Summary
Melanoma is a cancer with a poorly understood molecular pathobiology. We find the transcription factors PAX3, SOX10, MITF, and the tyrosine kinase receptor MET expressed in melanoma cell lines and primary tumors. Analysis for MET expression in primary tumor specimens showed 27/40 (68%) of the samples displayed an increased expression of MET, and this expression was highly correlated with parallel expression of PAX3, SOX10, and MITF. PAX3 and MITF bind to elements in the MET promoter independently, without evidence of either synergistic activation or inhibition. SOX10 does not directly activate the MET gene alone, but can synergistically activate MET expression with either PAX3 or MITF. In melanoma cells, there was evidence of two pathways for PAX3 mediated MET induction: 1) direct activation of the gene, and 2) indirect regulation through MITF. SK-MEL23 melanoma cells have both of these pathways intact, while SK-MEL28 melanoma cells only have the first pathway. In summary, we find that PAX3, SOX10 and MITF play an active role in melanoma cells by regulating the MET gene. In consequence, MET promotes the melanoma cancer phenotype by promoting migration, invasion, resistance to apoptosis, and tumor cell growth.
Keywords: PAX3, SOX10, MITF, MET, melanoma, transcription
Introduction
Melanoma is a highly lethal tumor type, with the incidence of disease increasing at a faster rate than most other tumor types over the past ten years (Garbe & Leiter, 2009). The transcription factor PAX3 is expressed in melanoma primary tumors and in cell lines (Plummer et al., 2008; Scholl et al., 2001). PAX3 is also expressed in developing murine melanoblasts in the embryo and in neonatal skin, and PAX3 expression can be upregulated in human neonatal foreskin cultures after ultraviolet radiation (Lang et al., 2005; Yang et al., 2008). In adult skin, PAX3 expression is restricted to the melanocyte progenitor/stem cells and is absent in mature and differentiated melanocytes. PAX3 functions as a transcriptional regulator during embryonic development and is likely to act in this capacity in the melanocyte progenitor/stem cells and in melanomas. A function for PAX3 in melanoma has not been described previously.
PAX3 can interact with other transcription factors to regulate gene expression. One of these proteins is SOX10, a transcription factor containing a HMG (High Mobility Group) DNA binding domain. SOX10 is expressed in migratory neural crest cells, melanoblasts, and melanocytes, but is down-regulated as cells differentiate (Cook et al., 2005; Kuhlbrodt et al., 1998; Southard-Smith et al., 1998). PAX3 and SOX10 directly bind to each other and synergistically activate downstream genes such as MITF and RET (Bondurand et al., 2000; Lang et al., 2000; Lang & Epstein, 2003; Watanabe et al., 1998). SOX10 is also overexpressed in melanomas (Nonaka et al., 2008). The activation of MITF in the embryonic neural crest is essential for the development and maintenance of melanoblasts and melanocytes (Hodgkinson et al., 1993). MITF expression is also commonly found in melanomas (King et al., 1999).
PAX3 activates several other down-stream genes, including the MET receptor. This pathway has been described only in embryonic muscle development (Epstein et al., 1996). The transcription factor MITF can also regulate expression of the MET gene (Beuret et al., 2007; McGill et al., 2006). MET is a transmembrane receptor tyrosine kinase that is activated by hepatocyte growth factor (HGF). MET is essential for normal development and plays a role in cell migration, growth, survival, differentiation, angiogenesis, and tube formation/branching morphogenesis (reviewed in (Gentile et al., 2008)). In normal melanocytes, HGF-MET signaling is required not only for survival, proliferation and differentiation of melanocyte precursors, but also for maintenance of melanocyte specific genes such as tyrosinase (Kos et al., 1999; Kunisada et al., 2000). MET has also been implicated in cancer progression and is directly involved in metastasis, resistance to apoptosis, and tumor growth. Transgenic mice constitutively expressing HGF are characterized by an increased number of melanomas and liver metastases resulting from the activation of MET signaling (Takayama et al., 1997). Further, expression of MET has been observed in melanoma and has been associated with tumor progression and metastasis (Natali et al., 1993; Puri et al., 2007; Saitoh et al., 1994).
Here, we find PAX3, SOX10, MITF and MET expressed in melanoma. PAX3 and MITF can independently activate MET expression, and both of these factors can synergistically activate MET expression with the addition of SOX10. PAX3 and SOX10 can induce MET expression indirectly through regulation of MITF and in a direct pathway independent of MITF. We present here in this report a cancer pathway, where atypical expression of PAX3 and SOX10, or MITF and SOX10, leads to expression of MET, a receptor protein directly involved in the cancer phenotype of metastasis, resistance to apoptosis, and tumor growth.
Results
PAX3, SOX10 and MET are expressed in melanoma cell lines
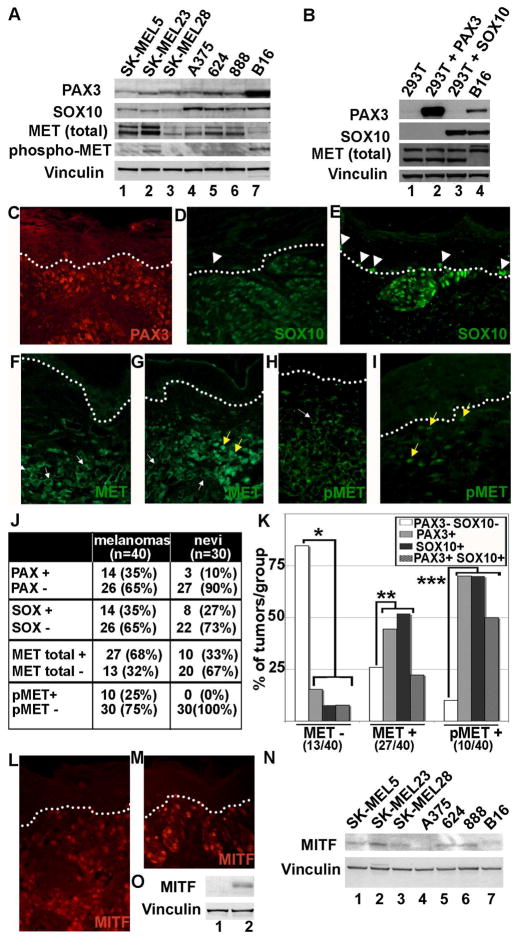
Expression of PAX3, SOX10 and MET in melanoma cell lines was detected by immunoblotting (Figure 1A). All melanoma cell lines showed expression of PAX3, SOX10 and MET with varying levels of protein. The expression of MET was relatively higher in the cell lines SK-MEL5, SK-MEL23, A375, 624, and 888 than in the cell lines SK-MEL28 and B16. In all these cell lines, expression of MET was accompanied by expression of both PAX3 and SOX10. Although the cell line B16 appeared to have higher levels of PAX3, this is likely due to a higher affinity of the PAX3 antibody to mouse versus human antigen rather than actual higher protein levels. In order to check for the presence of the activated form of MET protein, blots were re-probed with an antibody that recognizes specific phosporylated tyrosine residues (tyr 1234/1235, auto-catalytic site). A specific band corresponding to activated MET was detected only in cell lines B16 and SK-MEL23. These data demonstrate expression of PAX3, SOX10 and MET in all melanoma cell lines analyzed.
Figure 1.
PAX3, SOX10, MITF and MET are expressed in melanoma cell lines and in primary tumors. (A) Western blot analysis of melanoma cell lines. Human melanoma cell lines (lanes 1–6) and the mouse melanoma cell line B16 (lane 7) all have expression of PAX3, SOX10, and MET to variable degrees. Vinculin levels are shown as a loading control in both (A) and (B). (B) Control western analysis for antibody specificity. HEK-293T cells lack expression of both PAX3 and SOX10 (lane 1) and were transfected with constructs expressing either PAX3 (lane 2) or SOX10 (lane 3). (C–I) Immunofluorescence for PAX3, SOX10, and MET in primary tumor samples. These examples are representative samples for melanoma tissues with expression for PAX3 (C), SOX10 (D, E), MET (F,G) or phosphorylated MET (pMET) (H,I). The dotted line demarcates the dermo-epidermal junction layer, which separates the upper keratinocyte layer in the epidermis and the lower connective tissue of the dermis. Normal melanocytes are located on this layer, and when found next to melanocytic lesions (white arrow heads), have either low/absent SOX10 expression (D) or high levels (E). MET expression was expressed in the cell membrane/cytosol (white arrows) or in the nucleus (yellow arrows). (J) Summary of the number of melanocytic lesions that expressed PAX3, SOX10, MET, or pMET. Tissue specimens are superficial spreading melanomas (n=40) or dysplastic but non-cancerous pigmented lesions (nevi) (n=30). (K) A graphical summary of the correlation of PAX3 and/or SOX10 expression with the presence or absence of MET and pMET. The compared groups (*, **, and ***) showed a significant difference from the expected mean (p<0.05). (L, M) Immunofluorescent staining for MITF expression in primary tumor samples. Two representative samples are shown. (N) Western blot analysis for MITF expression in melanoma cell lines. Human melanoma cell lines (lanes 1–6) and murine cell line B16 (lane 7) express variable levels of MITF. (O) Control western analysis for antibody specificity. HEK-293T cells lack expression of MITF (lane 1) and were transfected with an expression contruct for MITF (lane 2).
