Abstract
The asymmetric unit of the title compound, C11H12N2O4S2, contains two independent molecules with similar dihedral angles of 76.7 (1) and 77.3 (1)° between the mean planes of the five- and six-membered rings. In both molecules, the amino groups are involved in intramolecular N—H⋯O hydrogen bonds. In the crystal structure, weak intermolecular C—H⋯O hydrogen bonds link molecules into ribbons extended along the a axis.
Related literature
For a related structure, see: Gowda et al. (2010 ▶). For details of the synthesis, see: Chen & Shen (2008 ▶). For the biological activity of related compounds, see: Fujimoto & Shimizu (1978 ▶); Liu et al. (2007 ▶, 2009 ▶).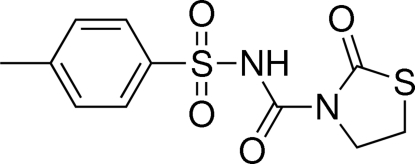
Experimental
Crystal data
C11H12N2O4S2
M r = 300.35
Triclinic,

a = 9.5560 (4) Å
b = 9.6722 (4) Å
c = 14.6352 (6) Å
α = 88.7232 (10)°
β = 86.3267 (11)°
γ = 76.5399 (11)°
V = 1312.80 (9) Å3
Z = 4
Mo Kα radiation
μ = 0.42 mm−1
T = 296 K
0.39 × 0.36 × 0.14 mm
Data collection
Rigaku R-AXIS RAPID diffractometer
Absorption correction: multi-scan (ABSCOR; Higashi, 1995 ▶) T min = 0.825, T max = 0.943
13020 measured reflections
5941 independent reflections
4475 reflections with F 2 > 2σ(F 2)
R int = 0.019
Refinement
R[F 2 > 2σ(F 2)] = 0.048
wR(F 2) = 0.153
S = 1.00
5941 reflections
345 parameters
H-atom parameters constrained
Δρmax = 0.52 e Å−3
Δρmin = −0.38 e Å−3
Data collection: PROCESS-AUTO (Rigaku, 2006 ▶); cell refinement: PROCESS-AUTO; data reduction: CrystalStructure (Rigaku, 2007 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810014467/cv2707sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810014467/cv2707Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N12—H12⋯O11 | 0.86 | 2.00 | 2.665 (3) | 134 |
| N32—H32⋯O31 | 0.86 | 1.91 | 2.608 (3) | 137 |
| C33—H332⋯O11 | 0.97 | 2.40 | 3.352 (3) | 166 |
| C11—H112⋯O33i | 0.96 | 2.55 | 3.500 (4) | 170 |
| C32—H321⋯O12ii | 0.97 | 2.60 | 3.384 (3) | 138 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
This work was funded by the National Natural Science Foundation (grant No. 30900959), the Natural Science Foundation of Zhejiang Province (grant No. Y3080096) and the Zhejiang University of Technology Research Fund (grant No. X 1017104).
supplementary crystallographic information
Comment
2-Thiazolidione derivatives are known as fungicides, insecticides and plant growth regulators (Chen et al. 2008; Fujimoto et al. 1978). Meanwhile, many pesticides contain amide fragments (Liu et al. 2007; Liu et al. 2009) . Herewith we present the title compound (I) - a new thiazolidione derivative synthesized by the reaction of 2-thiazolidione and 4-methyl-benzenesulfonylisocyanate.
In (I) (Fig. 1), the carboxamide moiety is nearly coplanar with the thiazole ring [dihedral angle 7.2 (2)°]. The C4-O12 bond length of 1.202 (3) Å is normal for C=O double bond. The conformation of C—SO2—NH—C(O) fragment is similar to that observed in N-Benzoyl-2-chlorobenzenesulfonamide (Gowda et al., 2010). Amino groups in two independent molecules are involved in intramolecular N—H···O hydrogen bonds (Table 1). In the crystal structure, weak intermolecular C—H···O hydrogen bonds (Table 1) link molecules into ribbons extended along the axis a.
