Abstract
Reports of novel emerging and resurging wildlife and zoonotic diseases have increased. Consequently, integration of pathogen sampling into wildlife monitoring programs has grown. Sampling frequency influences interpretations of coupled host–pathogen dynamics, with direct implication to human exposure risk, but has received little empirical attention. To address this, a 15-year study, based on monthly sampling, of deer mouse (Peromyscus maniculatus) populations and Sin Nombre virus (SNV; a virulent disease in humans) dynamics was evaluated. Estimates of deer mouse abundance, number infected with SNV, and SNV prevalence from sampling less frequently than each month (achieved by deletion of months and recalculation of these parameters) were compared to monthly sampling frequencies. Deer mouse abundance was underestimated (10%–20%), SNV prevalence was overestimated when prevalence was high (>15%), and fewer annual extremes of abundance and infection were detected when sampling frequency was less than monthly. Effort necessary to detect temporal dynamics of SNV differed from effort to detect demographic patterns in deer mouse abundance. Findings here are applicable to sampling strategies for other host–pathogen dynamics and have direct implications for allocation of public health resources and intervention programs.
Key Words: emerging infectious disease, hantavirus, Peromyscus maniculatus, population dynamics, sampling interval, Sin Nombre virus, wildlife diseases, zoonoses
Introduction
Reports of novel emerging and resurging wildlife and zoonotic infectious diseases have increased in recent decades (Berger et al. 1998, Jones et al. 2008, Blehert et al. 2009). Consequently, interest in these types of diseases among ecologists, wildlife biologists, and, in the case of zoonoses, public health scientists and practitioners has grown (Gortazar et al. 2007). In relevant cases, sampling of pathogens has been integrated into existing and newly established wildlife-monitoring programs: for example, Sin Nombre virus (SNV) (Mills et al. 1999a, 1999b), Ross River virus (Carver et al. 2008), and amphibian chytrid fungus (Berger et al. 1998, Bell et al. 2004). Despite direct implication to human exposure risk and health, however, the effect of sampling frequency on coupled interpretations of wildlife and zoonotic pathogen dynamics has received little empirical attention.
Here we evaluate how a reduction in sampling frequency influences our ability to detect fluctuations in abundance of deer mice, Peromyscus maniculatus; the infection dynamics of a directly transmitted zoonotic virus, SNV (Bunyaviridae: Hantavirus; an agent of hantavirus pulmonary syndrome [HPS] that causes severe morbidity and mortality in humans); and implications this may have to interpretation of human exposure risk. Populations of hosts that are capable of high levels of fecundity, such as deer mice, can exhibit extreme changes in abundance over brief periods (Krebs 1966, 1996, Singleton 1989). The use of infrequent sampling (i.e., seasonal or semiannual) to monitor population abundance of small mammals, such as deer mice, is common (e.g., Saitoh et al. 1999, Strann et al. 2002, Flowerdew et al. 2004, Larsen et al. 2007). In some instances studies using infrequent sampling of small mammals also extend to examination of wildlife (e.g., Cavanagh et al. 2004) or zoonotic pathogens (e.g., Carver et al. 2008) among individuals comprising reservoir populations, such as for hantaviruses in their rodent hosts (Biggs et al. 2000a, 2000b, Escutenaire et al. 2000, Olsson et al. 2003, Pearce-Duvet et al. 2006).
Potentially, the frequency of sampling necessary to describe dynamics of deer mouse populations may differ from the frequency needed to describe dynamics of SNV infection. For example, bi-monthly trapping of deer mice may adequately detect their population fluctuations [due to an approximate 1–2-month lag between the birth of pups and first detection by trapping (King et al. 1963, Kirkland and Layne 1989)], whereas a monthly trapping interval may be necessary to detect the dynamics of SNV transmission [due to a 2–4-week time interval between infection and detection of SNV IgG antibody in deer mice (Botten et al. 2000, 2002)]. Accordingly, it is possible that the sampling interval that is adequate to quantify deer mouse population demographics may be longer than what is necessary to capture dynamics of SNV infection. If the frequency of sampling does not adequately capture both deer mouse population demographics and SNV dynamics, it could lead to erroneous conclusions about host–pathogen relationships and, by extension, sub-optimal management and health interventions.
