Abstract
In the title compound, C19H24O4, the six-membered lactone ring adopts an envelope conformation with the tert-butoxycarbonyl and isopropyl substituents in axial positions, and the phenyl group in an equatorial position. In the crystal structure, weak intermolecular C—H⋯O hydrogen bonds link the molecules into centrosymmetric dimers.
Related literature
For the applications and synthesis of endocyclic enol lactones, see: Davies & Jin (2004 ▶); Evans et al. (2005 ▶); Krafft & Katzenellenbogen (1981 ▶); Li et al. (2007 ▶); Zeni et al. (2004 ▶); Zhao et al. (1997 ▶); Jimenez-Tenorio et al. (2001 ▶). For the synthesis, see: Li et al. (2009 ▶).
Experimental
Crystal data
C19H24O4
M r = 316.38
Triclinic,

a = 8.6163 (9) Å
b = 10.888 (1) Å
c = 11.261 (1) Å
α = 68.393 (2)°
β = 79.118 (2)°
γ = 67.998 (2)°
V = 909.09 (15) Å3
Z = 2
Mo Kα radiation
μ = 0.08 mm−1
T = 293 K
0.48 × 0.46 × 0.42 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2004 ▶) T min = 0.760, T max = 1.000
4986 measured reflections
3510 independent reflections
2759 reflections with I > 2σ(I)
R int = 0.051
Refinement
R[F 2 > 2σ(F 2)] = 0.055
wR(F 2) = 0.158
S = 1.04
3510 reflections
213 parameters
H-atom parameters constrained
Δρmax = 0.22 e Å−3
Δρmin = −0.21 e Å−3
Data collection: SMART (Bruker, 2001 ▶); cell refinement: SAINT (Bruker, 2001 ▶); data reduction: SHELXTL (Sheldrick, 2008 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶) and DIAMOND (Brandenburg, 1998 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053681001367X/lx2139sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053681001367X/lx2139Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C2—H2⋯O1i | 0.98 | 2.44 | 3.407 (2) | 170 |
Symmetry code: (i)  .
.
Acknowledgments
The authors thank the EPSRC for financial support.
supplementary crystallographic information
Comment
Endocyclic enol lactones are important structural elements of biologically active natural products (Zhao et al., 1997) and useful synthetic intermediates for organic synthesis (Evans et al., 2005, Davies et al., 2004). The cyclization of alkynoic acids under acidic conditions (Krafft et al., 1981) , employing transition-metal complexes as catalysts (Zeni et al., 2004, Valerga et al., 2001),and the carbonylation coupling of alkynes and 1,3-dicarbonyl compounds are main synthetic pathways for the preparation of Enol lactones (Li et al., 2007)
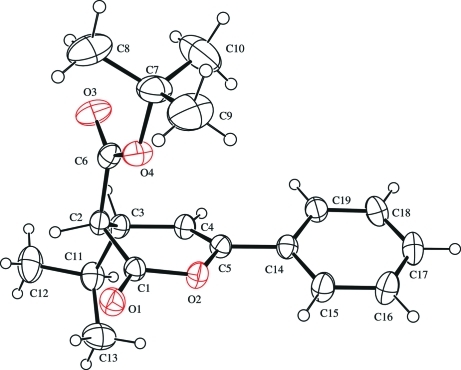
In the title compound as shown in Fig. 1, the six-membered lactone ring adopts an envelope conformation with the tert-butoxycarbonyl, isopropyl and phenyl groups attached to it. The tert-butoxycarbonyl and isopropyl groups occupy axial positions, and the phenyl group occupies equatorial position. The crystal packing (Fig. 2) is stabilized by weak intermolecular C—H···O hydrogen bonds between the pyran H atom and the oxygen of the C═O unit in pyran ring, with a C2—H2···O1i (Table 1).