To test the specificity of the antibodies we transfected HEK-293T cells, which lack endogenous PAX3 and SOX10, with constructs that express these factors. A control western analysis measuring PAX3, SOX10, MET, and vinculin protein levels showed the presence of PAX3 and SOX10 bands corresponding to transfections with PAX3 and SOX10 expression constructs (Figure 1B). The cell line B16 served as a positive control. These results demonstrate the specificity of the PAX3 and SOX10 antibodies.
MET expression in melanoma is correlated with PAX3 and SOX10 expression in primary melanoma tumors
Primary invasive melanoma tumors and benign nevi were tested for expression of PAX3, SOX10, and MET (Figure 1C-I). PAX3 was expressed in 14/40 (35%) of malignant melanoma primary tumor samples (Figure 1C, J), but only in 3/30 (10%) of benign melanocytic lesions (nevi) (Figure 1D, J). Expression of SOX10 was comparable in melanoma (14/40, 35%) and nevi (8/30, 27%) specimens. In addition, SOX10 was also expressed in a sub-set of normal melanocytes located in the basal layer (Figure 1E, arrowheads) but was absent in other samples (Figure 1D, arrowheads). Analysis for MET expression in these tumor specimens showed 27/40 (68%) displayed an increased expression of MET. In contrast, 10/30 (33%) of nevi expressed MET. Some tumors displayed predominantly cytoplasmic staining for MET (Figure 1F) while others displayed both nuclear and cytoplasmic expression (Figure 1G). Activation of the MET results in phosphorylation on a number of tyrosine residues. When stained with an antibody against phosphorylated and activated MET both nuclear and cytoplasmic staining was observed (Figure 1H, I). When the melanoma samples were analyzed for the presence of activated and phosphorylated MET (pMET), 25% of the samples showed positive staining for pMET. In contrast none of the nevi samples showed presence of activated MET.
To analyze the correlation between PAX3 and SOX10 on MET expression we classified the melanoma tumors into groups (Figure 1K). The criteria for classification were based on MET or pMET immunoreactivity. We found that tumors that were negative for MET expression were associated with a lack of both PAX3 and SOX10 expression (11/13 MET (-) tumors, 84.6%) (Figure 1K, *). The tumors that expressed MET also express PAX3 and/or SOX10 (20/27 MET (+) tumors, 74.1%) (Figure 1K, **). 22% of MET positive tumors (6/27 MET (+) tumors) showed expression of both PAX3 and SOX10. These data indicate that the occurrence of PAX3 and/or SOX10 is accompanied by an increase in MET expression. In addition, tumors with an activated form of MET (pMET) were highly correlated to tumors that express PAX3 and/or SOX10 (9/10 pMET (+) tumors, 90%) (Figure 1K, ***). Fifty percent of these pMET positive tumors showed expression of both PAX3 and SOX10 (5/10 pMET (+) tumors). These correlations (groups *, **, and ***) are statistically significant (p<0.05). Taken together, these data suggest that there is a relationship between the presence of PAX3 and/or SOX10 and the expression of MET and phosphorylated MET in melanoma.
Microphthalmia-associated transcription factor (MITF) is expressed in primary melanoma tissue and cell lines
Primary melanoma tumors and nevi were tested for the expression of MITF (Figure 1L,M). MITF was expressed in 31/34 (91.2%) of melanoma primary tumor samples and 23/27 (85.2%) benign melanocytic lesions (nevi). The three MITF non-expressing tumors were also PAX3 and SOX10 negative. For the four nevi lacking abundant MITF expression, 3/4 were also PAX3 negative, and 3/4 were SOX10 negative. None of the nevi were negative for all three factors.
Expression of MITF in melanoma cell lines was detected by immunoblotting (Figure 1N). All melanoma cell lines showed expression of MITF with varying levels of expression, with A375 showing nearly undetectable levels of MITF protein. To test the specificity of the antibodies, the MITF-non-expressing HEK-293T cells were transfected with an expression construct for MITF. A control western analysis measuring MITF and vinculin showed the presence of a MITF band in the transfected HEK-293T cells but not in untransfected controls. (Figure 1O). These results demonstrate the specificity of the MITF antibody. The data measuring the level of MITF in primary melanoma tumors and cell lines correspond to previously reported studies (Beuret et al., 2007).
PAX3 activates the MET promoter
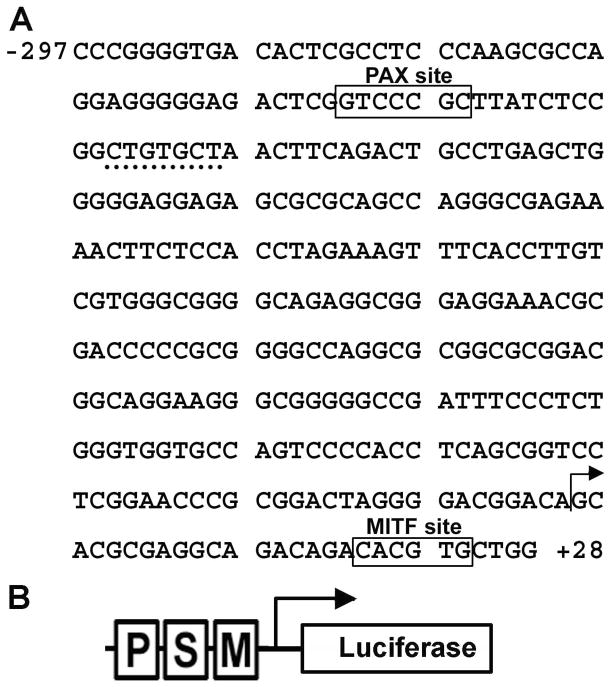
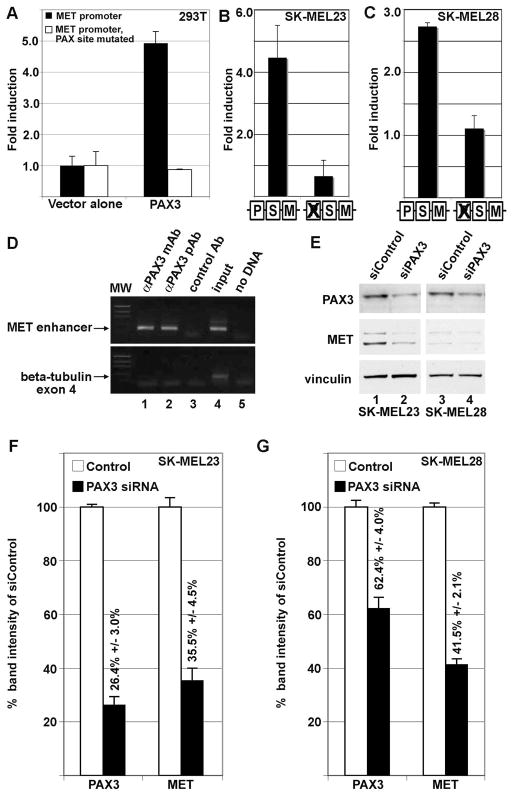
A previous study has shown the presence of a conserved PAX3 site within an enhancer in the MET locus (Figure 2A)(Epstein et al., 1996). PAX3 was identified to activate MET during development and in muscle cells, but it was unknown if this pathway was also active in melanoma tumor cells. A reporter construct was assembled containing 297 base pairs of 5′ proximal MET promoter and 28 base pairs of 5′ UTR sequence driving expression of the reporter gene luciferase (Figure 2). This vector was utilized to create a second reporter construct with the PAX site mutated. These constructs were transfected into HEK-293T cells with or without PAX3 expression construct. An advantage of using HEK-293T cells is the lack of endogenous PAX3, MITF and SOX10, and the ability to add these factors by transfection. Addition of PAX3 induced luciferase expression 4.9±0.4 fold over vector alone (Figure 3A, black bars). PAX3 was unable to drive expression of reporter when the PAX3 binding site in the MET promoter was mutated (Figure 3A, white bars).
Figure 2.
Sequence of the proximal human MET promoter and reporter construct. (A) The MET promoter segment contains the 297 base pairs directly 5′ directly upstream and 28 bases directly 3′ of the MET gene transcriptional start site. The previously described PAX and MITF sites are highlighted with boxes. A putative SOX site is underlined with a dotted line. The transcriptional start site is marked with an arrow. (B) Schematic of the MET promoter reporter construct. The sequence shown in (A) corresponds with the DNA segment contained in the MET promoter luciferase reporter construct utilized in Figures 3, 4, and 5. The PAX (P), SOX (S), and MITF (M) sites, transcriptional start site (arrow) and luciferase reporter gene cassette are shown in diagram form.