Experimental
The title compound was synthesized according to Chen et al. (2008). 2-Thiazolidiones (1.03 g, 0.01 mol) and 4-methyl- benzenesulfonylisocyanate (1.97 g,0.01 mol) were dissolved in anhydrous acetone(15 ml)with stirring. The mixture was then stirred at room temperature for 15 h. The solution was evaporated in a rotary evaporator at 30 degree under reduced pressure till 1.5 g of solide residue were obtained. This was washed three times with water and dried. Colourless single crystals suitable for crystallographic analysis were obtained by slow evaporation of an acetone-petroleum ether (1:20 v/v) solution.
Refinement
All H atoms were geometrically positioned (C—H 0.93-0.97 Å; N—H 0.86 Å), and refined as riding, with Uiso(H) = 1.2 Ueq(C, N).
Figures
Fig. 1.
The molecular structure of (I), showing displacement ellipsoids drawn at the 50% probability level.
Crystal data
| C11H12N2O4S2 | Z = 4 |
| Mr = 300.35 | F(000) = 624.00 |
| Triclinic, P1 | Dx = 1.519 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71075 Å |
| a = 9.5560 (4) Å | Cell parameters from 10300 reflections |
| b = 9.6722 (4) Å | θ = 3.1–27.4° |
| c = 14.6352 (6) Å | µ = 0.42 mm−1 |
| α = 88.7232 (10)° | T = 296 K |
| β = 86.3267 (11)° | Platelet, colourless |
| γ = 76.5399 (11)° | 0.39 × 0.36 × 0.14 mm |
| V = 1312.80 (9) Å3 |
Data collection
| Rigaku R-AXIS RAPID diffractometer | 4475 reflections with F2 > 2σ(F2) |
| Detector resolution: 10.00 pixels mm-1 | Rint = 0.019 |
| ω scans | θmax = 27.4° |
| Absorption correction: multi-scan (ABSCOR; Higashi, 1995) | h = −12→12 |
| Tmin = 0.825, Tmax = 0.943 | k = −10→12 |
| 13020 measured reflections | l = −18→18 |
| 5941 independent reflections |
Refinement
| Refinement on F2 | w = 1/[σ2(Fo2) + (0.0833P)2 + 0.8006P] where P = (Fo2 + 2Fc2)/3 |
| R[F2 > 2σ(F2)] = 0.048 | (Δ/σ)max = 0.001 |
| wR(F2) = 0.153 | Δρmax = 0.52 e Å−3 |
| S = 1.00 | Δρmin = −0.38 e Å−3 |
| 5941 reflections | Extinction correction: SHELXL97 (Sheldrick, 2008) |
| 345 parameters | Extinction coefficient: 0.0029 (10) |
| H-atom parameters constrained |
Special details
| Geometry. ENTER SPECIAL DETAILS OF THE MOLECULAR GEOMETRY |
| Refinement. Refinement using reflections with F2 > 2.0 σ(F2). The weighted R-factor(wR), goodness of fit (S) and R-factor (gt) are based on F, with F set to zero for negative F. The threshold expression of F2 > 2.0 σ(F2) is used only for calculating R-factor (gt). |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S11 | 0.70694 (8) | 0.42660 (8) | 0.60372 (5) | 0.0526 (2) | |
