 0.0271 (7) | −0.0007 (5) | −0.0021 (5) | 0.0038 (5) |
| C25 | 0.0238 (7) | 0.0245 (7) | 0.0183 (6) | −0.0010 (5) | −0.0008 (5) | 0.0009 (5) |
| C26 | 0.0195 (6) | 0.0186 (6) | 0.0178 (6) | −0.0004 (5) | 0.0025 (5) | −0.0003 (5) |
Geometric parameters (Å, °)
| S1—C2 | 1.7397 (13) | N12—C13 | 1.3067 (16) |
| S1—C8 | 1.7485 (13) | C13—C14 | 1.4303 (17) |
| C2—N3 | 1.3488 (16) | C13—C16 | 1.4896 (17) |
| C2—C14 | 1.3883 (18) | C14—C15 | 1.4402 (16) |
| N3—C9 | 1.3892 (16) | C15—O15 | 1.2486 (15) |
| N3—H3 | 0.8800 | C16—H16A | 0.9800 |
| C4—C5 | 1.3845 (19) | C16—H16B | 0.9800 |
| C4—C9 | 1.3924 (18) | C16—H16C | 0.9800 |
| C4—H4 | 0.9500 | C21—C26 | 1.3949 (17) |
| C5—C6 | 1.397 (2) | C21—C22 | 1.3969 (18) |
| C5—H5 | 0.9500 | C22—C23 | 1.3882 (19) |
| C6—C7 | 1.387 (2) | C22—H22 | 0.9500 |
| C6—H6 | 0.9500 | C23—C24 | 1.3861 (19) |
| C7—C8 | 1.3892 (18) | C23—H23 | 0.9500 |
| C7—H7 | 0.9500 | C24—C25 | 1.386 (2) |
| C8—C9 | 1.3956 (17) | C24—H24 | 0.9500 |
| N11—C15 | 1.3896 (16) | C25—C26 | 1.3921 (18) |
| N11—N12 | 1.4034 (14) | C25—H25 | 0.9500 |
| N11—C21 | 1.4158 (16) | C26—H26 | 0.9500 |
| C2—S1—C8 | 91.25 (6) | C14—C13—C16 | 127.71 (12) |
| N3—C2—C14 | 123.68 (11) | C2—C14—C13 | 130.85 (12) |
| N3—C2—S1 | 110.81 (9) | C2—C14—C15 | 123.10 (11) |
| C14—C2—S1 | 125.50 (10) | C13—C14—C15 | 106.05 (11) |
| C2—N3—C9 | 115.43 (10) | O15—C15—N11 | 126.94 (11) |
| C2—N3—H3 | 122.3 | O15—C15—C14 | 129.34 (12) |
| C9—N3—H3 | 122.3 | N11—C15—C14 | 103.72 (10) |
| C5—C4—C9 | 117.76 (12) | C13—C16—H16A | 109.5 |
| C5—C4—H4 | 121.1 | C13—C16—H16B | 109.5 |
| C9—C4—H4 | 121.1 | H16A—C16—H16B | 109.5 |
| C4—C5—C6 | 121.25 (13) | C13—C16—H16C | 109.5 |
| C4—C5—H5 | 119.4 | H16A—C16—H16C | 109.5 |
| C6—C5—H5 | 119.4 | H16B—C16—H16C | 109.5 |
| C7—C6—C5 | 121.13 (13) | C26—C21—C22 | 119.65 (12) |
| C7—C6—H6 | 119.4 | C26—C21—N11 | 121.59 (11) |
| C5—C6—H6 | 119.4 | C22—C21—N11 | 118.74 (11) |
| C6—C7—C8 | 117.66 (13) | C23—C22—C21 | 120.00 (12) |
| C6—C7—H7 | 121.2 | C23—C22—H22 | 120.0 |
| C8—C7—H7 | 121.2 | C21—C22—H22 | 120.0 |
| C7—C8—C9 | 121.27 (12) | C24—C23—C22 | 120.70 (13) |
| C7—C8—S1 | 128.27 (11) | C24—C23—H23 | 119.7 |
| C9—C8—S1 | 110.45 (10) | C22—C23—H23 | 119.7 |
| N3—C9—C4 | 127.02 (12) | C23—C24—C25 | 119.07 (12) |
| N3—C9—C8 | 112.04 (11) | C23—C24—H24 | 120.5 |
| C4—C9—C8 | 120.93 (12) | C25—C24—H24 | 120.5 |
| C15—N11—N12 | 112.38 (10) | C24—C25—C26 | 121.21 (12) |
| C15—N11—C21 | 129.86 (10) | C24—C25—H25 | 119.4 |
| N12—N11—C21 | 117.51 (10) | C26—C25—H25 | 119.4 |
| C13—N12—N11 | 106.02 (10) | C25—C26—C21 | 119.36 (12) |
| N12—C13—C14 | 111.83 (11) | C25—C26—H26 | 120.3 |
| N12—C13—C16 | 120.47 (11) | C21—C26—H26 | 120.3 |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N3—H3···O15 | 0.88 | 2.24 | 2.8483 (14) | 126 |
| N3—H3···O15i | 0.88 | 2.22 | 2.9161 (13) | 136 |
Symmetry codes: (i) −x+2, y, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: FK2016).
References
- Brandenburg, K. (2006). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Bruker (2008). APEX2, SADABS and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Chen, W. (1994). J. Organomet. Chem.471, 69–71.
- Chu, Q., Wang, Z., Huang, Q., Yan, C. & Zhu, S. (2003). New J. Chem.27, 1522–1527.
- Harnden, M. R., Bailey, S., Boyd, M. R., Taylor, D. R. & Wright, N. D. (1978). J. Med. Chem.21, 82–87. [DOI] [PubMed]
- Hatheway, G. J., Hansch, C., Kim, K. H., Milstein, S. R., Schmidt, C. L., Smith, R. N. & Quinn, F. R. (1978). J. Med. Chem.21, 563–574. [DOI] [PubMed]
- Liu, G. F., Liu, L., Jia, D.-Z. & Yu, K. B. (2004). J. Chem. Crystallogr.34, 835–841.
- Londershausen, M. (1996). Pestic. Sci.48, 269–292.
- Sawusch, S. & Schilde, U. (1999). Z. Naturforsch. Teil B, 54, 881–886.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Teo, S.-B., Teoh, S.-G., Okechukwu, R. C. & Fun, H.-K. (1993). J. Organomet. Chem.454, 67–71.
- Tewari, A. K. & Mishra, A. (2001). Bioorg. Med. Chem.9, 715–718. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810013176/fk2016sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810013176/fk2016Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report