Experimental
The title compound was obtained as a by-product in the copper-catalyzed tandem conjugate addition–cyclization–hydrolysis–decarboxylation reactions of alkynes and 5-alkylidene-Meldrum's acids (Jiao et al., 2009) acids as follows: To a mixture of CuBr (20 mg, 0.1 mmol), 1-ethynylbenzene (102 mg, 1 mmol) in H2O : t-BuOH = 10 : 1 (3 ml) was added and 2,2-dimethyl-5-(2-methylpropylidene)-1,3-dioxane-4,6-dione (99 mg, 0.5 mmol) at room temperature. The resulting mixture was refluxed for 10 h monitored by TLC. After evaporation, the residue was carefully purified by flash chromatography on silica gel. The title compound was obtained as a by-product (25% yield), which was crystallized from n-hexane-ethyl acetate.
Refinement
All H atoms were positioned geometrically and refined using a riding model, with C—H = 0.93 Å for aryl, 0.98 Å for methyne and 0.96 Å for methyl H atoms. Uiso(H) = 1.2Ueq(C) for aryl and methyne H atoms, and 1.5Ueq(C) for methyl H atoms.
Figures
Fig. 1.
The molecular structure of the title compound with the atom numbering scheme. Displacement ellipsoids are drawn at the 30% probability level. H atoms are presented as a small spheres of arbitrary radius.
Fig. 2.
C—H···O interactions (dotted lines) in the crystal structure of the title compound. [Symmetry codes: (i) - x, - y + 2, - z + 1.]
Crystal data
| C19H24O4 | Z = 2 |
| Mr = 316.38 | F(000) = 340 |
| Triclinic, P1 | Dx = 1.156 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.6163 (9) Å | Cell parameters from 2216 reflections |
| b = 10.888 (1) Å | θ = 4.8–55.3° |
| c = 11.261 (1) Å | µ = 0.08 mm−1 |
| α = 68.393 (2)° | T = 293 K |
| β = 79.118 (2)° | Prismatic, colorless |
| γ = 67.998 (2)° | 0.48 × 0.46 × 0.42 mm |
| V = 909.09 (15) Å3 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 3510 independent reflections |
| Radiation source: fine-focus sealed tube | 2759 reflections with I > 2σ(I) |
| graphite | Rint = 0.051 |
| Detector resolution: 10.0 pixels mm-1 | θmax = 26.0°, θmin = 2.0° |
| φ and ω scans | h = −10→10 |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2004) | k = −10→13 |
| Tmin = 0.760, Tmax = 1.000 | l = −13→12 |
| 4986 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.055 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.158 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0925P)2 + 0.0377P] where P = (Fo2 + 2Fc2)/3 |
| 3510 reflections | (Δ/σ)max < 0.001 |
| 213 parameters | Δρmax = 0.22 e Å−3 |
| 0 restraints | Δρmin = −0.21 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | −0.04144 (14) | 0.87317 (11) | 0.66284 (11) | 0.0535 (3) | |