| S12 | 0.33746 (6) | 0.09549 (6) | 0.40275 (4) | 0.03965 (16) | |
| S31 | 1.11065 (8) | −0.10590 (9) | 0.27249 (5) | 0.0583 (2) | |
| S32 | 0.72340 (8) | 0.38204 (6) | 0.06460 (5) | 0.04988 (18) | |
| O11 | 0.6604 (2) | 0.3008 (2) | 0.45563 (12) | 0.0524 (4) | |
| O12 | 0.2711 (2) | 0.2641 (2) | 0.57769 (12) | 0.0538 (4) | |
| O13 | 0.2613 (2) | 0.0224 (2) | 0.46669 (12) | 0.0489 (4) | |
| O14 | 0.4392 (2) | 0.0178 (2) | 0.33572 (12) | 0.0505 (4) | |
| O31 | 1.0687 (2) | 0.0667 (2) | 0.13083 (14) | 0.0704 (6) | |
| O32 | 0.6590 (2) | 0.2350 (2) | 0.24079 (13) | 0.0598 (5) | |
| O33 | 0.6406 (2) | 0.4885 (2) | 0.12455 (14) | 0.0596 (5) | |
| O34 | 0.8234 (2) | 0.4156 (2) | −0.00386 (16) | 0.0681 (6) | |
| N11 | 0.4892 (2) | 0.3182 (2) | 0.57686 (12) | 0.0380 (4) | |
| N12 | 0.4356 (2) | 0.1810 (2) | 0.45973 (13) | 0.0416 (4) | |
| N31 | 0.8833 (2) | 0.0924 (2) | 0.24434 (13) | 0.0440 (4) | |
| N32 | 0.8270 (2) | 0.2587 (2) | 0.12745 (16) | 0.0531 (5) | |
| C1 | 0.6163 (2) | 0.3371 (2) | 0.53329 (17) | 0.0390 (5) | |
| C2 | 0.5766 (3) | 0.4345 (4) | 0.6988 (2) | 0.0638 (8) | |
| C3 | 0.4517 (3) | 0.3829 (3) | 0.66798 (18) | 0.0528 (6) | |
| C4 | 0.3885 (2) | 0.2544 (2) | 0.54018 (17) | 0.0393 (5) | |
| C5 | 0.2141 (2) | 0.2307 (2) | 0.34841 (16) | 0.0382 (5) | |
| C6 | 0.0829 (2) | 0.2946 (3) | 0.39243 (18) | 0.0484 (6) | |
| C7 | −0.0075 (2) | 0.4065 (3) | 0.3507 (2) | 0.0514 (6) | |
| C8 | 0.0293 (2) | 0.4558 (2) | 0.26469 (18) | 0.0430 (5) | |
| C9 | 0.1602 (2) | 0.3878 (2) | 0.22135 (18) | 0.0492 (6) | |
| C10 | 0.2527 (2) | 0.2758 (2) | 0.26209 (17) | 0.0462 (5) | |
| C11 | −0.0668 (3) | 0.5805 (3) | 0.2195 (2) | 0.0579 (7) | |
| C31 | 1.0186 (2) | 0.0317 (2) | 0.20360 (18) | 0.0457 (5) | |
| C32 | 0.9758 (3) | −0.0719 (3) | 0.36468 (19) | 0.0549 (6) | |
| C33 | 0.8416 (3) | 0.0248 (3) | 0.32863 (18) | 0.0552 (7) | |
| C34 | 0.7786 (2) | 0.2013 (2) | 0.20679 (17) | 0.0431 (5) | |
| C35 | 0.6085 (2) | 0.2939 (2) | 0.01393 (17) | 0.0454 (5) | |
| C36 | 0.4716 (3) | 0.3001 (3) | 0.05305 (19) | 0.0519 (6) | |
| C37 | 0.3815 (3) | 0.2319 (3) | 0.0121 (2) | 0.0613 (7) | |
| C38 | 0.4260 (3) | 0.1557 (2) | −0.0680 (2) | 0.0594 (7) | |
| C39 | 0.5622 (4) | 0.1492 (3) | −0.1052 (2) | 0.0691 (9) | |
| C40 | 0.6540 (3) | 0.2172 (3) | −0.0651 (2) | 0.0640 (8) | |
| C41 | 0.3275 (4) | 0.0852 (3) | −0.1184 (3) | 0.0843 (11) | |