Figure 3.
PAX3 activates the MET promoter. (A) PAX3 activates the MET promoter in HEK-293T cells. PAX3 induced luciferase expression 4.9±0.4 fold over vector alone with MET promoter reporter constructs with wild-type sequence (black bars) but not when the PAX site is mutated (white bars). (B,C) PAX3 regulates MET expression in SK-MEL23 (B) and SK-MEL28 (C) melanoma cells. Luciferase reporter is expressed from a construct containing MET promoter when the PAX site is intact or with the PAX site mutated (as illustrated by schematic, as described in Figure 2B). For calculation of fold light units over basal promoter activity (y axis), luciferase activity is measured in arbitrary light units, normalized against beta-galactosidase activity, and divided by the measurements obtained for reporter vector alone. Each bar represents n=9, with standard error of the mean as shown. (D) PAX3 protein is located on the endogenous MET promoter in SK-MEL23 cells. Chromatin Immunoprecipitation (ChIP) analysis utilized primers specific for the MET promoter region (top gels) or for exon 4 of the beta tubulin gene (bottom gels, negative control). Antibodies used for immunoprecipitations are either against PAX3 (monoclonal antibody lanes 1, and polyclonal antibody 2), or normal mouse IgG (control Ab, negative control lane 3). Input DNA was collected for each sample after cell sonication but before immunoprecipitation (lane 4). The “no DNA” lanes lacked a template during PCR amplification (water only, negative control lane 5). (E,F,G) Inhibition of PAX3 protein expression reduces MET receptor levels. Graphs shown in (F) and (G) are quantified densitometry readings of the western analysis shown in (E) of SK-MEL23 cells (lanes 1 and 2) or SK-MEL28 cells (lanes 3 and 4). Cells were transfected with either scrambled siRNA (E, lanes 1,3, F,G white bars) or gene specific siRNA (E, lanes 2,4, F,G black bars). For densitometry readings, bars represent percent of band intensity of the experimental sample compared to the controls, and these percentages are also indicated above each bar. This experiment is a representative of three independent western analyses.
To further determine if endogenous PAX3 can activate the MET promoter in melanoma cell lines, the MET promoter luciferase constructs with or without intact PAX binding sites were transfected into SK-MEL23 and SK-MEL28 cells. Both of these cells lines express PAX3 (Figure 1A). Expression of luciferase reporter from constructs containing the wild-type MET proximal promoter was 4.5±1.0 (SK-MEL23, Figure 3B) and 2.7±0.1 (SK-MEL28, Figure 3C) fold over basal levels. However, luciferase levels dropped to levels comparable to empty pGL2 vector alone when the PAX site in the MET promoter was mutated.
To determine if PAX3 was located at the endogenous MET enhancer in these cells, Chromatin immunoprecipitation (ChIP) analysis was performed using monoclonal and polyclonal antibodies against PAX3. A control non-specific antibody was also included as a negative control. The resulting precipitated DNA fragments were amplified by PCR using specific primers to the MET enhancer or to exon 4 of the beta-tubulin gene (negative control). A specific amplification product for the MET promoter was detected in SK-MEL23 cells only when specific anti-PAX3 antibodies (Figure 3D, lanes 1 and 2) were used. However, no amplification product was detected with a control antibody (Figure 3D, lane 3) or when DNA template was absent (Figure 3D, lane 5).
We determined the loss-of-function effects of PAX3 on MET gene expression by gene specific knockdown in the melanoma cell lines SK-MEL23 and SK-MEL28. In the SK-MEL23 cell line, RNA interference down-regulated levels of PAX3 protein to 26.4%±3.0% of control levels, and reduced MET expression to 35.5%±4.5% of control (Figure 3E, lanes 1,2 and densitometry shown in Figure 3F). In a similar pattern, the SK-MEL28 cell line demonstrated a reduction of PAX3 and MET protein of 62.4%±4.0% and 41.5%±2.1% of controls after siRNA treatment, respectively (Figure 3E lanes 3,4 and densitometry shown in Figure 3G). These data demonstrate that, in melanoma cells, PAX3 is located on the MET promoter, PAX3 directly regulates the MET gene through the PAX enhancer site, and there is a direct relationship between PAX3 levels and the overall quantity of MET protein levels.
MITF regulates MET expression independently of PAX3
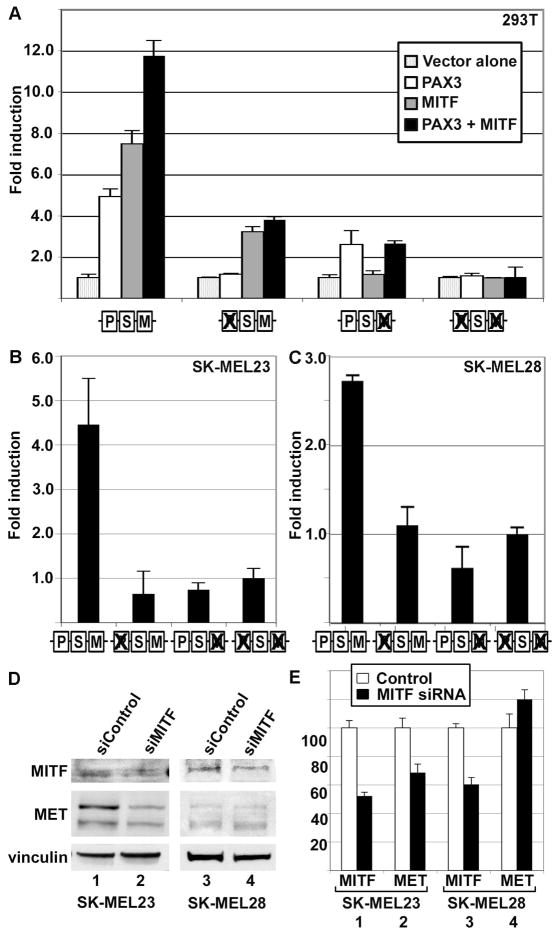
A previous study has shown the presence of a conserved MITF site on the MET enhancer (Figure 2A) (McGill et al., 2006). Several MITF response sites have been characterized in the MET promoter, but only one of these sites falls in the most proximal MET promoter sequence. Luciferase assays in HEK-293T cells, which lack endogenous MITF, showed 7.5±0.6 fold reporter expression over vector alone (Figure 4A, grey bar first set) in the presence of MITF protein, but not when either the MITF protein or the MITF binding site in the promoter was absent. Addition of both MITF and PAX3 to the assay increased the levels of reporter expressed by the wild-type MET promoter to 11.8±0.7 fold reporter expression over vector alone (Figure 4A, black bar first set). However, no evidence of synergistic activation between MITF and PAX3 was observed. Mutation of the PAX3 site reduced but did not eliminate the ability of MITF to activate the MET promoter (3.3±0.3 fold, Figure 4A grey bar second set). Mutation of the MITF binding site significantly attenuated the ability of MITF to drive expression of reporter (1.1±0.2 fold, grey bar, Figure 4A third set) but did not significantly reduce PAX3-dependent reporter expression (2.6±0.7 fold, Figure 4A white bar, third set). Mutation of both the PAX3 and MITF binding sites in the MET promoter blocked the ability of either protein to drive reporter expression (Figure 4A, fourth bar set).
Figure 4.
MITF activates MET, and PAX3 does not inhibit this activation. (A) MITF and PAX3 independently activate the MET promoter in HEK-293T cells. PAX3 induced luciferase expression 4.9±0.4 fold (white bar, first set), and MITF activated luciferase 7.5±0.6 fold over vector alone (grey bar, first set), and both proteins stimulated reporter expression 11.8±0.7 fold (black bar, second set). Ability of PAX3 and/or MITF to drive expression of reporter was eliminated by mutating the specific binding element (as illustrated by schematic, as described in Figure 2B) and reduced but did not eliminate the ability of the other factor to activate through un-mutated sites. (B,C) PAX3 and MITF regulate MET expression in SK-MEL23 (B) and SK-MEL28 (C) melanoma cells. Luciferase reporter is expressed from a construct containing MET promoter when the PAX and/or MITF site is intact or mutated (as illustrated by schematic, as described in Figure 2B). For calculation of fold light units over basal promoter activity (y axis), luciferase activity is measured in arbitrary light units, normalized against beta-galactosidase activity, and divided by the measurements obtained for reporter vector alone. Each bar represents n=9, with standard error of the mean as shown. (D,E) Inhibition of MITF protein expression reduces MET protein levels in SK-MEL23 but not in SK-MEL28 melanoma cells. Graph shown in (E) is the quantified densitometry readings of the western analysis shown in (D). Numbers of the bar sets shown in (E) correspond to the lane numbers shown in (D). Cells were transfected with either scrambled siRNA (D, lanes 1,3, E white bars) or gene specific siRNA (D, lanes 2,4, E black bars). For densitometry readings, graphs represent percent of band intensity of the experimental sample compared to the control. In SKMEL23 cells, inhibition of MITF expression (52.3%±2.5% of controls, bar set 1 in E) leads to an inhibition of MET expression of 68.6%±5.9% (bar set 2). In SK-MEL28 cells, inhibition of MITF expression (60.4%±4.8%, bar set 3) did not lead to a significant change in MET protein levels (119.%8±6.9%, bar set 4). This experiment is a representative of two independent western analyses.
