Abstract
In the title compound, [CuCl2(C11H17N3)], the CuII ion is five-coordinated with a distorted square-pyramidal configuration. The three N atoms of the Schiff base ligand and one Cl atom are located in the basal plane, whereas the other Cl atom is apically positioned.
Related literature
For the crystal structures of similar copper (II) complexes, see: Wang et al. (2009 ▶); Yuan & Zhang (2005 ▶); Zhang et al. (2009 ▶). For a description of the geometry of five-coordinated metal complexes, see: Addison et al. (1984 ▶).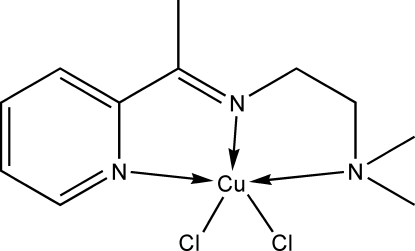
Experimental
Crystal data
[CuCl2(C11H17N3)]
M r = 325.72
Orthorhombic,

a = 9.81448 (12) Å
b = 9.90297 (13) Å
c = 14.21414 (18) Å
V = 1381.51 (2) Å3
Z = 4
Mo Kα radiation
μ = 1.95 mm−1
T = 100 K
0.30 × 0.23 × 0.07 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.592, T max = 0.876
10998 measured reflections
2439 independent reflections
2378 reflections with I > 2σ(I)
R int = 0.026
Refinement
R[F 2 > 2σ(F 2)] = 0.017
wR(F 2) = 0.043
S = 1.03
2439 reflections
157 parameters
H-atom parameters constrained
Δρmax = 0.21 e Å−3
Δρmin = −0.20 e Å−3
Absolute structure: Flack (1983 ▶), 1022 Friedel pairs
Flack parameter: 0.010 (9)
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: X-SEED (Barbour, 2001 ▶); software used to prepare material for publication: publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810011712/pv2266sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810011712/pv2266Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors thank the University of Malaya for funding this study (FRGS grant No. FP009/2008 C).
supplementary crystallographic information
Comment
The title compound was obtained by the reaction of N,N-dimethyl-N'-[methyl(2-pyridyl)methylene]ethane-1,2-diamine with copper(II) chloride. In the molecule of the complex, the metal ion is penta-coordinated by the tridentate Schiff base ligand and two chloride atoms (Fig. 1). The geometry of the complex can be determined by using the index τ = (β-α)/60, where β is the largest angle and α is the second one around the metal center. For an ideal square-pyramidal geometry τ is 0, while it is 1 in a perfect trigonal-bipyramid (Addison et al.,1984). The two largest angels in the title compound are 158.45 (6)° (N1—Cu—N3) and 154.98 (5)° (N2—Cu—Cl2) which give a τ value of 0.058. This value indicates a slightly distorted square pyramidal geometry in which the three N atoms of the Schiff base ligand and one chloride atom occupy the basal positions and the other chloride atom is placed in the apical position.
Experimental
The Schiff base ligand was prepared via condensation reaction of N,N-dimethylethyldiamine (0.44 g, 5 mmol) and 2-acetylpyridine (0.61 g, 5 mmol) by refluxing in ethanol (50 ml) for 2 h. For synthesis of the title complex a mixture of the Schiff base ligand (0.57 g, 3 mmol) and copper (II) chloride dihydrate (0.51 g, 3 mmol) in ethanol (50 ml) was stirred at room temperature for half an hour. The solvent was then evaporated partially to yield the title complex as a green solid. Suitable crystals for X-ray crystallography were obtained upon slow evaporation of an ethanolic solution at room temperature.
Refinement
Hydrogen atoms were placed at calculated positions (C—H 0.95-0.98 Å), and were treated as riding on their parent atoms, with Uiso(H) set to 1.2-1.5 times Ueq(C). An absolute structure was established using anomalous dispersion effects; 1021 Friedel pairs were not merged.
Figures
Fig. 1.
Thermal ellipsoid plot of the title compound at the 50% probability level.