| O2 | 0.07228 (13) | 0.64576 (11) | 0.71597 (10) | 0.0461 (3) | |
| O3 | 0.48770 (16) | 0.75686 (16) | 0.51105 (14) | 0.0721 (4) | |
| O4 | 0.33639 (14) | 0.77581 (13) | 0.69310 (12) | 0.0554 (3) | |
| C1 | 0.06511 (19) | 0.77682 (15) | 0.63769 (14) | 0.0408 (4) | |
| C2 | 0.19583 (19) | 0.78720 (16) | 0.52821 (14) | 0.0425 (4) | |
| H2 | 0.1560 | 0.8805 | 0.4644 | 0.051* | |
| C3 | 0.2282 (2) | 0.67697 (16) | 0.46303 (15) | 0.0441 (4) | |
| H3 | 0.3364 | 0.6694 | 0.4143 | 0.053* | |
| C4 | 0.2483 (2) | 0.53947 (16) | 0.56580 (15) | 0.0450 (4) | |
| H4 | 0.3113 | 0.4581 | 0.5464 | 0.054* | |
| C5 | 0.18081 (19) | 0.52752 (15) | 0.68321 (15) | 0.0412 (4) | |
| C6 | 0.3589 (2) | 0.77079 (16) | 0.57572 (16) | 0.0470 (4) | |
| C7 | 0.4744 (3) | 0.7625 (3) | 0.7630 (2) | 0.0739 (6) | |
| C8 | 0.5394 (3) | 0.8833 (3) | 0.6928 (3) | 0.0982 (9) | |
| H8A | 0.5896 | 0.8764 | 0.6108 | 0.147* | |
| H8B | 0.6216 | 0.8799 | 0.7418 | 0.147* | |
| H8C | 0.4481 | 0.9703 | 0.6812 | 0.147* | |
| C9 | 0.3851 (4) | 0.7727 (4) | 0.8901 (2) | 0.1092 (10) | |
| H9A | 0.2958 | 0.8610 | 0.8764 | 0.164* | |
| H9B | 0.4630 | 0.7658 | 0.9448 | 0.164* | |
| H9C | 0.3398 | 0.6978 | 0.9296 | 0.164* | |
| C10 | 0.6078 (4) | 0.6211 (3) | 0.7773 (4) | 0.1282 (12) | |
| H10A | 0.5556 | 0.5509 | 0.8000 | 0.192* | |
| H10B | 0.6796 | 0.5992 | 0.8432 | 0.192* | |
| H10C | 0.6730 | 0.6233 | 0.6979 | 0.192* | |
| C11 | 0.0973 (2) | 0.7132 (2) | 0.36787 (17) | 0.0589 (5) | |
| H11 | 0.1194 | 0.6283 | 0.3467 | 0.071* | |
| C12 | 0.1187 (4) | 0.8256 (3) | 0.2436 (2) | 0.0887 (8) | |
| H12A | 0.0410 | 0.8410 | 0.1845 | 0.133* | |
| H12B | 0.2312 | 0.7957 | 0.2076 | 0.133* | |
| H12C | 0.0976 | 0.9111 | 0.2601 | 0.133* | |
| C13 | −0.0809 (3) | 0.7518 (3) | 0.4235 (2) | 0.0799 (6) | |
| H13A | −0.1104 | 0.8386 | 0.4398 | 0.120* | |
| H13B | −0.0909 | 0.6793 | 0.5021 | 0.120* | |
| H13C | −0.1549 | 0.7622 | 0.3639 | 0.120* | |
| C14 | 0.1969 (2) | 0.39882 (16) | 0.79280 (14) | 0.0423 (4) | |
| C15 | 0.0804 (2) | 0.39429 (19) | 0.89627 (17) | 0.0575 (5) | |
| H15 | −0.0092 | 0.4750 | 0.8975 | 0.069* | |
| C16 | 0.0954 (3) | 0.2717 (2) | 0.99749 (19) | 0.0686 (6) | |
| H16 | 0.0167 | 0.2701 | 1.0665 | 0.082* | |
| C17 | 0.2272 (3) | 0.1519 (2) | 0.99593 (19) | 0.0678 (6) | |
| H17 | 0.2381 | 0.0692 | 1.0643 | 0.081* | |
| C18 | 0.3423 (3) | 0.15430 (19) | 0.89392 (19) | 0.0637 (5) | |
| H18 | 0.4300 | 0.0726 | 0.8925 | 0.076* | |
| C19 | 0.3295 (2) | 0.27597 (18) | 0.79372 (17) | 0.0531 (4) | |
| H19 | 0.4099 | 0.2766 | 0.7258 | 0.064* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0522 (7) | 0.0401 (7) | 0.0579 (7) | −0.0079 (5) | 0.0052 (5) | −0.0165 (5) |