Mutation of the MITF site in the MET proximal promoter also inhibited the expression of reporter in melanoma cell lines. The MET promoter luciferase constructs described were transfected into SK-MEL23 and SK-MEL28 cells. These cells lines both express MITF (Figure 1N). Expression of luciferase reporter from constructs containing the wild-type MET proximal promoter was 4.5±1.0 (SK-MEL23, Figure 4B) and 2.7±0.1 (SK-MEL28, Figure 4C) fold over basal levels. However, luciferase levels dropped to levels comparable to empty pGL2 vector alone when the MITF and/or PAX site was mutated.
To determine if inhibiting MITF expression would alter MET protein levels, SK-MEL23 and SK-MEL28 cells were transfected with siControl (scrambled siRNA, negative control) or an siRNA specific for the MITF RNA (siMITF). In the SK-MEL23 cell line, RNA interference down-regulated levels of MITF protein to 52.3%±2.5% of control levels, and reduced MET expression to 68.6%±5.9% of control (Figure 4D, lanes 1,2 and densitometry shown in Figure 4E). However, inhibition of MITF had little effect on MET protein levels in SK-MEL28 cells. The SK-MEL28 cell line demonstrated a reduction of MITF levels to 60.4%±4.8%, and an insignificant change in MET protein levels to 119.8%±6.9% of controls after siRNA treatment, respectively (Figure 4D lanes 3,4 and densitometry shown in Figure 4E). These data demonstrate that MITF can activate the MET promoter from a cis regulatory site located in the 5′UTR of the MET gene, and that inhibition of MITF expression can reduce MET expression in SK-MEL23 but not in SK-MEL28 melanoma cell lines.
SOX10 does not activate the MET gene independently through a predicted response element
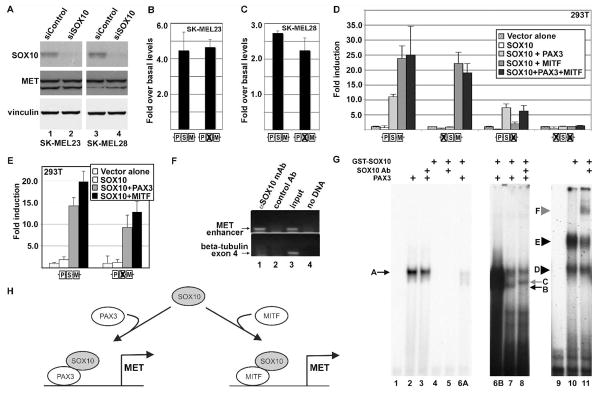
Analysis of the proximal 3 Kb of the MET promoter revealed only one potential SOX10 site in the sequence located in islands of homology between murine and human genomes. This site, CTGTGCT, is located just 3′ to the PAX site and −235 to −229 5′ upstream of the transcriptional start (Figure 2A, dotted underline). This sequence is similar to previously described SOX10 binding sites (Kuhlbrodt et al., 1998). However, inhibition of SOX10 in SK-MEL23 and SK-MEL28 melanoma cells did not significantly alter the levels of MET protein (Figure 5A) and reporter constructs transfected into these cells with the potential SOX site mutated showed no change in the levels of reporter generated (Figure 5B,C). In addition, transfecting SOX10 into non-expressing HEK-293T cells with reporter constructs with or without intact putative SOX10 cis response sites resulted in no increase of reporter induction compared to control levels (Figure 5D, white bars). This suggests that SOX10 does not regulate the MET promoter through the predicted SOX site shown in Figure 2A.
Figure 5.
SOX10 does not appear to directly activate the MET gene, but synergistically activates expression with either PAX3 or MITF. (A) Inhibition of SOX10 protein expression does not affect MET protein levels. Western analysis of cells transfected with either scrambled siRNA (lanes 1,3) or gene specific siRNA (lanes 2,4) show >90% reduction of SOX10 protein but no significant change in MET levels. (B,C) Alteration of a putative SOX site in the MET promoter does not alter reporter expression in SK-MEL23 (B) and SK-MEL28 (C) melanoma cells. (D) SOX10 does not activate the MET promoter in HEK-293T cells alone (white bars), but synergistically activates expression with either PAX3 (light grey) or MITF (dark grey). There is no significant increase in reporter expression when SOX10 is present with both PAX3 and MITF (black bars) than with MITF alone. Activation of the wild-type reporter construct (first bar set) with SOX10 alone is 0.9±0.7 fold over vector alone (white bar), 11.0±0.8 fold with PAX3 (light grey bar), 23.8±4.2 fold with MITF (grey bar), and 25.0±9.6 fold with both PAX3 and MITF (black bar). (E) Synergistic activation of the MET promoter reporter construct by SOX10 and PAX3 or MITF is unaffected when the putative SOX10 site is mutated. Reporter expression is not significantly reduced from wild-type reporter levels (bar set 1) when the putative SOX10 site shown in Figure 2 is mutated (bar set 2). For calculation of fold light units over basal promoter activity (y axis), luciferase activity is measured in arbitrary light units, normalized against beta-galactosidase activity, and divided by the measurements obtained for reporter vector alone. Each bar represents n=9, with standard error of the mean as shown. (F) SOX10 protein is located on the endogenous MET promoter in SK-MEL23 cells. Chromatin Immunoprecipitation (ChIP) analysis utilized primers specific for the MET promoter region (top gels) or for exon 4 of the beta tubulin gene (bottom gels, negative control). Antibodies used for the immunoprecipitation are either against SOX10 (lane 1), or normal mouse IgG (control Ab, negative control lane 2). Input DNA was collected for each sample after cell sonication but before immunoprecipitation (positive control lane 3). The no DNA lanes lacked a template during PCR amplification (water only, negative control lane 4). (G) SOX10 does not directly bind to the enhancer, but is in complex with PAX3. Two probes were utilized for EMSA analysis, one containing the MET enhancer (lanes 1–8) or the P0 probe (SOX10 positive control, lanes 9–11). Lanes 1-6A are a short film exposure and lanes 6B-8 are a long exposure of the same gel. Lanes 6A and 6B are the same lane with different exposure times. Lane 1 is probe alone without the addition of PAX3 or SOX10 proteins. PAX3 binds to the MET enhancer and produces a slow migrating band (lane 2, arrow A) and this migration is not altered by the addition of SOX10 antibody (lane 3). SOX10 does not bind to the probe on its own (lane 4) or with SOX10 antibody (lane 5). PAX3 and SOX10 together results in a band migrating at the same level as PAX3 alone (arrow A) and an additional band (arrow B) with high levels (lane 6A,6B) or low levels (lane 7) of SOX10 protein. Addition of a SOX10-specific antibody alters the migration of this second band (lane 8, grey arrow C). A probe comprising sequence from the P0 promoter is utilized as a positve control for SOX10 binding (lanes 9–11). Lanes include probe without SOX10 protein (lane 9), or with SOX10 protein, resulting in two slow migrating bands (lane 10, arrowheads D and E). The addition of SOX10-specific antibody produces a slower migrating band (lane 11, grey arrowhead F). (H) Model for SOX10 synergistic activation of the MET promoter. SOX10 directly interacts with PAX3 or MITF, and this complex is recruited to an enhancer in the 5′ proximal MET promoter to drive gene expression.
SOX10 synergistically activates MET expression with either PAX3 or MITF
While SOX10 does not activate MET expression alone, it can synergistically activate MET with either PAX3 or MITF. In HEK-293T cells, PAX3 or MITF, with the addition of SOX10, activated reporter expression 11.0±0.8 and 23.8±4.2 fold over controls, respectively (Figure 5D, first bar set). In the presence of SOX10, the addition of both PAX3 and MITF did not increase reporter levels significantly over levels obtained with MITF alone (25.0±9.6, Figure 5D first bar set, black bar). The synergistic activation of reporter with SOX10 and PAX3 was dependent on an intact PAX3 site (Figure 5D, second and fourth bar set). Similarly, the activation seen with both SOX10 and MITF is dependent on the presence of a MITF response element (Figure 5D, third and fourth bar set). However, the predicted SOX10 site was dispensable for the synergistic activation of SOX10 with either PAX3 or MITF (Figure 5E).