Crystal data
| [CuCl2(C11H17N3)] | F(000) = 668 |
| Mr = 325.72 | Dx = 1.566 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 7155 reflections |
| a = 9.81448 (12) Å | θ = 2.5–30.4° |
| b = 9.90297 (13) Å | µ = 1.95 mm−1 |
| c = 14.21414 (18) Å | T = 100 K |
| V = 1381.51 (2) Å3 | Block, green |
| Z = 4 | 0.30 × 0.23 × 0.07 mm |
Data collection
| Bruker APEXII CCD diffractometer | 2439 independent reflections |
| Radiation source: fine-focus sealed tube | 2378 reflections with I > 2σ(I) |
| graphite | Rint = 0.026 |
| φ and ω scans | θmax = 25.0°, θmin = 2.5° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −11→11 |
| Tmin = 0.592, Tmax = 0.876 | k = −11→11 |
| 10998 measured reflections | l = −16→16 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.017 | H-atom parameters constrained |
| wR(F2) = 0.043 | w = 1/[σ2(Fo2) + (0.0245P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max = 0.001 |
| 2439 reflections | Δρmax = 0.21 e Å−3 |
| 157 parameters | Δρmin = −0.20 e Å−3 |
| 0 restraints | Absolute structure: Flack (1983), 1022 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: 0.010 (9) |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cu | 0.80028 (2) | 0.16329 (2) | 0.099598 (15) | 0.01175 (7) | |
| Cl1 | 0.56577 (5) | 0.08033 (5) | 0.13120 (3) | 0.01619 (11) | |
| Cl2 | 0.92633 (5) | 0.14346 (5) | 0.23208 (3) | 0.01790 (12) | |
| N1 | 0.86920 (16) | −0.00757 (17) | 0.03434 (12) | 0.0139 (4) | |
| N2 | 0.76828 (16) | 0.20963 (16) | −0.03353 (11) | 0.0124 (4) | |
| N3 | 0.75938 (15) | 0.36500 (16) | 0.11931 (11) | 0.0132 (4) | |
| C1 | 0.9300 (2) | −0.11396 (19) | 0.07379 (15) | 0.0169 (4) | |
| H1 | 0.9448 | −0.1136 | 0.1398 | 0.020* | |
| C2 | 0.9722 (2) | −0.2245 (2) | 0.02225 (15) | 0.0185 (5) | |
| H2 | 1.0174 | −0.2976 | 0.0522 | 0.022* | |
| C3 | 0.9479 (2) | −0.2274 (2) | −0.07327 (15) | 0.0200 (5) | |
| H3 | 0.9737 | −0.3036 | −0.1098 | 0.024* | |
| C4 | 0.8846 (2) | −0.1161 (2) | −0.11553 (15) | 0.0175 (4) | |
| H4 | 0.8666 | −0.1155 | −0.1812 | 0.021* | |
| C5 | 0.84897 (19) | −0.0075 (2) | −0.06026 (13) | 0.0131 (4) | |
| C6 | 0.79321 (18) | 0.12257 (18) | −0.09696 (13) | 0.0129 (4) | |
| C7 | 0.7771 (2) | 0.1485 (2) | −0.20048 (13) | 0.0200 (5) | |
| H7A | 0.6843 | 0.1805 | −0.2132 | 0.030* | |
| H7B | 0.7935 | 0.0648 | −0.2354 | 0.030* | |
| H7C | 0.8428 | 0.2173 | −0.2204 | 0.030* | |
| C8 | 0.71958 (19) | 0.3470 (2) | −0.05216 (13) | 0.0139 (4) | |
| H8A | 0.6464 | 0.3460 | −0.1001 | 0.017* | |
| H8B | 0.7949 | 0.4047 | −0.0751 | 0.017* | |
| C9 | 0.6657 (2) | 0.3992 (2) | 0.04119 (14) | 0.0158 (4) | |
| H9A | 0.6548 | 0.4985 | 0.0377 | 0.019* | |