We use a longitudinal study of deer mouse populations and prevalence of antibody to SNV, which we have conducted monthly (1994–2008) at three trapping grids near Cascade, Montana (Douglass et al. 1996, 2001). We simulate variable sampling frequencies by selective deletion of sampling occasions from this study, and recalculation of deer mouse abundance and prevalence of antibody to SNV. We ask three questions: (1) How does a reduction in sampling frequency influence our ability to detect fluctuations in deer mouse populations and SNV infection dynamics in the field? (2) Can a sampling schedule, which adequately represents the dynamics of a host (deer mice), be generalized to a pathogen (SNV)? (3) How frequently are annual peaks and troughs in population abundance and infection prevalence missed by sampling less frequently? The direct implication of these results to investment in public health campaigns or interventions and human HPS exposure risk is discussed.
Materials and Methods
This investigation was conducted on three live trapping grids (grid numbers 10, 11, and 12) located near Cascade (46°59.3′ N, 111°35.3′ W, 1408 m Average Mean Sea Level (AMS)), Montana. The trapping grids were situated in grassland habitat supporting an active cattle ranch (Douglass et al. 1996). Deer mice were live trapped for three consecutive nights each month on all three grids for 174 consecutive months between June 1994 and November 2008. Trapping grids were 1 hectare in area and consisted of 100 equally spaced Sherman live traps (H.B. Sherman Traps., Tallahassee, FL), baited with rolled oats and peanut butter and provisioned with polyester Fiberfil bedding. Upon capture, each rodent was given a uniquely numbered model #1005-1 ear tag (National Band and Tag Co., Newport, KY), and their species, sex, body mass, reproductive condition, and presence of scars or wounds were recorded. Blood samples, which were later tested for antibodies to SNV, were routinely collected from grids 11 and 12 only. We followed animal handling, blood collection, and safety precautions described by Mills et al. (1995) and approved by the University of Montana Institutional Animal Care and Use Committee. Serological analyses were performed at the Montana State Public Health Laboratory and at Special Pathogens Branch, U.S. Centers for Disease Control and Prevention.
We used the minimum number of individuals known to be alive (MNA) during a 3-day trapping session as an index of population abundance for each month (cf. Krebs 1966). The minimum number of deer mice antibody positive to SNV (MNI), during each trap session, was calculated in the same way as MNA for grids 11 and 12. Estimated standing prevalence (ESP) of deer mice for a given month was calculated by dividing MNI by MNA (Mills et al. 1999a). Less frequent sampling (e.g, bi-annual or annual), and consequently a lack of recaptures, precluded the use of more complex population estimates of deer mouse abundance and the number of deer mice infected. Because hantaviruses cause chronic (likely life-time) infection in their hosts, antibody is considered a marker of infection (Mills et al. 1999a). We consider antibody-positive deer mice to be actively infected and likely infectious (shedding infectious virus).
Analyses
To examine how well estimates of MNA, MNI, and ESP from less frequent sampling intervals predict the same estimates from monthly sampling, we recalculated MNA, MNI, and ESP at sampling intervals of bi-monthly, quarterly, semiannually, and annually. To calculate/simulate MNA, MNI, and ESP from these less frequent sampling regimes, we temporarily deleted trapping sessions from our dataset and recalculated MNA, MNI, and ESP in each instance.
Dates for less frequent sampling, particularly semiannually and annually, were focused on times of year that were of relevance to researchers studying the population dynamics of deer mice and the transmission of SNV. For bi-monthly sampling, every 2 months from February was chosen. For quarterly sampling the middle month of each season was chosen: January, April, July, and October. Months chosen for semiannual sampling were April, when deer mouse ESP is high, and October, when deer mouse abundance is high. April was chosen as a relevant month for annual sampling, because of elevated ESP around this time of the year.
To examine how much a reduction in sampling frequency underestimated monthly values of MNA and MNI, we evaluated the percent difference between less frequent and monthly estimates:
 |
where n is the number of sampling occasions derived from our database (bi-monthly 86, quarterly 58, semiannually 29, and annually 14), l is the estimate of MNA or MNI derived from less frequent sampling, and m is the monthly estimate of MNA or MNI.
To evaluate the effect of less frequent sampling on estimates of ESP, we compared ESP estimates calculated by sampling less frequently to three measures of ESP from monthly sampling: ESP derived from monthly sampling, mean ESP over the period not captured by sampling less frequently, and weighted mean. Mean ESP for bi-monthly sampling was calculated over the month the less frequent sample was obtained and the month previous. Mean ESP for quarterly sampling was calculated over the 3 months of each season (December–February, March–May, June–August, September–November). Mean ESP for semiannual values consisted of the previous and current season (December–May and June–November). Mean annual ESP was calculated over the two seasons previous through to the end of the season following (September–August). Weighted mean ESP was the total MNI across the months (as described for mean ESP) divided by the total MNA. Paired t-tests were employed to determine if ESP from less frequent sampling differed from our three measures of monthly ESP.