SOX10 interacts with PAX3 on the MET promoter
To determine if SOX10 was located at the endogenous MET enhancer in melanoma cells, ChIP analysis was performed using antibodies against SOX10. A control non-specific antibody was also included as a negative control. The resulting precipitated DNA fragments were amplified by PCR using specific primers to the MET enhancer or to exon 4 of the beta-tubulin gene (negative control). A specific amplification product for the MET promoter was detected in SK-MEL23 cells only when a specific anti-SOX10 antibody (Figure 5F, lane 1) was used. However, no amplification product was detected with a control antibody (Figure 5F, lane 2) or when DNA template was absent (Figure 5F, lane 4).
Since SOX10 is located on the enhancer element in SK-MEL23 melanoma cells, but no direct enhancer element in the proximal promoter has been found, it is possible that SOX10 is being recruited to the promoter by other factors. PAX3 has the ability to recruit SOX10 to the MITF promoter without direct DNA binding by SOX10 (Lang & Epstein, 2003). To test the possibility that PAX3 is recruiting SOX10 to the MET enhancer, an electrophoretic mobility shift assay (EMSA) was performed. Using a radioactive probe comprised of the sequence from the MET promoter containing the PAX binding site, a slower migrating band was seen in the presence of cellular lysate containing PAX3 protein (Figure 5G, lane 2, arrow A). This slower migrating band was not altered by the addition of SOX10 antibody (Figure 5G, lane 3). SOX10 protein was unable to produce any slower migrating bands either alone or with SOX10 antibody (Figure 5G, lanes 4,5). However, this recombinant SOX10 protein was competent to produce a band shift with a probe containing a portion of the P0 promoter (Figure 5G, lane 10, arrowheads D,E), and this band can be supershifted with the addition of a SOX10-specific antibody (lane 11, grey arrowhead F). The ability of SOX10 to bind to a probe comprised of P0 promoter sequence has been previously described (Peirano et al., 2000). While SOX10 protein alone does not cause the presence of slower migrating bands, the addition of both PAX3 and SOX10 proteins together yielded both the band found with PAX3 alone (Figure 5G, lanes 6A, 6B, 7, arrow A) and an additional band (arrow B). This second band can be supershifted with the addition of SOX10 antibody (Figure 5G, lane 8, grey arrow C). These experiments support a model where SOX10 can interact with PAX3 to regulate the expression of the MET gene. Another possibility is that MITF can regulate MET expression with SOX10 (shown schematically in Figure 5H).
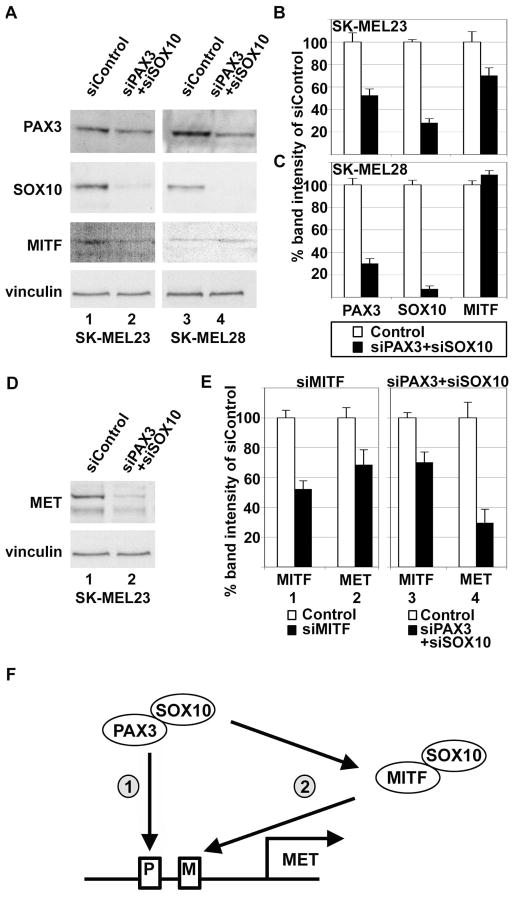
Inhibition of PAX3 and SOX10 inhibits the expression of MITF in SK-MEL23 but not in SK-MEL28 melanoma cells
PAX3 and SOX10 are known to regulate the expression of MITF during neural crest and melanocyte development (Bondurand et al., 2000; Potterf et al., 2000). It is not known if this pathway is conserved in melanoma cells. To determine the consequences of inhibiting PAX3 and SOX10 expression on MITF gene expression, SK-MEL23 and SK-MEL28 melanoma cells were transfected with siRNA against both the transcripts for PAX3 and SOX10, or with a scrambled non-specific siRNA (siControl) as a negative control. In the SK-MEL23 cell line, RNA interference down-regulated levels of PAX3, SOX10 and MITF proteins to 52.5%±5.7%, 28.2%±3.7%, and 70.2%±6.87%, respectively, of control levels (Figure 6A, lanes 1,2 and densitometry shown in Figure 6B). The SK-MEL28 cell line demonstrated a reduction of PAX3 and SOX10 protein to 30.0%±4.3% and 7.5%±2.6% of controls after siRNA treatment, respectively (Figure 6A lanes 3,4 and densitometry shown in Figure 6C). However, there was no significant change in MITF expression in the SK-MEL28 cell line after the inhibition of both PAX3 and SOX10 (109.0%±3.7%).
Figure 6.
PAX3 and SOX10 activate MET expression either directly, or directly and indirectly, in melanoma cells. (A,B,C) Inhibition of both PAX3 and SOX10 protein expression reduces MITF protein levels in SK-MEL23 but not in SK-MEL28 melanoma cells. Western analysis of cells transfected with either scrambled siRNA (lanes 1,3) or gene specific siRNA (lanes 2,4). Graph shown in (B) is the quantified densitometry readings of the western analysis of (A), lanes 1 and 2, and graph shown in (C) is densitometry of lanes 3 and 4. In SK-MEL23 cells (B), inhibition of both PAX3 and SOX10 leads to a reduction of PAX3, SOX10, and MITF 52.5%±5.7%, 28.2%±3.7%, and 70.2%±6.8%, respectively. In SK-MEL28 cells (C), inhibition of both PAX3 and SOX10 leads to a reduction of PAX3 (30.0%±4.3%) and SOX10 (7.5%±2.6%), but no significant reduction of MITF (109.1%±3.7%). (D) Western analysis for MET expression in SK-MEL23 cells transfected with either scrambled siRNA (lanes 1,3) or gene specific siRNA (lanes 2,4). These samples are the same as those shown in lanes 1 and 2 of panel A. (E) Comparison of the inhibition of MET protein expression between targeting MITF (bar sets 1,2) and targeting both PAX3 and SOX10 (bar sets 3,4) in SK-MEL23 cells. Bar sets 1 and 2 show densitometry reading of western analyses shown in Figure 4D,E (lanes 1,2) where inhibition of MITF expression (52.3%±2.5% of controls, bar set 1) leads to an inhibition of MET expression of 68.6%±5.9% (bar set 2). Bar sets 3 and 4 show densitometry of the western analysis shown in (D), where inhibition of both PAX3 and SOX10 expression lead to a reduction of MITF (70.2%±6.8% of controls, bar set 3) and MET (29.6%±9.1%, bar set 4) protein expression. (F) A model for direct and indirect upstream regulation of MET by PAX3 and SOX10. PAX3 and SOX10 can bind directly to upstream enhancer elements in the MET gene locus (arrow 1), or can work through an indirect pathway (arrow 2), where PAX3 and SOX10 regulates the expression of MITF, and MITF with or without SOX10 can activate MET expression.
Reduction of MET protein levels in SK-MEL23 melanoma cells by the inhibition of PAX3 and SOX10 expression may be partially, but not fully, through the downregulation of MITF
Since inhibition of PAX3 and SOX10 also decreased MITF expression in SK-MEL23 cells, the reduction of MET in these cells may be totally or partially due to a loss of MITF rather than direct MET gene regulation by PAX3 and SOX10. To test this hypothesis, MET levels were measured and compared from lysates from SK-MEL23 cells transfected with both PAX3 and SOX10 or MITF siRNAs. In the SK-MEL23 cell line transfected with siRNAs targeted against transcripts for both PAX3 and SOX10, RNA interference down-regulated levels of MITF and MET protein to 70.2%±6.8% and 29.6%±9.1%, respectively, of control levels (Figure 6D, and densitometry shown in Figure 6E, lanes 3,4). In comparison, SK-MEL23 cells transfected with siRNAs targeted against MITF transcripts resulted in a down-regulation of MITF and MET levels to 52.3%±5.4% and 68.6%±9.9%, respectively, of control levels (Figure 4D, and densitometry shown in Figure 6E, lanes 1,2). This suggests that down-regulation of MET by both PAX3 and SOX10 is partially but not fully through inhibition of the MITF gene.