| H9B | 0.5751 | 0.3592 | 0.0536 | 0.019* | |
| C10 | 0.6923 (2) | 0.3973 (2) | 0.20995 (13) | 0.0195 (4) | |
| H10A | 0.7533 | 0.3737 | 0.2620 | 0.029* | |
| H10B | 0.6076 | 0.3456 | 0.2154 | 0.029* | |
| H10C | 0.6718 | 0.4941 | 0.2124 | 0.029* | |
| C11 | 0.8874 (2) | 0.4429 (2) | 0.11176 (15) | 0.0200 (5) | |
| H11A | 0.9314 | 0.4230 | 0.0514 | 0.030* | |
| H11B | 0.9486 | 0.4176 | 0.1633 | 0.030* | |
| H11C | 0.8671 | 0.5396 | 0.1155 | 0.030* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cu | 0.01269 (12) | 0.01237 (12) | 0.01020 (12) | 0.00077 (10) | −0.00026 (10) | 0.00045 (10) |
| Cl1 | 0.0134 (2) | 0.0185 (3) | 0.0166 (2) | −0.0024 (2) | −0.00053 (18) | 0.0027 (2) |
| Cl2 | 0.0177 (2) | 0.0229 (3) | 0.0130 (2) | 0.0021 (2) | −0.00368 (19) | 0.0008 (2) |
| N1 | 0.0115 (8) | 0.0159 (9) | 0.0143 (8) | −0.0012 (7) | 0.0026 (7) | 0.0014 (7) |
| N2 | 0.0107 (9) | 0.0133 (8) | 0.0131 (8) | 0.0003 (6) | −0.0012 (7) | 0.0033 (7) |
| N3 | 0.0137 (8) | 0.0146 (8) | 0.0114 (8) | 0.0001 (6) | 0.0009 (6) | 0.0000 (7) |
| C1 | 0.0181 (10) | 0.0146 (10) | 0.0180 (11) | −0.0018 (8) | 0.0028 (9) | 0.0036 (8) |
| C2 | 0.0203 (11) | 0.0136 (11) | 0.0217 (12) | 0.0004 (8) | 0.0034 (9) | 0.0057 (9) |
| C3 | 0.0229 (11) | 0.0144 (10) | 0.0227 (11) | −0.0009 (9) | 0.0024 (9) | −0.0051 (9) |
| C4 | 0.0170 (10) | 0.0184 (11) | 0.0171 (11) | −0.0034 (8) | −0.0009 (9) | −0.0014 (9) |
| C5 | 0.0101 (10) | 0.0148 (10) | 0.0143 (10) | −0.0034 (8) | 0.0011 (8) | −0.0012 (8) |
| C6 | 0.0093 (9) | 0.0160 (9) | 0.0134 (9) | −0.0037 (7) | −0.0012 (9) | −0.0009 (8) |
| C7 | 0.0223 (11) | 0.0237 (11) | 0.0139 (10) | 0.0061 (10) | −0.0012 (8) | −0.0014 (9) |
| C8 | 0.0145 (10) | 0.0136 (10) | 0.0137 (10) | 0.0008 (9) | −0.0019 (7) | 0.0020 (8) |
| C9 | 0.0146 (10) | 0.0144 (10) | 0.0185 (10) | 0.0017 (8) | −0.0009 (8) | 0.0019 (9) |
| C10 | 0.0272 (11) | 0.0176 (10) | 0.0136 (10) | 0.0018 (10) | 0.0029 (10) | −0.0021 (8) |
| C11 | 0.0213 (11) | 0.0183 (11) | 0.0204 (12) | −0.0064 (8) | −0.0018 (9) | −0.0015 (10) |
Geometric parameters (Å, °)
| Cu—N2 | 1.9723 (16) | C4—C5 | 1.377 (3) |
| Cu—N1 | 2.0447 (17) | C4—H4 | 0.9500 |
| Cu—N3 | 2.0567 (16) | C5—C6 | 1.494 (3) |
| Cu—Cl2 | 2.2617 (5) | C6—C7 | 1.502 (3) |
| Cu—Cl1 | 2.4848 (5) | C7—H7A | 0.9800 |
| N1—C1 | 1.334 (3) | C7—H7B | 0.9800 |
| N1—C5 | 1.359 (2) | C7—H7C | 0.9800 |
| N2—C6 | 1.271 (2) | C8—C9 | 1.519 (3) |
| N2—C8 | 1.466 (3) | C8—H8A | 0.9900 |
| N3—C11 | 1.478 (2) | C8—H8B | 0.9900 |
| N3—C10 | 1.482 (2) | C9—H9A | 0.9900 |
| N3—C9 | 1.481 (2) | C9—H9B | 0.9900 |
| C1—C2 | 1.381 (3) | C10—H10A | 0.9800 |
| C1—H1 | 0.9500 | C10—H10B | 0.9800 |
