Abstract
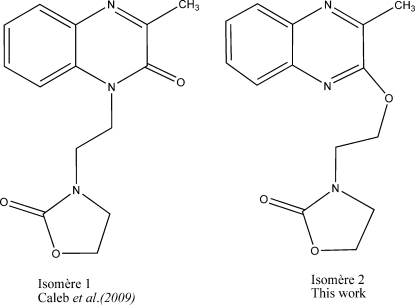
Two isomers were isolated during the reaction between 3-methylquinoxalin-2-one and bis(2-chloroethyl)amine hydrochloride. The crystal structure of one isomer has already been reported [Caleb, Bouhfid, Essassi & El Ammari (2009). Acta Cryst. E65, o2024–o2025], while that of the second isomer is the subject of this work. The title compound, C14H15N3O3, has a new structure containing oxazolidine and quinoxaline rings linked by an ethoxy group. The main difference between the two isomers is the position of the oxazolidine group with respect to the quinoxaline system. The dihedral angle between the fused planar rings and the oxazolidin-2-one ring is 41.63 (8)° in the title molecule.
Related literature
For the biological activity of 3-[2-(3-methyl-1,2-dihydroquinoxalin-2-yloxy)ethyl]oxazolidin-2-one, see: Madhusudhan et al. (2004 ▶); Soad et al. (2006 ▶); Sriharsha & Shashikanth (2006 ▶); Menoret et al. (2009 ▶); Wilhelmsson et al. (2008 ▶). For the structure of the isomer of the title compound, see: Caleb et al. (2009 ▶). For related structures, see: Doubia et al. (2007 ▶); Mamedov et al. (2007 ▶); Aschwanden et al.(1976 ▶)
Experimental
Crystal data
C14H15N3O3
M r = 273.29
Triclinic,
a = 6.9936 (3) Å
b = 7.6916 (3) Å
c = 13.3709 (6) Å
α = 86.649 (2)°
β = 77.044 (2)°
γ = 71.141 (2)°
V = 663.23 (5) Å3
Z = 2
Mo Kα radiation
μ = 0.10 mm−1
T = 296 K
0.41 × 0.33 × 0.20 mm
Data collection
Bruker X8 APEXII CCD area-detector diffractometer
15358 measured reflections
3030 independent reflections
2358 reflections with I > 2σ(I)
R int = 0.023
Refinement
R[F 2 > 2σ(F 2)] = 0.040
wR(F 2) = 0.121
S = 1.06
3030 reflections
198 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.21 e Å−3
Δρmin = −0.17 e Å−3
Data collection: APEX2 (Bruker, 2005 ▶); cell refinement: APEX2; data reduction: APEX2; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia,1997 ▶) and PLATON (Spek, 2009 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810012687/dn2552sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810012687/dn2552Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors thank the Unit of Support for Technical and Scientific Research (UATRS, CNRST) for the X-ray measurements.
supplementary crystallographic information
Comment
Oxazolidin-2-ones and quinoxalines are subjets of numerous articles in scientific journals concerning the development of new molecules as drug candidates such as antibacterials (Madhusudhan et al. 2004); (Sriharsha & Shashikanth, 2006), anti-viral (Wilhelmsson, et al. 2008), anti-tumor (Soad et al.2006),and anti-inflammatory (Menoret et al. 2009). Our investigation is intended to increase the biological activity of such molecules. During the synthesis, two isomers were isolated, and the structure of isomer 1 has already been published (Caleb et al. 2009) while that of ismer 2 is the subject of the present work.
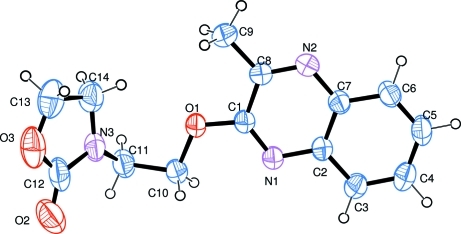
The structure of the 3-(2-(3-methyl-1,2-dihydro-quinoxalin-2-yloxy)ethoxy) oxazolidin-2-one molecule is also built up from two fused six-membered rings linked to a five-membered ring (oxazolidin-2-one) by an ethoxy group, as shown in Fig.1. It would be interesting to compare the crystal structures of both isomers of this compound (scheme 1). Actually, the geometric parameters (bond lenghths and angles) of the two isomers are very similar to those observed in other heterocyclic structures (Aschwanden et al., 1976; Doubia et al., 2007; Mamedov et al., 2007). However, the main difference between the two isomers is the position of the oxazolidine group with respect to the quinoxalin. Moreover, the dihedral angle between the fused six-membered rings and the five cycles measuring 20.04 (9)° in the isomer 1 instead of 41.63 (8)° in the isomer 2.