Discussion
In this report, MET is shown to be a direct down-stream target of PAX3 and SOX10 in melanoma cells. In addition, the expression of MET in melanoma primary tissues and cell lines is highly correlated with the expression of PAX3 and/or SOX10. All three factors have been reported in melanoma. MET is commonly over-expressed in melanoma and is associated with a more aggressive phenotype in terms of invasion and metastasis (Cruz et al., 2003; Natali et al., 1993; Puri et al., 2007; Saitoh et al., 1994). PAX3 and SOX10 have also been reported to be over-expressed in melanomas (Nonaka et al., 2008; Plummer et al., 2008; Scholl et al., 2001). This is the first report to find a relationship between the expression of PAX3 and/or SOX10 with the expression of MET protein (Figure 1). Tumors that expressed PAX3, SOX10, or both often express MET (20/27 MET expressing melanomas) and an absence of both of these factors is correlated with a lack of MET (11/13 MET non-expressing melanomas).
We find that there is a relationship with PAX3 or SOX10 expression with phosphorylated MET (Figure 1). This form of MET is considered an active form of the receptor, and phosphorylation occurs either through HGF-ligand binding or by an HGF-independent mechanism. A subset of the melanomas expressed phosphorylated MET, 25% of the total number of tumors analyzed (10/40) or 37% of the number of melanomas that expressed any form of MET (10/27). Only a small minority of the phospho-MET expressing tumors lacked both PAX3 and SOX10 expression (1/10, 10%). Of interest, we were unable to find any benign nevi lesions expressing the phosphorylated form of MET. It is unclear whether PAX3 and/or SOX10 are directly or indirectly involved with MET phosphorylation and activation. Future studies will focus on the role of PAX3 and/or SOX10 on the phosphorylation status of the MET protein.
We also find expression of MET on the membrane and in the nucleus. Expression of MET in the nucleus has been previously reported in a number of tumor types, including melanoma (Pozner-Moulis et al., 2006). While canonical MET signaling is initiated at the plasma membrane and is continued through intercellular intermediate factors, MET can form cleavage products and translocate to the nucleus before initiating a signaling cascade. Functions for this nuclear MET moiety include initiating calcium signaling (Gomes et al., 2008). While it is unclear what are the implications of the sub-cellular localization for the MET protein in melanoma, we found that most tumors had some examples of both membrane and nuclear versions.
This report establishes a mechanistic pathway in melanoma cells, where the MET gene is a functional transcriptional target of PAX3 and SOX10. The PAX3-MET pathway has been described previously in normal muscle development. PAX3 deficient mice are characterized by the absence of forelimb muscles resulting from an absence of MET transcription (Epstein et al., 1996). In rhabdomyosarcoma, a translocation results in a fusion of PAX3 with FOX01 (also known as FKHR) which converts PAX3 into a more potent transcription factor and a stronger activator for PAX3 down-stream genes such as MET (Ginsberg et al., 1998). Here, we demonstrate that a parallel pathway of PAX3 activation of MET also exists in melanoma.
MITF has been previously shown to regulate MET expression in melanoma cells (Beuret et al., 2007; McGill et al., 2006). The MITF response element (shown in Figure 2A) has been shown to be sufficient for MITF to bind directly to the enhancer element in vitro and in vivo in 501mel melanoma cell lines and to drive MET gene expression (McGill et al., 2006). While this site is sufficient, there are redundant MITF response elements located 3′ proximal to this element suggesting that this site is not necessary for MITF activation (Beuret et al., 2007). Our data agrees with these earlier reports, where MITF can activate a reporter construct containing the response element located at +19 to +24 in the human 5′ UTR. In addition, we find that inhibition of MITF expression with siRNA leads to a parallel reduction of MET levels in SK-MEL23 cells (Figure 4D, E), and this response has also been seen for MEL501 and B16 cells (Beuret et al., 2007). However, we did not see this pathway intact in SK-MEL28 cells, where reduction of MITF has no effect on MET protein levels (figure 4D, E).
MITF has also been described to be a down-stream target of PAX3 and SOX10 in neural crest cells and embryonic melanocytes (Bondurand et al., 2000; Potterf et al., 2000; Verastegui et al., 2000; Watanabe et al., 1998). It was not known if this pathway was conserved in melanoma cells. Although PAX3 and SOX10 appear to initiate MITF expression in melanoblasts, it is unclear whether these factors are essential for the maintenance of MITF expression. We find that inhibition of both PAX3 and SOX10 leads to a reduction of MITF in one melanoma cell line, SK-MEL23, but not in another, SK-MEL28 (Figure 6A, B, C). In addition to being a down-stream target of PAX3, MITF is also a molecular antagonist of PAX proteins, either by competing with PAX3 for DCT (dopachrome tautomerase) promoter binding sites, or by directly binding to PAX6 thereby blocking the ability of either protein to bind to DNA (Lang et al., 2005; Planque et al., 2001). In terms of the MET gene, we did not see any evidence of inhibition by either protein to function on the promoter (Figure 4A).
PAX3 directly interacts with SOX10, and, in addition to the MITF and MET promoters, synergistically activates other down-stream targets such RET (Lang et al., 2000; Lang & Epstein, 2003). Although we analyzed the MET promoter region for potential SOX binding sites and defined one putative site (Figure 2A), this site was insufficient for SOX10 to drive expression of luciferase in reporter assays (Figure 5). Our data suggest that SOX10 is recruited to the enhancer by PAX3 or MITF. Earlier studies have also shown that SOX10 is also recruited by PAX3 to an enhancer in the RET gene, and by Sp1 to the myelin basic protein gene promoter (Lang & Epstein, 2003; Wei et al., 2004). Since Sp1 also activates the expression of MET (Liang et al., 2004; Zhang et al., 2003), it is possible that Sp1 (in addition to PAX3 or MITF) can act as a chaperone for SOX10 for transcriptional regulation of the MET locus. In addition, SOX10 and MITF directly interact with the DCT promoter and together these proteins synergistically activate gene expression (Jiao et al., 2004; Ludwig et al., 2004). Our data support a model where PAX3 or MITF independently and directly activate the MET gene, and SOX10 is recruited to the enhancer to synergistically increase the expression of MET (Figure 5H).
Here, we present a model for direct and indirect upstream regulation of the MET gene by PAX3 and SOX10 in melanoma cells (Figure 6F). In the direct pathway (Figure 6F, arrow 1), PAX3 binds directly to a response element in the MET promoter, recruits SOX10, and activates MET expression. The indirect pathway (Figure 6F, arrow 2), PAX3 and SOX10 regulate the expression of MITF, and MITF (with or without the recruitment of SOX10) can directly activate the MET gene. In our work, we see examples of both pathways. The SK-MEL28 cells do not lose MITF expression when both PAX3 and SOX10 expression are inhibited (Figure 6A), and also do not reduce MET levels when MITF transcript is repressed (Figure 4D,E). However, these cells do have a reduction of MET levels when PAX3 expression is blocked (Figure 3E, G). Therefore, we propose that in SK-MEL28 cells only the direct pathway exists. In comparison, SK-MEL23 cells show a decrease in both MITF and MET protein levels when PAX3 and SOX10 transcripts were inhibited (Figure 6A, D). A direct inhibition of MITF expression to 52.3%±5.4% leads to a reduction of MET expression to 68.6%±9.9% of controls (Figure 6E, lanes 1,2), while indirect inhibition of MITF through the repression of PAX3 and SOX10 led to a reduction of MITF to 70.2%±6.8% and a corresponding decrease in MET levels to 29.6%±9.1% of controls (Figure 6E, lanes 3,4). For the SK-MEL23 cells, some of the decrease in MET levels by PAX3 and SOX10 may be due to the decrease in MITF levels. However, the levels of MET expression after PAX3 and SOX10 inhibition is much lower than in direct MITF siRNA targeting, so this reduction in MET cannot be fully attributable to MITF alone. Consequently, we propose that both the direct and indirect pathways are functional in SK-MEL23 cells.
Here, we present a model of melanoma progression. Dysregulation of transcription factors, such as PAX3, SOX10, and MITF, lead to the activation of a number of down-stream targets. The MET tyrosine kinase receptor is one such target, and this protein aids in the normal cellular functions of migration and growth. These normal cell functions can be subverted in cancer cells for invasion, metastases, resistance of apoptosis, and tumor cell expansion.
Materials and Methods
Cell Lines
The melanoma cell lines SK-MEL-5, SK-MEL-28, B16, A375, SK-MEL-23, 888, 624 and the human kidney cell line HEK-293T were obtained from American Type culture Collection, Manassas, VA. Cell lines were maintained in DMEM (Sigma-Alrich, St. Louis, MO, USA) supplemented with 10% FBS in a humidified atmosphere of 5% CO2 at 37°C.