| H6 | 0.0562 | 0.2622 | 0.4497 | 0.058* | |
| H7 | −0.0950 | 0.4499 | 0.3806 | 0.062* | |
| H9 | 0.1861 | 0.4187 | 0.1634 | 0.059* | |
| H10 | 0.3397 | 0.2313 | 0.2320 | 0.055* | |
| H12 | 0.5215 | 0.1796 | 0.4378 | 0.050* | |
| H32 | 0.9161 | 0.2290 | 0.1092 | 0.064* | |
| H36 | 0.4407 | 0.3504 | 0.1069 | 0.062* | |
| H37 | 0.2893 | 0.2368 | 0.0385 | 0.074* | |
| H39 | 0.5935 | 0.0978 | −0.1586 | 0.083* | |
| H40 | 0.7464 | 0.2113 | −0.0913 | 0.077* | |
| H111 | −0.0947 | 0.6578 | 0.2619 | 0.069* | |
| H112 | −0.1513 | 0.5536 | 0.2011 | 0.069* | |
| H113 | −0.0158 | 0.6098 | 0.1667 | 0.069* | |
| H201 | 0.6199 | 0.3745 | 0.7483 | 0.077* | |
| H202 | 0.5440 | 0.5317 | 0.7198 | 0.077* | |
| H301 | 0.4291 | 0.3126 | 0.7112 | 0.063* | |
| H302 | 0.3687 | 0.4622 | 0.6647 | 0.063* | |
| H321 | 0.9545 | −0.1605 | 0.3867 | 0.066* | |
| H322 | 1.0094 | −0.0257 | 0.4143 | 0.066* | |
| H331 | 0.7725 | −0.0307 | 0.3161 | 0.066* | |
| H332 | 0.7986 | 0.0968 | 0.3737 | 0.066* | |
| H411 | 0.3605 | −0.0162 | −0.1140 | 0.101* | |
| H412 | 0.3288 | 0.1137 | −0.1816 | 0.101* | |
| H413 | 0.2310 | 0.1135 | −0.0915 | 0.101* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S11 | 0.0476 (3) | 0.0585 (4) | 0.0574 (4) | −0.0234 (3) | −0.0027 (2) | −0.0058 (3) |
| S12 | 0.0403 (3) | 0.0369 (2) | 0.0419 (3) | −0.0088 (2) | −0.0039 (2) | −0.0024 (2) |
| S31 | 0.0489 (4) | 0.0606 (4) | 0.0561 (4) | 0.0039 (3) | 0.0030 (3) | 0.0065 (3) |
| S32 | 0.0506 (3) | 0.0487 (3) | 0.0487 (3) | −0.0095 (2) | −0.0010 (2) | 0.0084 (2) |
| O11 | 0.0514 (11) | 0.0589 (11) | 0.0493 (10) | −0.0207 (9) | 0.0110 (8) | −0.0083 (8) |
| O12 | 0.0405 (10) | 0.0667 (12) | 0.0561 (11) | −0.0180 (9) | 0.0075 (8) | −0.0143 (9) |
| O13 | 0.0557 (11) | 0.0470 (9) | 0.0482 (9) | −0.0205 (8) | −0.0062 (8) | 0.0069 (8) |
| O14 | 0.0501 (10) | 0.0454 (9) | 0.0516 (10) | −0.0023 (8) | 0.0004 (8) | −0.0113 (8) |
| O31 | 0.0567 (12) | 0.0821 (15) | 0.0581 (12) | 0.0059 (11) | 0.0174 (10) | 0.0146 (11) |
| O32 | 0.0471 (11) | 0.0673 (13) | 0.0561 (11) | 0.0019 (9) | 0.0060 (9) | 0.0031 (9) |
| O33 | 0.0660 (13) | 0.0463 (10) | 0.0644 (12) | −0.0075 (9) | −0.0076 (10) | −0.0037 (9) |
| O34 | 0.0636 (13) | 0.0741 (14) | 0.0673 (13) | −0.0216 (11) | 0.0032 (10) | 0.0216 (11) |
| N11 | 0.0359 (10) | 0.0406 (10) | 0.0373 (10) | −0.0087 (8) | 0.0007 (7) | −0.0039 (8) |
| N12 | 0.0341 (10) | 0.0474 (11) | 0.0433 (10) | −0.0096 (8) | 0.0000 (8) | −0.0083 (9) |