Experimental
In a 100 ml flask, is reacted 0.0125 moles of quinoxalin-2-one with 2.66 moles of dichloroethylamine hydrochloride in 40 ml of dimethyl formamide in presence of 2.87 moles of potassium carbonate and a few milligrams of tetran-butyl ammonium bromide. The mixture was brought to reflux in a sand bath, magnetic stirring and the reaction progress was monitored by thin layer chromatography. After evaporation of solvent under reduced pressure, the residue obtained is chromatographed on silica column (hexane / acetate: 4 / 6). Thus we have isolated two compounds. Recrystallization occurred in the same eluent. This compound was obtained in 38% and his melting point is 169°C.
Refinement
H atoms were located in a difference map and treated as riding with C—H = 0.96 Å for methyl groups and C—H = 0.93 Å for all other hydrogens with Uiso(H) = 1.2 Ueq(aromatic, methine ) or Uiso(H) = 1.5 Ueq(methyl). All other H atoms were located from difference Fourier maps and refined without any distance restraints.
Figures
Fig. 1.
: Molecular structure of the title compound with the atom-labelling scheme. Displacement ellipsoids are drawn at the 50% probability level. H atoms are represented as small circles.
Fig. 2.
The structures of the two isomers.
Crystal data
| C14H15N3O3 | Z = 2 |
| Mr = 273.29 | F(000) = 288 |
| Triclinic, P1 | Dx = 1.368 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 6.9936 (3) Å | Cell parameters from 15358 reflections |
| b = 7.6916 (3) Å | θ = 2.8–27.5° |
| c = 13.3709 (6) Å | µ = 0.10 mm−1 |
| α = 86.649 (2)° | T = 296 K |
| β = 77.044 (2)° | Prism, colourless |
| γ = 71.141 (2)° | 0.41 × 0.33 × 0.20 mm |
| V = 663.23 (5) Å3 |
Data collection
| Bruker X8 APEXII CCD area-detector diffractometer | 2358 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.023 |
| graphite | θmax = 27.5°, θmin = 2.8° |
| φ and ω scans | h = −9→9 |
| 15358 measured reflections | k = −9→9 |
| 3030 independent reflections | l = −17→17 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.040 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.121 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0643P)2 + 0.0716P] where P = (Fo2 + 2Fc2)/3 |
| 3030 reflections | (Δ/σ)max < 0.001 |
| 198 parameters | Δρmax = 0.21 e Å−3 |
| 0 restraints | Δρmin = −0.17 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.31122 (17) | 0.72867 (15) | 0.95763 (8) | 0.0378 (3) | |
| C2 | 0.08813 (17) | 0.80823 (15) | 1.11146 (8) | 0.0382 (3) | |
| C3 | −0.11007 (19) | 0.83829 (18) | 1.17492 (10) | 0.0470 (3) | |
| C4 | −0.1442 (2) | 0.8837 (2) | 1.27638 (10) | 0.0531 (3) | |