Results
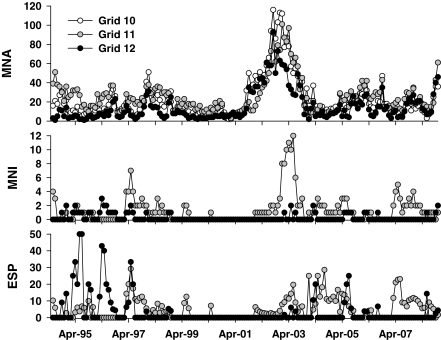
Monthly deer mouse MNA, MNI, and ESP fluctuated throughout the study period (Fig. 1). A large increase in abundance of deer mice was recorded between late 2001 and 2003, and the MNI of deer mice on grid 11 was also higher in 2003 (Fig. 1). There was a greater abundance of infected deer mice detected on grid 11 than on grid 12 (Fig. 1).
FIG. 1.
Monthly abundance (MNA), number of infected individuals (MNI), and estimated standing prevalence (ESP) of deer mice, Peromyscus maniculatus, between June 1994 and November 2008.
Across all three grids, a reduction in the frequency of sampling (even bi-monthly sampling) of deer mice resulted in an underestimation of monthly MNA by at least 10% (Table 1). The underestimation of monthly MNA grew to 13%–20% when the sampling frequency was reduced beyond bi-monthly. Monthly MNI of deer mice at grid 11 was also underestimated as the frequency of sampling was reduced, but by only a small amount, compared to MNA (Table 1). Monthly MNI was never underestimated at grid 12, due to a high frequency of months where no infected deer mice were detected (Fig. 1, Table 1).
Table 1.
Mean Percent Difference (±1 Standard Error) Between Monthly Estimate Values of Deer Mouse Abundance (MNA) and Number of Infected Deer Mice (MNI) and Values from Less Frequent Sampling
| Grid | 10 MNA | 11 MNA | 12 MNA | 11 MNI | 12 MNI |
|---|---|---|---|---|---|
| Bi-monthly | −10.34 (0.14) | −10.64 (0.14) | −11.48 (0.18) | −1.80 (0.13) | 0.00 (0.00) |
| Quarterly | −18.13 (0.32) | −16.77 (0.33) | −20.58 (0.42) | −1.08 (0.12) | 0.00 (0.00) |
| Semiannually | −14.32 (0.42) | −13.60 (0.49) | −15.97 (0.61) | −1.46 (0.22) | 0.00 (0.00) |
| Annually | −15.00 (0.58) | −14.30 (0.64) | −17.30 (0.82) | −0.65 (0.17) | 0.00 (0.00) |
Negative values indicate that deer mouse abundance or number of deer mice infected (MNI) were underestimated.
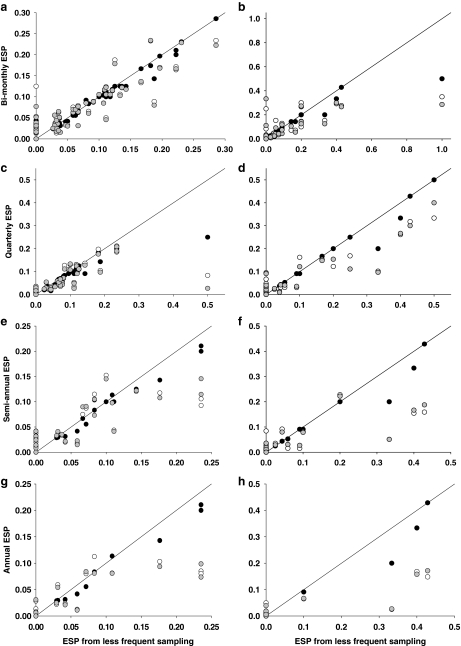
Values of ESP from less frequent sampling tended to overestimate measures of ESP from monthly sampling (Table 2, Fig. 2). Overestimation of ESP resulted from relatively stable MNI values, but a smaller MNA values (i.e., the numerator was relatively constant, but the denominator was generally smaller; Table 1) when sampling frequency was less than monthly. This pattern was particularly evident at quarterly, semiannual, and annual sampling intervals (Fig. 2c–h), especially when ESP was >0.15. These overestimates were only statistically significant, however, for monthly estimates at grid 11 (Table 2), which produced a higher number of infected mice throughout the study.
Table 2.