| N31 | 0.0410 (11) | 0.0481 (11) | 0.0408 (10) | −0.0073 (9) | 0.0009 (8) | 0.0011 (9) |
| N32 | 0.0419 (12) | 0.0627 (14) | 0.0506 (12) | −0.0056 (10) | 0.0013 (9) | 0.0092 (11) |
| C1 | 0.0360 (11) | 0.0334 (11) | 0.0471 (13) | −0.0078 (9) | −0.0001 (9) | 0.0014 (10) |
| C2 | 0.0552 (17) | 0.084 (2) | 0.0548 (16) | −0.0200 (16) | −0.0016 (13) | −0.0195 (15) |
| C3 | 0.0529 (16) | 0.0683 (17) | 0.0407 (13) | −0.0223 (13) | 0.0061 (11) | −0.0132 (12) |
| C4 | 0.0350 (11) | 0.0391 (11) | 0.0430 (12) | −0.0073 (9) | −0.0013 (9) | −0.0002 (10) |
| C5 | 0.0366 (11) | 0.0393 (11) | 0.0398 (11) | −0.0103 (9) | −0.0036 (9) | −0.0008 (9) |
| C6 | 0.0381 (13) | 0.0599 (16) | 0.0454 (13) | −0.0104 (11) | 0.0060 (10) | 0.0061 (12) |
| C7 | 0.0326 (12) | 0.0616 (16) | 0.0557 (15) | −0.0044 (11) | 0.0051 (11) | 0.0016 (13) |
| C8 | 0.0392 (12) | 0.0450 (13) | 0.0462 (13) | −0.0111 (10) | −0.0072 (10) | −0.0010 (10) |
| C9 | 0.0491 (14) | 0.0543 (15) | 0.0412 (13) | −0.0074 (12) | 0.0018 (11) | 0.0050 (11) |
| C10 | 0.0421 (13) | 0.0502 (14) | 0.0419 (12) | −0.0041 (11) | 0.0065 (10) | −0.0002 (11) |
| C11 | 0.0495 (16) | 0.0558 (16) | 0.0660 (18) | −0.0064 (13) | −0.0095 (13) | 0.0051 (14) |
| C31 | 0.0377 (12) | 0.0492 (14) | 0.0464 (13) | −0.0036 (10) | 0.0021 (10) | −0.0010 (11) |
| C32 | 0.0491 (15) | 0.0696 (18) | 0.0451 (14) | −0.0122 (13) | −0.0052 (11) | 0.0085 (13) |
| C33 | 0.0472 (15) | 0.0683 (18) | 0.0440 (14) | −0.0040 (13) | 0.0049 (11) | 0.0050 (13) |
| C34 | 0.0393 (13) | 0.0457 (13) | 0.0424 (12) | −0.0057 (10) | −0.0036 (10) | −0.0025 (10) |
| C35 | 0.0516 (14) | 0.0437 (13) | 0.0363 (12) | −0.0022 (11) | −0.0018 (10) | 0.0036 (10) |
| C36 | 0.0560 (16) | 0.0554 (15) | 0.0430 (13) | −0.0113 (13) | 0.0034 (11) | −0.0048 (11) |
| C37 | 0.0659 (19) | 0.0620 (18) | 0.0563 (17) | −0.0150 (15) | −0.0058 (14) | −0.0024 (14) |
| C38 | 0.076 (2) | 0.0405 (14) | 0.0592 (17) | −0.0052 (14) | −0.0201 (15) | 0.0057 (12) |
| C39 | 0.088 (2) | 0.0600 (18) | 0.0517 (16) | −0.0011 (17) | −0.0036 (16) | −0.0183 (14) |
| C40 | 0.0616 (19) | 0.0688 (19) | 0.0528 (16) | 0.0001 (15) | 0.0099 (14) | −0.0099 (14) |
| C41 | 0.086 (2) | 0.0507 (17) | 0.121 (3) | −0.0136 (17) | −0.050 (2) | −0.0125 (19) |
Geometric parameters (Å, °)
| S11—C1 | 1.750 (2) | C32—C33 | 1.518 (3) |
| S11—C2 | 1.797 (3) | C35—C36 | 1.383 (4) |
| S12—O13 | 1.424 (2) | C35—C40 | 1.379 (3) |
| S12—O14 | 1.4295 (18) | C36—C37 | 1.373 (5) |
| S12—N12 | 1.658 (2) | C37—C38 | 1.389 (4) |