| C5 | 0.0143 (2) | 0.90272 (19) | 1.31801 (10) | 0.0540 (3) | |
| C6 | 0.2068 (2) | 0.87655 (18) | 1.25750 (10) | 0.0503 (3) | |
| C7 | 0.24799 (18) | 0.82807 (16) | 1.15335 (9) | 0.0403 (3) | |
| C8 | 0.47691 (17) | 0.75240 (16) | 0.99773 (9) | 0.0412 (3) | |
| C9 | 0.68526 (19) | 0.7217 (2) | 0.92895 (11) | 0.0546 (3) | |
| H9A | 0.7371 | 0.5987 | 0.9015 | 0.082* | |
| H9B | 0.7784 | 0.7393 | 0.9674 | 0.082* | |
| H9C | 0.6743 | 0.8075 | 0.8737 | 0.082* | |
| C10 | 0.21267 (19) | 0.63937 (19) | 0.81444 (9) | 0.0465 (3) | |
| H10A | 0.1105 | 0.7551 | 0.8041 | 0.056* | |
| H10B | 0.1429 | 0.5643 | 0.8593 | 0.056* | |
| C11 | 0.3226 (2) | 0.54186 (19) | 0.71328 (10) | 0.0523 (3) | |
| H11A | 0.4218 | 0.4258 | 0.7258 | 0.063* | |
| H11B | 0.2221 | 0.5146 | 0.6826 | 0.063* | |
| C12 | 0.3726 (3) | 0.7066 (2) | 0.55269 (11) | 0.0625 (4) | |
| C13 | 0.6852 (3) | 0.7454 (3) | 0.54185 (14) | 0.0820 (5) | |
| H13A | 0.6957 | 0.8634 | 0.5570 | 0.098* | |
| H13B | 0.8151 | 0.6739 | 0.4986 | 0.098* | |
| C14 | 0.6363 (3) | 0.6450 (3) | 0.63974 (11) | 0.0682 (4) | |
| H14A | 0.7319 | 0.5209 | 0.6375 | 0.082* | |
| H14B | 0.6394 | 0.7098 | 0.6991 | 0.082* | |
| N1 | 0.12438 (14) | 0.75658 (13) | 1.01023 (7) | 0.0406 (2) | |
| N2 | 0.44386 (15) | 0.80020 (15) | 1.09343 (8) | 0.0462 (3) | |
| N3 | 0.43028 (18) | 0.64507 (16) | 0.64066 (8) | 0.0534 (3) | |
| O1 | 0.36793 (12) | 0.67107 (12) | 0.85906 (6) | 0.0460 (2) | |
| O2 | 0.2172 (2) | 0.70974 (18) | 0.52691 (9) | 0.0879 (4) | |
| O3 | 0.5186 (2) | 0.76983 (16) | 0.49204 (8) | 0.0829 (4) | |
| H3 | −0.220 (2) | 0.8300 (19) | 1.1449 (11) | 0.052 (4)* | |
| H4 | −0.280 (2) | 0.908 (2) | 1.3175 (13) | 0.068 (4)* | |
| H5 | −0.009 (2) | 0.934 (2) | 1.3883 (14) | 0.064 (4)* | |
| H6 | 0.322 (3) | 0.889 (2) | 1.2864 (13) | 0.071 (4)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0389 (6) | 0.0433 (6) | 0.0313 (6) | −0.0140 (5) | −0.0066 (4) | 0.0011 (4) |
| C2 | 0.0413 (6) | 0.0410 (6) | 0.0317 (6) | −0.0135 (5) | −0.0066 (4) | 0.0011 (4) |
| C3 | 0.0439 (7) | 0.0559 (7) | 0.0406 (7) | −0.0189 (6) | −0.0031 (5) | −0.0016 (5) |
| C4 | 0.0526 (7) | 0.0592 (8) | 0.0396 (7) | −0.0161 (6) | 0.0039 (6) | −0.0019 (6) |
| C5 | 0.0656 (8) | 0.0568 (8) | 0.0317 (7) | −0.0106 (6) | −0.0064 (6) | −0.0048 (5) |
| C6 | 0.0545 (8) | 0.0564 (8) | 0.0388 (7) | −0.0113 (6) | −0.0162 (6) | −0.0043 (5) |
| C7 | 0.0415 (6) | 0.0427 (6) | 0.0354 (6) | −0.0106 (5) | −0.0097 (5) | −0.0003 (5) |
| C8 | 0.0366 (6) | 0.0476 (6) | 0.0391 (6) | −0.0127 (5) | −0.0083 (5) | −0.0005 (5) |