| O2 | 0.0517 (6) | 0.0357 (6) | 0.0438 (6) | −0.0129 (5) | 0.0106 (5) | −0.0134 (5) |
| O3 | 0.0540 (8) | 0.0982 (11) | 0.0823 (10) | −0.0365 (7) | 0.0214 (7) | −0.0508 (9) |
| O4 | 0.0482 (7) | 0.0722 (9) | 0.0523 (7) | −0.0250 (6) | 0.0004 (5) | −0.0241 (6) |
| C1 | 0.0431 (8) | 0.0347 (8) | 0.0416 (8) | −0.0115 (7) | −0.0013 (6) | −0.0111 (6) |
| C2 | 0.0481 (9) | 0.0342 (8) | 0.0390 (8) | −0.0142 (7) | 0.0015 (7) | −0.0068 (6) |
| C3 | 0.0499 (9) | 0.0437 (9) | 0.0368 (8) | −0.0174 (7) | 0.0050 (7) | −0.0129 (7) |
| C4 | 0.0528 (9) | 0.0372 (8) | 0.0431 (9) | −0.0126 (7) | 0.0014 (7) | −0.0156 (7) |
| C5 | 0.0432 (8) | 0.0351 (8) | 0.0437 (9) | −0.0113 (6) | 0.0007 (6) | −0.0145 (7) |
| C6 | 0.0498 (9) | 0.0401 (9) | 0.0508 (10) | −0.0178 (7) | 0.0057 (7) | −0.0159 (7) |
| C7 | 0.0590 (12) | 0.1008 (17) | 0.0697 (13) | −0.0321 (12) | −0.0105 (10) | −0.0277 (12) |
| C8 | 0.0908 (18) | 0.139 (2) | 0.108 (2) | −0.0700 (17) | 0.0064 (15) | −0.0615 (18) |
| C9 | 0.110 (2) | 0.184 (3) | 0.0641 (15) | −0.076 (2) | −0.0044 (14) | −0.0459 (18) |
| C10 | 0.094 (2) | 0.116 (3) | 0.146 (3) | 0.0030 (18) | −0.057 (2) | −0.026 (2) |
| C11 | 0.0786 (13) | 0.0560 (11) | 0.0459 (10) | −0.0254 (9) | −0.0091 (9) | −0.0156 (8) |
| C12 | 0.133 (2) | 0.0810 (16) | 0.0502 (12) | −0.0445 (15) | −0.0187 (13) | −0.0041 (11) |
| C13 | 0.0670 (13) | 0.1032 (18) | 0.0811 (15) | −0.0288 (12) | −0.0189 (11) | −0.0356 (13) |
| C14 | 0.0498 (9) | 0.0409 (9) | 0.0378 (8) | −0.0173 (7) | −0.0034 (7) | −0.0123 (7) |
| C15 | 0.0672 (11) | 0.0478 (10) | 0.0471 (10) | −0.0165 (8) | 0.0072 (8) | −0.0120 (8) |
| C16 | 0.0870 (14) | 0.0654 (13) | 0.0459 (10) | −0.0332 (11) | 0.0097 (10) | −0.0086 (9) |
| C17 | 0.0943 (15) | 0.0496 (11) | 0.0497 (11) | −0.0275 (11) | −0.0122 (10) | 0.0020 (8) |
| C18 | 0.0752 (13) | 0.0427 (10) | 0.0580 (12) | −0.0073 (9) | −0.0148 (10) | −0.0071 (8) |
| C19 | 0.0539 (10) | 0.0487 (10) | 0.0482 (10) | −0.0123 (8) | −0.0030 (8) | −0.0112 (8) |
Geometric parameters (Å, °)
| O1—C1 | 1.1927 (18) | C9—H9C | 0.9600 |
| O2—C1 | 1.3584 (18) | C10—H10A | 0.9600 |
| O2—C5 | 1.4091 (17) | C10—H10B | 0.9600 |
| O3—C6 | 1.198 (2) | C10—H10C | 0.9600 |
| O4—C6 | 1.318 (2) | C11—C13 | 1.508 (3) |
| O4—C7 | 1.482 (2) | C11—C12 | 1.517 (3) |
| C1—C2 | 1.507 (2) | C11—H11 | 0.9800 |
| C2—C6 | 1.523 (2) | C12—H12A | 0.9600 |
| C2—C3 | 1.542 (2) | C12—H12B | 0.9600 |
| C2—H2 | 0.9800 | C12—H12C | 0.9600 |
| C3—C4 | 1.489 (2) | C13—H13A | 0.9600 |
| C3—C11 | 1.545 (2) | C13—H13B | 0.9600 |
| C3—H3 | 0.9800 | C13—H13C | 0.9600 |