| S12—C5 | 1.757 (2) | C38—C39 | 1.366 (5) |
| S31—C31 | 1.752 (2) | C38—C41 | 1.517 (5) |
| S31—C32 | 1.786 (2) | C39—C40 | 1.378 (5) |
| S32—O33 | 1.424 (2) | N12—H12 | 0.860 |
| S32—O34 | 1.425 (2) | N32—H32 | 0.860 |
| S32—N32 | 1.663 (2) | C2—H201 | 0.970 |
| S32—C35 | 1.747 (3) | C2—H202 | 0.970 |
| O11—C1 | 1.217 (3) | C3—H301 | 0.970 |
| O12—C4 | 1.202 (3) | C3—H302 | 0.970 |
| O31—C31 | 1.213 (3) | C6—H6 | 0.930 |
| O32—C34 | 1.192 (3) | C7—H7 | 0.930 |
| N11—C1 | 1.384 (3) | C9—H9 | 0.930 |
| N11—C3 | 1.473 (3) | C10—H10 | 0.930 |
| N11—C4 | 1.397 (3) | C11—H111 | 0.960 |
| N12—C4 | 1.383 (3) | C11—H112 | 0.960 |
| N31—C31 | 1.391 (3) | C11—H113 | 0.960 |
| N31—C33 | 1.460 (3) | C32—H321 | 0.970 |
| N31—C34 | 1.402 (3) | C32—H322 | 0.970 |
| N32—C34 | 1.376 (3) | C33—H331 | 0.970 |
| C2—C3 | 1.494 (4) | C33—H332 | 0.970 |
| C5—C6 | 1.387 (3) | C36—H36 | 0.930 |
| C5—C10 | 1.383 (3) | C37—H37 | 0.930 |
| C6—C7 | 1.376 (3) | C39—H39 | 0.930 |
| C7—C8 | 1.389 (3) | C40—H40 | 0.930 |
| C8—C9 | 1.390 (3) | C41—H411 | 0.960 |
| C8—C11 | 1.504 (3) | C41—H412 | 0.960 |
| C9—C10 | 1.380 (3) | C41—H413 | 0.960 |
| C1—S11—C2 | 93.82 (15) | C39—C38—C41 | 118.6 (2) |
| O13—S12—O14 | 120.18 (11) | C38—C39—C40 | 121.3 (2) |
| O13—S12—N12 | 108.77 (11) | C35—C40—C39 | 119.9 (3) |
| O13—S12—C5 | 109.42 (11) | S12—N12—H12 | 117.8 |
| O14—S12—N12 | 103.25 (11) | C4—N12—H12 | 117.8 |
| O14—S12—C5 | 109.48 (10) | S32—N32—H32 | 118.0 |
| N12—S12—C5 | 104.49 (11) | C34—N32—H32 | 118.0 |
| C31—S31—C32 | 93.95 (12) | S11—C2—H201 | 109.7 |
| O33—S32—O34 | 120.96 (14) | S11—C2—H202 | 109.7 |
| O33—S32—N32 | 108.22 (12) | C3—C2—H201 | 109.7 |
| O33—S32—C35 | 109.34 (13) | C3—C2—H202 | 109.7 |
| O34—S32—N32 | 102.86 (12) | H201—C2—H202 | 109.5 |
| O34—S32—C35 | 109.16 (13) | N11—C3—H301 | 109.5 |
| N32—S32—C35 | 105.02 (13) | N11—C3—H302 | 109.5 |
| C1—N11—C3 | 116.0 (2) | C2—C3—H301 | 109.5 |
| C1—N11—C4 | 126.5 (2) | C2—C3—H302 | 109.5 |
| C3—N11—C4 | 117.2 (2) | H301—C3—H302 | 109.5 |
| S12—N12—C4 | 124.46 (18) | C5—C6—H6 | 120.3 |
| C31—N31—C33 | 116.3 (2) | C7—C6—H6 | 120.3 |
| C31—N31—C34 | 125.9 (2) | C6—C7—H7 | 119.3 |
| C33—N31—C34 | 117.4 (2) | C8—C7—H7 | 119.3 |
| S32—N32—C34 | 123.93 (18) | C8—C9—H9 | 119.2 |
| S11—C1—O11 | 123.1 (2) | C10—C9—H9 | 119.2 |
| S11—C1—N11 | 111.07 (17) | C5—C10—H10 | 120.5 |
| O11—C1—N11 | 125.8 (2) | C9—C10—H10 | 120.5 |
| S11—C2—C3 | 108.6 (2) | C8—C11—H111 | 109.5 |
| N11—C3—C2 | 109.4 (2) | C8—C11—H112 | 109.5 |