| C9 | 0.0394 (6) | 0.0745 (9) | 0.0505 (8) | −0.0210 (6) | −0.0050 (5) | −0.0066 (6) |
| C10 | 0.0446 (6) | 0.0629 (8) | 0.0351 (6) | −0.0211 (6) | −0.0079 (5) | −0.0038 (5) |
| C11 | 0.0608 (8) | 0.0600 (8) | 0.0381 (7) | −0.0217 (6) | −0.0093 (6) | −0.0066 (5) |
| C12 | 0.0842 (11) | 0.0549 (8) | 0.0358 (7) | −0.0028 (8) | −0.0139 (7) | −0.0097 (6) |
| C13 | 0.0972 (13) | 0.0874 (12) | 0.0545 (10) | −0.0363 (10) | 0.0076 (9) | 0.0021 (8) |
| C14 | 0.0723 (10) | 0.0929 (11) | 0.0440 (8) | −0.0373 (9) | −0.0061 (7) | 0.0036 (7) |
| N1 | 0.0385 (5) | 0.0515 (6) | 0.0331 (5) | −0.0169 (4) | −0.0062 (4) | −0.0017 (4) |
| N2 | 0.0400 (5) | 0.0571 (6) | 0.0425 (6) | −0.0140 (5) | −0.0120 (4) | −0.0036 (5) |
| N3 | 0.0602 (7) | 0.0644 (7) | 0.0316 (5) | −0.0157 (5) | −0.0071 (5) | −0.0024 (5) |
| O1 | 0.0409 (4) | 0.0667 (6) | 0.0316 (4) | −0.0203 (4) | −0.0035 (3) | −0.0063 (4) |
| O2 | 0.1019 (9) | 0.0914 (9) | 0.0633 (8) | −0.0034 (7) | −0.0421 (7) | −0.0077 (6) |
| O3 | 0.1211 (10) | 0.0797 (8) | 0.0399 (6) | −0.0288 (7) | −0.0081 (6) | 0.0096 (5) |
Geometric parameters (Å, °)
| C1—N1 | 1.2932 (14) | C9—H9C | 0.9600 |
| C1—O1 | 1.3457 (13) | C10—O1 | 1.4398 (13) |
| C1—C8 | 1.4458 (15) | C10—C11 | 1.5041 (18) |
| C2—N1 | 1.3777 (14) | C10—H10A | 0.9700 |
| C2—C3 | 1.4083 (16) | C10—H10B | 0.9700 |
| C2—C7 | 1.4099 (15) | C11—N3 | 1.4523 (17) |
| C3—C4 | 1.3695 (18) | C11—H11A | 0.9700 |
| C3—H3 | 0.966 (14) | C11—H11B | 0.9700 |
| C4—C5 | 1.397 (2) | C12—O2 | 1.2046 (19) |
| C4—H4 | 0.949 (16) | C12—N3 | 1.3394 (19) |
| C5—C6 | 1.3646 (19) | C12—O3 | 1.357 (2) |
| C5—H5 | 0.948 (17) | C13—O3 | 1.424 (2) |
| C6—C7 | 1.4045 (17) | C13—C14 | 1.510 (2) |
| C6—H6 | 0.999 (17) | C13—H13A | 0.9700 |
| C7—N2 | 1.3793 (15) | C13—H13B | 0.9700 |
| C8—N2 | 1.3013 (15) | C14—N3 | 1.4379 (19) |
| C8—C9 | 1.4923 (16) | C14—H14A | 0.9700 |
| C9—H9A | 0.9600 | C14—H14B | 0.9700 |
| C9—H9B | 0.9600 | ||
| N1—C1—O1 | 121.60 (10) | C11—C10—H10A | 110.3 |
| N1—C1—C8 | 124.34 (10) | O1—C10—H10B | 110.3 |
| O1—C1—C8 | 114.06 (9) | C11—C10—H10B | 110.3 |
| N1—C2—C3 | 119.79 (10) | H10A—C10—H10B | 108.6 |
| N1—C2—C7 | 120.95 (10) | N3—C11—C10 | 114.20 (11) |
| C3—C2—C7 | 119.25 (11) | N3—C11—H11A | 108.7 |
| C4—C3—C2 | 119.75 (12) | C10—C11—H11A | 108.7 |
| C4—C3—H3 | 121.5 (9) | N3—C11—H11B | 108.7 |
| C2—C3—H3 | 118.7 (8) | C10—C11—H11B | 108.7 |
| C3—C4—C5 | 121.01 (12) | H11A—C11—H11B | 107.6 |
| C3—C4—H4 | 118.6 (10) | O2—C12—N3 | 127.96 (16) |
| C5—C4—H4 | 120.3 (10) | O2—C12—O3 | 122.31 (14) |
| C6—C5—C4 | 120.14 (12) | N3—C12—O3 | 109.73 (14) |