| C4—C5 | 1.320 (2) | C14—C15 | 1.386 (2) |
| C4—H4 | 0.9300 | C14—C19 | 1.394 (2) |
| C5—C14 | 1.467 (2) | C15—C16 | 1.380 (3) |
| C7—C10 | 1.510 (4) | C15—H15 | 0.9300 |
| C7—C8 | 1.512 (4) | C16—C17 | 1.375 (3) |
| C7—C9 | 1.513 (3) | C16—H16 | 0.9300 |
| C8—H8A | 0.9600 | C17—C18 | 1.368 (3) |
| C8—H8B | 0.9600 | C17—H17 | 0.9300 |
| C8—H8C | 0.9600 | C18—C19 | 1.370 (2) |
| C9—H9A | 0.9600 | C18—H18 | 0.9300 |
| C9—H9B | 0.9600 | C19—H19 | 0.9300 |
| C1—O2—C5 | 120.35 (11) | H9B—C9—H9C | 109.5 |
| C6—O4—C7 | 122.54 (14) | C7—C10—H10A | 109.5 |
| O1—C1—O2 | 117.45 (14) | C7—C10—H10B | 109.5 |
| O1—C1—C2 | 125.81 (14) | H10A—C10—H10B | 109.5 |
| O2—C1—C2 | 116.73 (12) | C7—C10—H10C | 109.5 |
| C1—C2—C6 | 109.81 (13) | H10A—C10—H10C | 109.5 |
| C1—C2—C3 | 112.32 (12) | H10B—C10—H10C | 109.5 |
| C6—C2—C3 | 109.70 (13) | C13—C11—C12 | 111.18 (19) |
| C1—C2—H2 | 108.3 | C13—C11—C3 | 113.38 (15) |
| C6—C2—H2 | 108.3 | C12—C11—C3 | 111.50 (16) |
| C3—C2—H2 | 108.3 | C13—C11—H11 | 106.8 |
| C4—C3—C2 | 107.53 (12) | C12—C11—H11 | 106.8 |
| C4—C3—C11 | 112.76 (13) | C3—C11—H11 | 106.8 |
| C2—C3—C11 | 115.19 (14) | C11—C12—H12A | 109.5 |
| C4—C3—H3 | 107.0 | C11—C12—H12B | 109.5 |
| C2—C3—H3 | 107.0 | H12A—C12—H12B | 109.5 |
| C11—C3—H3 | 107.0 | C11—C12—H12C | 109.5 |
| C5—C4—C3 | 123.13 (14) | H12A—C12—H12C | 109.5 |
| C5—C4—H4 | 118.4 | H12B—C12—H12C | 109.5 |
| C3—C4—H4 | 118.4 | C11—C13—H13A | 109.5 |
| C4—C5—O2 | 121.22 (13) | C11—C13—H13B | 109.5 |
| C4—C5—C14 | 127.91 (14) | H13A—C13—H13B | 109.5 |
| O2—C5—C14 | 110.81 (12) | C11—C13—H13C | 109.5 |
| O3—C6—O4 | 126.45 (17) | H13A—C13—H13C | 109.5 |
| O3—C6—C2 | 122.48 (16) | H13B—C13—H13C | 109.5 |
| O4—C6—C2 | 111.06 (13) | C15—C14—C19 | 118.15 (15) |
| O4—C7—C10 | 108.8 (2) | C15—C14—C5 | 121.59 (15) |
| O4—C7—C8 | 109.19 (19) | C19—C14—C5 | 120.24 (14) |
| C10—C7—C8 | 113.0 (2) | C16—C15—C14 | 120.91 (17) |
| O4—C7—C9 | 101.57 (16) | C16—C15—H15 | 119.5 |
| C10—C7—C9 | 111.9 (2) | C14—C15—H15 | 119.5 |
| C8—C7—C9 | 111.7 (2) | C17—C16—C15 | 119.78 (19) |
| C7—C8—H8A | 109.5 | C17—C16—H16 | 120.1 |
| C7—C8—H8B | 109.5 | C15—C16—H16 | 120.1 |
| H8A—C8—H8B | 109.5 | C18—C17—C16 | 120.00 (17) |
| C7—C8—H8C | 109.5 | C18—C17—H17 | 120.0 |
| H8A—C8—H8C | 109.5 | C16—C17—H17 | 120.0 |
| H8B—C8—H8C | 109.5 | C17—C18—C19 | 120.62 (18) |
| C7—C9—H9A | 109.5 | C17—C18—H18 | 119.7 |
| C7—C9—H9B | 109.5 | C19—C18—H18 | 119.7 |
| H9A—C9—H9B | 109.5 | C18—C19—C14 | 120.52 (17) |
| C7—C9—H9C | 109.5 | C18—C19—H19 | 119.7 |
| H9A—C9—H9C | 109.5 | C14—C19—H19 | 119.7 |
| C5—O2—C1—O1 | 172.04 (14) | C3—C2—C6—O4 | −134.60 (13) |