Mean% Difference (±1 Standard Error) and Paired t-Test of Estimated Standing Prevalence from Comparison of Less Frequent Sampling with Monthly Estimates of Estimated Standing Prevalence (Monthly, Mean, and Weighted Mean)
| |
Grid 11 |
Grid 12 |
||||
|---|---|---|---|---|---|---|
| Monthly | Mean | Weighted mean | Monthly | Mean | Weighted mean | |
| Bi-monthly | ||||||
| % difference | 0.221 | −0.021 | 0.098 | 0.872 | 1.036 | 1.033 |
| SE | 0.092 | 0.294 | 0.277 | 0.604 | 0.914 | 1.010 |
| t1,84 | 2.413 | 0.073 | 0.355 | 1.435 | 1.127 | 1.017 |
| P | 0.018 | 0.942 | 0.724 | 0.155 | 0.263 | 0.312 |
| Quarterly | ||||||
| % difference | 1.048 | 1.233 | 1.394 | 0.371 | 1.050 | 0.893 |
| SE | 0.445 | 0.772 | 0.863 | 0.255 | 0.650 | 0.661 |
| t1,56 | 2.355 | 1.597 | 1.615 | 1.440 | 1.601 | 1.340 |
| p | 0.022 | 0.116 | 0.112 | 0.155 | 0.115 | 0.186 |
| Semiannually | ||||||
| % difference | 0.652 | 0.752 | 0.683 | 0.742 | 2.303 | 2.335 |
| SE | 0.251 | 0.842 | 0.734 | 0.505 | 1.637 | 1.516 |
| t1,26 | 2.555 | 0.879 | 0.913 | 1.443 | 1.382 | 1.514 |
| p | 0.016 | 0.387 | 0.369 | 0.160 | 0.178 | 0.141 |
| Annually | ||||||
| % difference | 0.965 | 2.602 | 2.887 | 1.494 | 5.439 | 5.319 |
| SE | 0.361 | 1.633 | 1.576 | 1.027 | 3.230 | 3.167 |
| t1,12 | 2.579 | 1.535 | 1.765 | 1.402 | 1.622 | 1.618 |
| p | 0.023 | 0.149 | 0.101 | 0.184 | 0.129 | 0.130 |
Positive and negative values indicate that ESP was on average overestimated and underestimated, respectively. Significant differences are shown in bold. ESP, estimated standing prevalence; SE, standard error.
FIG. 2.
The relationship of Estimated Standing Prevalence (ESP) calculated from less frequent sampling (X-axis) to ESP values derived from monthly sampling (Y-axis: closed circles, monthly ESP; open circles, mean ESP; shaded circle, weighted mean ESP) for grids 11 (a, c, e, g) and 12 (b, d, f, h). The solid line represents a perfect one-to-one linear relationship between less frequent and monthly ESP.
On the basis of monthly sampling, the highest annual MNA of deer mice was recorded most frequently between August and October and least frequently between April and June (Table 3). The most common months for which lows in MNA were observed were less consistent among trapping grids (Table 3). We did not detect any infection in deer mice in one of the 14 years of this study at grid 11, and 4 of the 14 years at grid 12. The maximum annual MNI was most frequently observed between March and June at grids 11 and 12, but annual highs in MNI were also observed in other months, including winter (Table 3). Lows in MNI (often zero) were recorded more frequently in the autumn and winter months, although annual lows in MNI were often recorded in other months as well (Table 3). Maximum annual ESP was also observed in many months, including winter (Table 3). The highest annual ESP, at both grids, was most commonly observed in May or June (Table 3). Similar to MNI, lows in ESP were frequently observed across most months, but were most common in autumn and winter (Table 3).
Table 3.