| O12—C4—N11 | 121.0 (2) | C8—C11—H113 | 109.5 |
| O12—C4—N12 | 124.1 (2) | H111—C11—H112 | 109.5 |
| N11—C4—N12 | 114.9 (2) | H111—C11—H113 | 109.5 |
| S12—C5—C6 | 120.75 (18) | H112—C11—H113 | 109.5 |
| S12—C5—C10 | 118.64 (17) | S31—C32—H321 | 109.9 |
| C6—C5—C10 | 120.6 (2) | S31—C32—H322 | 109.9 |
| C5—C6—C7 | 119.5 (2) | C33—C32—H321 | 109.9 |
| C6—C7—C8 | 121.3 (2) | C33—C32—H322 | 109.9 |
| C7—C8—C9 | 118.0 (2) | H321—C32—H322 | 109.5 |
| C7—C8—C11 | 121.7 (2) | N31—C33—H331 | 109.8 |
| C9—C8—C11 | 120.3 (2) | N31—C33—H332 | 109.8 |
| C8—C9—C10 | 121.7 (2) | C32—C33—H331 | 109.8 |
| C5—C10—C9 | 119.0 (2) | C32—C33—H332 | 109.8 |
| S31—C31—O31 | 123.45 (19) | H331—C33—H332 | 109.5 |
| S31—C31—N31 | 110.43 (18) | C35—C36—H36 | 120.1 |
| O31—C31—N31 | 126.1 (2) | C37—C36—H36 | 120.1 |
| S31—C32—C33 | 107.76 (18) | C36—C37—H37 | 119.5 |
| N31—C33—C32 | 108.3 (2) | C38—C37—H37 | 119.5 |
| O32—C34—N31 | 121.5 (2) | C38—C39—H39 | 119.4 |
| O32—C34—N32 | 125.0 (2) | C40—C39—H39 | 119.4 |
| N31—C34—N32 | 113.5 (2) | C35—C40—H40 | 120.1 |
| S32—C35—C36 | 120.2 (2) | C39—C40—H40 | 120.1 |
| S32—C35—C40 | 120.2 (2) | C38—C41—H411 | 109.5 |
| C36—C35—C40 | 119.6 (2) | C38—C41—H412 | 109.5 |
| C35—C36—C37 | 119.8 (2) | C38—C41—H413 | 109.5 |
| C36—C37—C38 | 120.9 (3) | H411—C41—H412 | 109.5 |
| C37—C38—C39 | 118.5 (3) | H411—C41—H413 | 109.5 |
| C37—C38—C41 | 122.8 (3) | H412—C41—H413 | 109.5 |
| C1—S11—C2—C3 | −7.7 (2) | S12—N12—C4—N11 | 179.73 (15) |
| C2—S11—C1—O11 | −179.1 (2) | C31—N31—C33—C32 | 15.7 (3) |
| C2—S11—C1—N11 | 2.52 (19) | C33—N31—C31—S31 | −4.9 (3) |
| O13—S12—N12—C4 | −41.4 (2) | C33—N31—C31—O31 | 175.1 (3) |
| O13—S12—C5—C6 | 28.0 (2) | C31—N31—C34—O32 | 170.4 (2) |
| O13—S12—C5—C10 | −154.4 (2) | C31—N31—C34—N32 | −7.4 (4) |
| O14—S12—N12—C4 | −170.14 (19) | C34—N31—C31—S31 | −177.4 (2) |
| O14—S12—C5—C6 | 161.6 (2) | C34—N31—C31—O31 | 2.6 (4) |
| O14—S12—C5—C10 | −20.7 (2) | C33—N31—C34—O32 | −2.0 (4) |
| N12—S12—C5—C6 | −88.3 (2) | C33—N31—C34—N32 | −179.9 (2) |
| N12—S12—C5—C10 | 89.3 (2) | C34—N31—C33—C32 | −171.2 (2) |
| C5—S12—N12—C4 | 75.4 (2) | S32—N32—C34—O32 | −1.8 (4) |
| C31—S31—C32—C33 | 14.4 (2) | S32—N32—C34—N31 | 175.9 (2) |
| C32—S31—C31—O31 | 174.0 (2) | S11—C2—C3—N11 | 10.7 (3) |
| C32—S31—C31—N31 | −6.0 (2) | S12—C5—C6—C7 | 175.8 (2) |
| O33—S32—N32—C34 | 45.7 (2) | S12—C5—C10—C9 | −176.0 (2) |
| O33—S32—C35—C36 | −19.4 (2) | C6—C5—C10—C9 | 1.6 (4) |