Western Analysis
To examine PAX3, SOX10, MITF and MET expression in melanoma, cells were washed with ice-cold PBS and lysed in RIPA buffer. Equal amounts of protein were analyzed on NUPAGE (4–12%) gels (Invitrogen Corporation, Carlsbad, CA, USA), blotted onto PVDF or nitrocellulose membranes, and analyzed with specific antibodies. Antibodies used were against PAX3 (1:200 dilution, University of Iowa Hybridoma Bank, Iowa City, IA, USA), SOX10 (sc17342; 1:1000 dilution, Santa Cruz Biotechnology), MET (sc10; 1:200 dilution, Santa Cruz Biotechnology, Santa Cruz, CA, USA), pMET Tyr1234/1235 (3126; 1:1000 dilution, Cell signaling technology, Danvers, MA), and MITF (MS-771-P, C5, 1:100 dilution, Thermo Fisher Scientific, CA, USA). Blots were stripped using Restore Western Blot Stripping Buffer (Pierce Biotechnology, Rockford, IL, USA) and reprobed for vinculin (clone hVIN1; 1:5000 dilution, Sigma-Alrich) as a loading control.
Immunohistochemistry on primary tissue blocks
Archival blocks of formalin-fixed, paraffin-embedded skin specimens were obtained in accordance with University of Chicago Institutional Review Board and Clinical Trials Committee from the University of Chicago Medical Center Dermatology tissue bank. All samples were previously diagnosed by a board certified dermatopathologist as either a “ benign nevus” or “ primary melanoma” (Plummer et al., 2008). A staining protocol based on fluorescence was used to avoid melanin interference in interpretation of the results. Tissue sections of 5 μm were boiled in citrate buffer as a method of antigen retrieval and blocked in ImmunoPure Normal Goat Serum (Pierce Biotechnology), then washed and incubated with primary antibodies overnight at 4°C. Antibodies utilized in these studies were against PAX3 (University of Iowa Hybridoma Bank), SOX10 (sc17342, Santa Cruz), MET (sc-10, Santa Cruz), phosphorylated MET (pMET) (44888G, Invitrogen) and MITF (MS-772-P, D5, Thermo Fisher Scientific). The secondary antibody was conjugated to DyLight 547 (red emission, PAX3, MITF) or fluorescein (green, SOX10, MET, pMET) (Pierce Biotechnology). Observations were recorded in a blinded manner and scored as ‘positive for expression’ (equal or greater intensity of staining of positive controls, equal or more than 10% of cells in the lesion) or ‘negative for expression’ (equal or lesser intensity of staining of normal adjacent tissue and negative controls). All tissue samples are of sufficient quality for immunohistochemical analysis, and have all been stained successfully previously with Ki67 and/or S100 specific antibodies (data not shown). Sections of mid-gestation mouse embryos served as a control, where neural crest and neural tube is a positive control, and non-PAX3 expressing embryonic tissue functions as a negative control.
Statistical Analysis
Difference in sample sets and categorical variables were determined by Student’s t-test and chi square distribution (X2) using statistical software (Graphpad Prism, version 5.0, Graphpad software, Inc., La Jolla, CA, USA). A p-value of less than 0.05 was considered statistically significant.
Luciferase Assays and Vectors
A fragment of the human MET promoter that included 297 bp upstream of the transcriptional start site and 22 bp of exon 1 was amplified from genomic DNA (Epstein et al., 1996; Gambarotta et al., 1996). The fragment was subcloned into pGL2-basic vector (Promega Biosciences, Inc., Madison, WI, USA) to generate the reporter plasmid wt hMET pm, as described (Mascarenhas et al., 2009). This construct was subsequently altered by site directed mutagenesis to extend the MET promoter to include a consensus MITF site to bp +28 as shown in Figure 2A. Mutation of putatative PAX, SOX, and MITF elements were mutated by site directed mutagenesis. The PAX site was mutated by replacing the sequence GTCCCGC to ACTAGTC (creating a SpeI site), the SOX10 site was mutated from CTGTGCT to ACTCGAG creating a XhoI site, and the MITF site from a CACGTG to GAGGTG. Cells were transfected with a MET promoter luciferase reporter construct, an internal control (beta-galactosidase expressing construct pCMV – Clontech Laboratories, Inc., Palo Alto, CA, USA) and the presence or absence of PAX3, SOX10 and/or MITF expression constructs. PAX3, MITF and SOX10 were constructed as described (Lang & Epstein, 2003). Total amount of DNA transfected was kept constant by the addition of pBluescript plasmid (Stratagene, La Jolla, CA, USA). Cells were transfected according to manufacturer’s protocols (Effectene reagent, Qiagen, Valencia, CA, USA). After 48 hours post transfection, luciferase and beta-galactosidase assays (Promega) were performed. Luciferase activity was normalized to the internal control beta-galactosidase. All experiments were performed thrice, with triplicates of each variable.
ChIP assays
SK-MEL-23 cells were fixed in 1% formaldehyde and quenched in 0.125 M glycine, then processed according to manufacturers protocol (Upstate Biotechnology, Temecula, CA). For immunoprecipitation of PAX3/DNA complexes, 1 ul of antibody against PAX3 (University of Iowa Hybridoma Bank, monoclonal or SantaCruz Biotechnology sc25409 polyclonal antibody) or SOX10 (Santa Cruz Biotechnology sc17342) was added per experimental reaction. A nonspecific antibody, normal IgG (Sigma-Alrich), was used as a negative control against non-specific DNA precipitation by an antibody. Nested PCR was performed with primers to the MET enhancer, first with TCC GCC TCT AAC AAT GAA CTC C (F) and AAG GTG AAA CTT TCT AGG TGG (R), then a second PCR reaction with primers TGC CCA AAT CTC TCT AAA CCC (F) and AAG TTT TCT CGC CCT GGC TGC G (R). All ChIP samples were tested for false positive PCR amplification using primers that amplify a sequence from within the fourth large coding exon of the beta-tubulin gene to control against genomic DNA contamination. The nested primer set for this control was AAA GGC CAC TAC ACA GAG GG (F) and TAC CAA CTG ATG GAC GGA GAG G (R) for the first PCR reaction, then TTG ATT CTG TCC TGG ATG TGG (F) and TCA GAC ACT TTG GGT GAA GGC(R) for the second PCR round.
RNA inhibition with small interfering RNA (siRNA)
SK-MEL23 and SK-MEL28 cells were seeded in 6 well dishes and transfected with PAX3, SOX10, and/or MITF transcript specific or GFP-labeled control siRNA (100 pmol, Dharmacon, Lafayette, CO, USA) using Lipofectamine 2000 (Invitrogen) following manufacturer’s protocols. Control experiments showed that the transfection efficiency was greater than 80%, as determined by the number of fluorescing cells successfully transfected with labeled control siRNA. Cell lysates were collected 48 hr post transfection and analyzed by western analysis. Densitometry measurements were performed digitally using NIH ImageJ.
Electrophoretic Mobitlity Shift Assay (EMSA)
Nuclear extracts were prepared from HEK-293T cells transfected with PAX3. GST-SOX10 was expressed in bacteria and purified on Glutathione-Sepharose 4B following manufacturers protocol (GE Healthcare Biosciences, Uppsala, Sweden). The reaction was performed in 50 mM Tris. HCl pH 7.5, 50 mM NaCl, 4% Glycerol, 1mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT, 50ug/ml polydeoxy (Inosinate-Cytidylate) (USB, Cleveland, OH, USA) and 500ug/ml BSA. Primers to create probes follow the sequence GGG AGA CTC GGT CCC GCT TAT CTC CGG CTG TGC TAA CTT CAG A for MET promoter sequence, and AGG TCA GTA TAC ACA AAG CCC TCT GTG TAA GGG GTG for P0 promoter sequence as described previously (Mascarenhas et al., 2009; Peirano et al., 2000). Annealed probes were labeled by end filing using T4 polynucleotide kinase (New England Biolabs, Ipswich, MA, USA) and purified on microspin G-50 columns (GE healthcare Biosciences, Buckinghamshire, UK). Nuclear extracts were incubated for 30 minutes with GST-SOX10 (kindly provided by Myung K. Shin, and as described (Zhu et al., 2004)) on ice for 1.5 hr followed by addition of labeled probe. For band supershift experiments, an antibody specific for SOX10 (SC17342, SantaCruz Biotechnology) was added to the reaction for an additional 10 minutes on ice. Electrophoresis was performed on 4% native gels. Gels were dried and exposed to Kodak Biomax MS film (Carestream Health, Rochester, NY, USA).
Significance.
PAX3 is essential for melanocyte development. PAX3 is also expressed in melanoma although its function in these tumor cells had not been defined. Here, we find PAX3, together with SOX10, is frequently co-expressed with the tyrosine kinase receptor MET and directly upregulates MET expression in melanoma. PAX3 and SOX10 can also activate MITF expression, and MITF can also upregulate the MET gene. We find evidence of MITF-dependent and independent MET regulation in melanoma cells. This is the first report to define a role of PAX3 in melanoma through the direct up-regulation of MET.