| C5—O2—C1—C2 | −9.4 (2) | C6—O4—C7—C10 | −61.0 (3) |
| O1—C1—C2—C6 | 97.55 (18) | C6—O4—C7—C8 | 62.8 (2) |
| O2—C1—C2—C6 | −80.92 (16) | C6—O4—C7—C9 | −179.16 (19) |
| O1—C1—C2—C3 | −140.10 (16) | C4—C3—C11—C13 | 72.2 (2) |
| O2—C1—C2—C3 | 41.43 (19) | C2—C3—C11—C13 | −51.7 (2) |
| C1—C2—C3—C4 | −47.02 (17) | C4—C3—C11—C12 | −161.41 (17) |
| C6—C2—C3—C4 | 75.39 (15) | C2—C3—C11—C12 | 74.6 (2) |
| C1—C2—C3—C11 | 79.62 (17) | C4—C5—C14—C15 | −158.33 (18) |
| C6—C2—C3—C11 | −157.96 (13) | O2—C5—C14—C15 | 18.9 (2) |
| C2—C3—C4—C5 | 25.9 (2) | C4—C5—C14—C19 | 20.1 (3) |
| C11—C3—C4—C5 | −102.21 (19) | O2—C5—C14—C19 | −162.68 (14) |
| C3—C4—C5—O2 | 5.5 (2) | C19—C14—C15—C16 | 0.2 (3) |
| C3—C4—C5—C14 | −177.58 (15) | C5—C14—C15—C16 | 178.69 (17) |
| C1—O2—C5—C4 | −15.6 (2) | C14—C15—C16—C17 | −0.3 (3) |
| C1—O2—C5—C14 | 166.99 (13) | C15—C16—C17—C18 | −0.4 (3) |
| C7—O4—C6—O3 | −0.8 (3) | C16—C17—C18—C19 | 1.2 (3) |
| C7—O4—C6—C2 | 179.96 (15) | C17—C18—C19—C14 | −1.3 (3) |
| C1—C2—C6—O3 | 170.02 (16) | C15—C14—C19—C18 | 0.6 (3) |
| C3—C2—C6—O3 | 46.1 (2) | C5—C14—C19—C18 | −177.89 (16) |
| C1—C2—C6—O4 | −10.70 (17) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C2—H2···O1i | 0.98 | 2.44 | 3.407 (2) | 170 |
Symmetry codes: (i) −x, −y+2, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: LX2139).
References
- Brandenburg, K. (1998). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Bruker (2001). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Davies, H. M. L. & Jin, Q. H. (2004). Proc. Natl. Acad. Sci.101, 5472–5475. [DOI] [PMC free article] [PubMed]
- Evans, D. A., Thomson, R. J. & Franco, F. (2005). J. Am. Chem. Soc.127, 10816–10817. [DOI] [PubMed]
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Jimenez-Tenorio, M., Puerta, M. C., Valerga, P., Moreno-Dorado, F. J., Guerra, F. M. & Massanet, G. M. (2001). Chem. Commun. pp. 2324–2325. [DOI] [PubMed]
- Krafft, G. A. & Katzenellenbogen, J. A. (1981). J. Am. Chem. Soc.103, 5459–5466.
- Li, S., Jia, W. & Jiao, N. (2009). Adv. Synth. Catal.351, 569–575.
- Li, Y., Yu, Z. & Alper, H. (2007). Org. Lett.9, 1647–1649. [DOI] [PubMed]
- Sheldrick, G. M. (2004). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Zeni, G. & Larock, R. C. (2004). Chem. Rev.104, 2285–2309. [DOI] [PubMed]
- Zhao, H., Neamati, N., Hong, H., Mazumder, A., Wang, S., Sunder, S., Milne, G. W. A., Pommier, Y. & Burke, T. R. Jr (1997). J. Med. Chem.40, 242–249. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053681001367X/lx2139sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053681001367X/lx2139Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report