| O33—S32—C35—C40 | 160.9 (2) | C10—C5—C6—C7 | −1.8 (4) |
| O34—S32—N32—C34 | 174.9 (2) | C5—C6—C7—C8 | 0.7 (4) |
| O34—S32—C35—C36 | −153.8 (2) | C6—C7—C8—C9 | 0.6 (4) |
| O34—S32—C35—C40 | 26.5 (2) | C6—C7—C8—C11 | −178.3 (2) |
| N32—S32—C35—C36 | 96.5 (2) | C7—C8—C9—C10 | −0.8 (4) |
| N32—S32—C35—C40 | −83.2 (2) | C11—C8—C9—C10 | 178.1 (2) |
| C35—S32—N32—C34 | −71.0 (2) | C8—C9—C10—C5 | −0.3 (4) |
| C1—N11—C3—C2 | −9.6 (3) | S31—C32—C33—N31 | −18.7 (3) |
| C3—N11—C1—S11 | 3.8 (2) | S32—C35—C36—C37 | 179.2 (2) |
| C3—N11—C1—O11 | −174.5 (2) | S32—C35—C40—C39 | −179.3 (2) |
| C1—N11—C4—O12 | −167.6 (2) | C36—C35—C40—C39 | 1.0 (4) |
| C1—N11—C4—N12 | 12.8 (3) | C40—C35—C36—C37 | −1.0 (4) |
| C4—N11—C1—S11 | 178.29 (17) | C35—C36—C37—C38 | 0.4 (4) |
| C4—N11—C1—O11 | 0.0 (3) | C36—C37—C38—C39 | 0.4 (4) |
| C3—N11—C4—O12 | 6.9 (3) | C36—C37—C38—C41 | −176.7 (2) |
| C3—N11—C4—N12 | −172.8 (2) | C37—C38—C39—C40 | −0.5 (4) |
| C4—N11—C3—C2 | 175.3 (2) | C41—C38—C39—C40 | 176.7 (2) |
| S12—N12—C4—O12 | 0.1 (2) | C38—C39—C40—C35 | −0.2 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N12—H12···O11 | 0.86 | 2.00 | 2.665 (3) | 134 |
| N32—H32···O31 | 0.86 | 1.91 | 2.608 (3) | 137 |
| C33—H332···O11 | 0.97 | 2.40 | 3.352 (3) | 166 |
| C11—H112···O33i | 0.96 | 2.55 | 3.500 (4) | 170 |
| C32—H321···O12ii | 0.97 | 2.60 | 3.384 (3) | 138 |
Symmetry codes: (i) x−1, y, z; (ii) −x+1, −y, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: CV2707).
References
- Chen, Q. W. & Shen, D. L. (2008). J. Zhejiang Univ. Technol.36, 562–564.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Fujimoto, E. & Shimizu, T. (1978). Jpn Patent JP 53127466.
- Gowda, B. T., Foro, S., Suchetan, P. A. & Fuess, H. (2010). Acta Cryst. E66, o794. [DOI] [PMC free article] [PubMed]
- Higashi, T. (1995). ABSCOR Rigaku Corporation, Tokyo, Japan.
- Liu, X. H., Chen, P. Q., Wang, B. L., Li, Y. H., Wang, S. H. & Li, Z. M. (2007). Bioorg. Med. Chem. Lett.17, 3784–3788. [DOI] [PubMed]
- Liu, X. H., Shi, Y. X., Ma, Y., He, G. R., Dong, W. L., Zhang, C. Y., Wang, B. L., Wang, S. H., Li, B. J. & Li, Z. M. (2009). Chem. Biol. Drug Des.73, 320–327. [DOI] [PubMed]
- Rigaku (2006). PROCESS-AUTO Rigaku Corporation, Tokyo, Japan.
- Rigaku (2007). CrystalStructure Rigaku Americas Corporation, The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810014467/cv2707sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810014467/cv2707Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