Acknowledgments
The authors thank Christopher R. Shea (University of Chicago) for providing support for this research. This work was supported by The Schweppe Foundation, University of Chicago Cancer Center Pilot program (P30 CA014599), the American Skin Association, the Dermatology Foundation, Friends of Dermatology-University of Chicago, Outrun the Sun, Inc., and the National Institutes of Health R01CA130202 (D.L.) and R01CA100750, R01CA125541, R01CA129501, and R21CA140003 (R.S.).
References
- Beuret L, Flori E, Denoyelle C, Bille K, Busca R, Picardo M, Bertolotto C, Ballotti R. J Biol Chem. 2007;282:14140–7. doi: 10.1074/jbc.M611563200. [DOI] [PubMed] [Google Scholar]
- Bondurand N, Pingault V, Goerich DE, Lemort N, Sock E, Caignec CL, Wegner M, Goossens M. Hum Mol Genet. 2000;9:1907–17. doi: 10.1093/hmg/9.13.1907. [DOI] [PubMed] [Google Scholar]
- Cook AL, Smith AG, Smit DJ, Leonard JH, Sturm RA. Exp Cell Res. 2005;308:222–35. doi: 10.1016/j.yexcr.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Cruz J, Reis-Filho JS, Silva P, Lopes JM. Oncology. 2003;65:72–82. doi: 10.1159/000071207. [DOI] [PubMed] [Google Scholar]
- Epstein JA, Shapiro DN, Cheng J, Lam PY, Maas RL. Proc Natl Acad Sci U S A. 1996;93:4213–8. doi: 10.1073/pnas.93.9.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambarotta G, Boccaccio C, Giordano S, Ando M, Stella MC, Comoglio PM. Oncogene. 1996;13:1911–7. [PubMed] [Google Scholar]
- Garbe C, Leiter U. Clin Dermatol. 2009;27:3–9. doi: 10.1016/j.clindermatol.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Gentile A, Trusolino L, Comoglio PM. Cancer Metastasis Rev. 2008;27:85–94. doi: 10.1007/s10555-007-9107-6. [DOI] [PubMed] [Google Scholar]
- Ginsberg JP, Davis RJ, Bennicelli JL, Nauta LE, Barr FG. Cancer Res. 1998;58:3542–6. [PubMed] [Google Scholar]
- Gomes DA, Rodrigues MA, Leite MF, Gomez MV, Varnai P, Balla T, Bennett AM, Nathanson MH. J Biol Chem. 2008;283:4344–51. doi: 10.1074/jbc.M706550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- Jiao Z, Mollaaghababa R, Pavan WJ, Antonellis A, Green ED, Hornyak TJ. Pigment Cell Res. 2004;17:352–62. doi: 10.1111/j.1600-0749.2004.00154.x. [DOI] [PubMed] [Google Scholar]
- King R, Weilbaecher KN, McGill G, Cooley E, Mihm M, Fisher DE. Am J Pathol. 1999;155:731–8. doi: 10.1016/S0002-9440(10)65172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos L, Aronzon A, Takayama H, Maina F, Ponzetto C, Merlino G, Pavan W. Pigment Cell Res. 1999;12:13–21. doi: 10.1111/j.1600-0749.1999.tb00503.x. [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. J Neurosci. 1998;18:237–50. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisada T, Yamazaki H, Hirobe T, Kamei S, Omoteno M, Tagaya H, Hemmi H, Koshimizu U, Nakamura T, Hayashi SI. Mech Dev. 2000;94:67–78. doi: 10.1016/s0925-4773(00)00308-7. [DOI] [PubMed] [Google Scholar]
- Lang D, Chen F, Milewski R, Li J, Lu MM, Epstein JA. J Clin Invest. 2000;106:963–71. doi: 10.1172/JCI10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Epstein JA. Hum Mol Genet. 2003;12:937–45. doi: 10.1093/hmg/ddg107. [DOI] [PubMed] [Google Scholar]
- Lang D, Lu MM, Huang L, Engleka KA, Zhang M, Chu EY, Lipner S, Skoultchi A, Millar SE, Epstein JA. Nature. 2005;433:884–7. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- Liang H, O’Reilly S, Liu Y, Abounader R, Laterra J, Maher VM, McCormick JJ. Int J Oncol. 2004;24:1057–67. [PubMed] [Google Scholar]
- Ludwig A, Rehberg S, Wegner M. FEBS Lett. 2004;556:236–44. doi: 10.1016/s0014-5793(03)01446-7. [DOI] [PubMed] [Google Scholar]
- Mascarenhas JB, Young KP, Littlejohn EL, Yoo BK, Salgia R, Lang D. J Biol Chem. 2009;284:27524–32. doi: 10.1074/jbc.M109.047209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill GG, Haq R, Nishimura EK, Fisher DE. J Biol Chem. 2006;281:10365–73. doi: 10.1074/jbc.M513094200. [DOI] [PubMed] [Google Scholar]
- Natali PG, Nicotra MR, Di Renzo MF, Prat M, Bigotti A, Cavaliere R, Comoglio PM. Br J Cancer. 1993;68:746–50. doi: 10.1038/bjc.1993.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka D, Chiriboga L, Rubin BP. Am J Surg Pathol. 2008;32:1291–8. doi: 10.1097/PAS.0b013e3181658c14. [DOI] [PubMed] [Google Scholar]
- Peirano RI, Goerich DE, Riethmacher D, Wegner M. Mol Cell Biol. 2000;20:3198–209. doi: 10.1128/mcb.20.9.3198-3209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planque N, Leconte L, Coquelle FM, Martin P, Saule S. J Biol Chem. 2001;276:29330–7. doi: 10.1074/jbc.M101812200. [DOI] [PubMed] [Google Scholar]
- Plummer RS, Shea CR, Nelson M, Powell SK, Freeman DM, Dan CP, Lang D. Mod Pathol. 2008;21:525–30. doi: 10.1038/modpathol.3801019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterf SB, Furumura M, Dunn KJ, Arnheiter H, Pavan WJ. Hum Genet. 2000;107:1–6. doi: 10.1007/s004390000328. [DOI] [PubMed] [Google Scholar]
- Pozner-Moulis S, Pappas DJ, Rimm DL. Cancer Res. 2006;66:7976–82. doi: 10.1158/0008-5472.CAN-05-4335. [DOI] [PubMed] [Google Scholar]
- Puri N, Ahmed S, Janamanchi V, Tretiakova M, Zumba O, Krausz T, Jagadeeswaran R, Salgia R. Clin Cancer Res. 2007;13:2246–53. doi: 10.1158/1078-0432.CCR-06-0776. [DOI] [PubMed] [Google Scholar]
- Saitoh K, Takahashi H, Sawada N, Parsons PG. J Pathol. 1994;174:191–9. doi: 10.1002/path.1711740308. [DOI] [PubMed] [Google Scholar]
- Scholl FA, Kamarashev J, Murmann OV, Geertsen R, Dummer R, Schafer BW. Cancer Res. 2001;61:823–6. [PubMed] [Google Scholar]
- Southard-Smith EM, Kos L, Pavan WJ. Nat Genet. 1998;18:60–4. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- Takayama H, LaRochelle WJ, Sharp R, Otsuka T, Kriebel P, Anver M, Aaronson SA, Merlino G. Proc Natl Acad Sci U S A. 1997;94:701–6. doi: 10.1073/pnas.94.2.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verastegui C, Bille K, Ortonne JP, Ballotti R. J Biol Chem. 2000;275:30757–60. doi: 10.1074/jbc.C000445200. [DOI] [PubMed] [Google Scholar]
- Watanabe A, Takeda K, Ploplis B, Tachibana M. Nat Genet. 1998;18:283–6. doi: 10.1038/ng0398-283. [DOI] [PubMed] [Google Scholar]
- Wei Q, Miskimins WK, Miskimins R. J Neurosci Res. 2004;78:796–802. doi: 10.1002/jnr.20342. [DOI] [PubMed] [Google Scholar]
- Yang G, Li Y, Nishimura EK, Xin H, Zhou A, Guo Y, Dong L, Denning MF, Nickoloff BJ, Cui R. Mol Cell. 2008;32:554–63. doi: 10.1016/j.molcel.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Zhang X, Li Y, Dai C, Yang J, Mundel P, Liu Y. Am J Physiol Renal Physiol. 2003;284:F82–94. doi: 10.1152/ajprenal.00200.2002. [DOI] [PubMed] [Google Scholar]
- Zhu L, Lee HO, Jordan CS, Cantrell VA, Southard-Smith EM, Shin MK. Nat Genet. 2004;36:732–7. doi: 10.1038/ng1371. [DOI] [PubMed] [Google Scholar]