| C6—C5—H5 | 118.9 (9) | O3—C13—C14 | 106.05 (14) |
| C4—C5—H5 | 120.9 (9) | O3—C13—H13A | 110.5 |
| C5—C6—C7 | 120.42 (12) | C14—C13—H13A | 110.5 |
| C5—C6—H6 | 120.9 (10) | O3—C13—H13B | 110.5 |
| C7—C6—H6 | 118.6 (10) | C14—C13—H13B | 110.5 |
| N2—C7—C6 | 119.74 (10) | H13A—C13—H13B | 108.7 |
| N2—C7—C2 | 120.85 (10) | N3—C14—C13 | 101.80 (14) |
| C6—C7—C2 | 119.41 (11) | N3—C14—H14A | 111.4 |
| N2—C8—C1 | 119.95 (10) | C13—C14—H14A | 111.4 |
| N2—C8—C9 | 120.35 (10) | N3—C14—H14B | 111.4 |
| C1—C8—C9 | 119.70 (11) | C13—C14—H14B | 111.4 |
| C8—C9—H9A | 109.5 | H14A—C14—H14B | 109.3 |
| C8—C9—H9B | 109.5 | C1—N1—C2 | 115.96 (9) |
| H9A—C9—H9B | 109.5 | C8—N2—C7 | 117.92 (10) |
| C8—C9—H9C | 109.5 | C12—N3—C14 | 112.13 (13) |
| H9A—C9—H9C | 109.5 | C12—N3—C11 | 122.09 (13) |
| H9B—C9—H9C | 109.5 | C14—N3—C11 | 123.42 (12) |
| O1—C10—C11 | 106.91 (10) | C1—O1—C10 | 117.34 (9) |
| O1—C10—H10A | 110.3 | C12—O3—C13 | 109.59 (12) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: DN2552).
References
- Aschwanden, W., Kyburz, E. & Schonholzer, P. (1976). Helv. Chim. Acta, 59, 1245–1252. [DOI] [PubMed]
- Bruker (2005). APEX2 Bruker AXS Inc., Madison, Wisconsin, USA.
- Caleb, A. A., Bouhfid, R., Essassi, E. M. & El Ammari, L. (2009). Acta Cryst. E65, o2024–o2025. [DOI] [PMC free article] [PubMed]
- Doubia, M. L., Bouhfid, R., Ahabchane, N. H., Essassi, E. M. & El Ammari, L. (2007). Acta Cryst. E63, o3305.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Madhusudhan, G., Om Reddy, G., Ramanathan, J. & Dubey, P. K. (2004). Indian J. Chem. Sect. B, 43, 957–963.
- Mamedov, V. A., Khafizova, E. A., Beschastnova, T. N., Zhukova, N. A., Gubaidullin, A. T., Rizvanov, I. Kh., Berdnikov, E. A. & Litvinov, I. A. (2007). Izv. Akad. Nauk SSSR Ser. Khim. (Russ.) (Russ. Chem. Bull.), 5, 1047–1048
- Menoret, A., Mcaleer, J. P., Ngoi, S. M., Swagatam, R., Eddy, N. A., Fenteany, G., Lee, S. J., Rossi, R. J., Mukherji, B., Allen, D. L., Chakraborty, N. G. & Vella, A. T. (2009). J. Immunol.183, 7489–7496. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Soad, A. M., El-Hawash, S. A., Abeer, E. & Wahab, A. (2006). Arch. Pharm. (Weinheim), 339, 437–447. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Sriharsha, S. N. & Shashikanth, S. J. (2006). Heterocycl. Commun 12, 213–218.
- Wilhelmsson, L. M., Kingi, N. & Bergman, J. (2008). J. Med. Chem.51, 7744–7750. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810012687/dn2552sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810012687/dn2552Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report