| C2—C3 | 1.379 (3) | C10—H10C | 0.9800 |
| C2—H2 | 0.9500 | C11—H11A | 0.9800 |
| C3—C4 | 1.401 (3) | C11—H11B | 0.9800 |
| C3—H3 | 0.9500 | C11—H11C | 0.9800 |
| N2—Cu—N1 | 79.04 (7) | N1—C5—C6 | 113.54 (17) |
| N2—Cu—N3 | 82.74 (6) | C4—C5—C6 | 124.56 (17) |
| N1—Cu—N3 | 158.45 (6) | N2—C6—C5 | 114.06 (16) |
| N2—Cu—Cl2 | 154.98 (5) | N2—C6—C7 | 123.94 (18) |
| N1—Cu—Cl2 | 97.17 (5) | C5—C6—C7 | 121.90 (16) |
| N3—Cu—Cl2 | 94.45 (4) | C6—C7—H7A | 109.5 |
| N2—Cu—Cl1 | 95.91 (5) | C6—C7—H7B | 109.5 |
| N1—Cu—Cl1 | 96.59 (5) | H7A—C7—H7B | 109.5 |
| N3—Cu—Cl1 | 96.65 (4) | C6—C7—H7C | 109.5 |
| Cl2—Cu—Cl1 | 109.113 (18) | H7A—C7—H7C | 109.5 |
| C1—N1—C5 | 118.77 (18) | H7B—C7—H7C | 109.5 |
| C1—N1—Cu | 127.64 (14) | N2—C8—C9 | 105.76 (15) |
| C5—N1—Cu | 113.59 (13) | N2—C8—H8A | 110.6 |
| C6—N2—C8 | 124.33 (16) | C9—C8—H8A | 110.6 |
| C6—N2—Cu | 119.47 (13) | N2—C8—H8B | 110.6 |
| C8—N2—Cu | 116.17 (12) | C9—C8—H8B | 110.6 |
| C11—N3—C10 | 109.14 (15) | H8A—C8—H8B | 108.7 |
| C11—N3—C9 | 110.72 (15) | N3—C9—C8 | 111.17 (16) |
| C10—N3—C9 | 109.07 (15) | N3—C9—H9A | 109.4 |
| C11—N3—Cu | 109.33 (12) | C8—C9—H9A | 109.4 |
| C10—N3—Cu | 114.53 (12) | N3—C9—H9B | 109.4 |
| C9—N3—Cu | 103.96 (12) | C8—C9—H9B | 109.4 |
| N1—C1—C2 | 122.50 (19) | H9A—C9—H9B | 108.0 |
| N1—C1—H1 | 118.8 | N3—C10—H10A | 109.5 |
| C2—C1—H1 | 118.8 | N3—C10—H10B | 109.5 |
| C3—C2—C1 | 119.1 (2) | H10A—C10—H10B | 109.5 |
| C3—C2—H2 | 120.4 | N3—C10—H10C | 109.5 |
| C1—C2—H2 | 120.4 | H10A—C10—H10C | 109.5 |
| C2—C3—C4 | 118.9 (2) | H10B—C10—H10C | 109.5 |
| C2—C3—H3 | 120.6 | N3—C11—H11A | 109.5 |
| C4—C3—H3 | 120.6 | N3—C11—H11B | 109.5 |
| C5—C4—C3 | 118.86 (19) | H11A—C11—H11B | 109.5 |
| C5—C4—H4 | 120.6 | N3—C11—H11C | 109.5 |
| C3—C4—H4 | 120.6 | H11A—C11—H11C | 109.5 |
| N1—C5—C4 | 121.81 (19) | H11B—C11—H11C | 109.5 |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: PV2266).
References
- Addison, A. W., Rao, T. N., Reedijk, J., Rijn, V. J. & Verschoor, G. C. (1984). J. Chem. Soc. Dalton Trans pp. 1349–1356.
- Barbour, L. J. (2001). J. Supramol. Chem 1, 189–191.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wang, Q., Bi, C.-F., Wang, D.-Q. & Fan, Y.-H. (2009). Acta Cryst. E65, m439. [DOI] [PMC free article] [PubMed]
- Westrip, S. P. (2010). publCIF In preparation.
- Yuan, W.-B. & Zhang, Q. (2005). Acta Cryst. E61, m1883–m1884.
- Zhang, X.-Q., Li, C.-Y., Bian, H.-D., Yu, Q. & Liang, H. (2009). Acta Cryst. E65, m1610. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810011712/pv2266sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810011712/pv2266Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