Percentage of Annual Highs and Lows in Deer Mouse Abundance (MNA), Number of Infected Deer Mice (MNI) and Estimated Standing Prevalence of Deer Mice, Peromyscus maniculatus, Detected at Each Month, for Each Grid Between June 1994 and January 2009
| Grid | 10 MNA | 11 MNA | 12 MNA | 11 MNI | 12 MNI | 11 ESP | 12 ESP |
|---|---|---|---|---|---|---|---|
| Highs | |||||||
| January | 7 | 21 | 0 | 7 | 29 | 14 | 29 |
| February | 7 | 0 | 0 | 7 | 36 | 14 | 29 |
| March | 7 | 7 | 7 | 14 | 43 | 7 | 29 |
| April | 0 | 0 | 0 | 21 | 43 | 7 | 43 |
| May | 0 | 0 | 0 | 64 | 50 | 29 | 43 |
| June | 0 | 0 | 7 | 29 | 43 | 29 | 43 |
| July | 0 | 7 | 7 | 7 | 36 | 14 | 43 |
| August | 21 | 21 | 21 | 14 | 43 | 14 | 36 |
| September | 21 | 21 | 14 | 14 | 29 | 7 | 29 |
| October | 21 | 21 | 14 | 29 | 36 | 21 | 36 |
| November | 7 | 0 | 7 | 7 | 36 | 7 | 29 |
| December | 7 | 14 | 21 | 21 | 43 | 14 | 29 |
| Lows | |||||||
| January | 7 | 14 | 7 | 64 | 100 | 57 | 100 |
| February | 14 | 14 | 7 | 71 | 86 | 79 | 86 |
| March | 0 | 0 | 0 | 43 | 64 | 36 | 64 |
| April | 7 | 7 | 0 | 29 | 71 | 29 | 71 |
| May | 0 | 0 | 0 | 14 | 71 | 14 | 64 |
| June | 7 | 0 | 21 | 36 | 64 | 36 | 64 |
| July | 29 | 14 | 29 | 36 | 71 | 29 | 71 |
| August | 21 | 7 | 14 | 43 | 79 | 43 | 79 |
| September | 21 | 7 | 21 | 57 | 93 | 50 | 93 |
| October | 7 | 7 | 0 | 64 | 71 | 57 | 71 |
| November | 0 | 21 | 14 | 79 | 86 | 71 | 86 |
| December | 0 | 29 | 0 | 64 | 86 | 43 | 86 |
When sampling frequency was reduced, fewer annual highs and lows in MNA were detected (Table 4). Similarly, the percentage of highs detected in annual MNI and ESP also declined with less frequent sampling (Tables 3 and 4). The capacity to detect annual lows in MNI and ESP, however, remained high, regardless of reduced sampling frequency, reflecting a relatively high number of months in which no infected deer mice were captured (Tables 3 and 4).
Table 4.
Percentage of Annual Highs and Lows in Abundance (MNA), Number Infected (MNI), and Estimated Standing Prevalence of Deer Mice, Peromyscus maniculatus, Detected by Less Frequent Sampling, for Each Grid from June 1994 to January 2009
| Grid | 10 MNA | 11 MNA | 12 MNA | 11 MNI | 12 MNI | 11 ESP | 12 ESP |
|---|---|---|---|---|---|---|---|
| Highs | |||||||
| Bi-monthly | 57 | 64 | 64 | 71 | 79 | 64 | 79 |
| Quarterly | 29 | 50 | 21 | 43 | 57 | 36 | 64 |
| Semiannual | 21 | 21 | 14 | 43 | 50 | 21 | 50 |
| Annual | 0 | 0 | 0 | 21 | 43 | 7 | 43 |
| Low | |||||||
| Bi-monthly | 50 | 64 | 43 | 100 | 100 | 100 | 100 |
| Quarterly | 50 | 43 | 36 | 79 | 100 | 71 | 100 |
| Semiannual | 14 | 14 | 0 | 79 | 79 | 71 | 79 |
| Annual | 7 | 7 | 0 | 29 | 71 | 29 | 71 |
Discussion
In general, greater sampling effort or frequency is perceived to result in more accurate quantification of organism dynamics over time than sampling less frequently. Here we used a longitudinal study of deer mice and SNV to examine how a reduction in the frequency of sampling can influence detection of coupled host population demographics and pathogen infection. In general, sampling less frequently (bi-monthly, quarterly, semiannually, and annually) produced underestimates (10%–20%) of deer mouse abundance (MNA) derived from monthly sampling. Overall estimates of the number of infected deer mice (MNI) and standing infection prevalence (ESP), detected by less frequent sampling, were similar to monthly MNI and ESP values (monthly, mean, and weighted mean). However, monthly values for ESP were predominantly overestimated when values of ESP from less frequent sampling were high (particularly when >0.15). As such the effect of sampling less frequently than each month had a nonlinear effect on estimates of infection (i.e., less frequent sampling produced similar estimates of ESP as monthly when values were ≤0.10, but monthly ESP was frequently overestimated above this value).
Overestimation of high levels of ESP is of direct interest and application to human HPS exposure risk. Human incidence of HPS in Montana is not detected when ESP of deer mice, from monthly sampling, is below 10% (Madhav et al. 2007), suggesting that investment in public health awareness programs or interventions are best targeted when deer mouse ESP exceeds this value. This study demonstrates that if this knowledge were currently applied to direct health awareness programs or interventions under a less frequent sampling regime (such as would be the case with semiannual and annual sampling frequencies), values of deer mouse ESP exceeding 10% would be overestimated and would result in premature investment in public health campaigns or interventions. Conversely, intervention when it may not be necessary is far superior, in terms of public health, to inaction. Of course, less frequent sampling is also likely to delay detection of epizootic conditions.
Resources to invest in host–pathogen monitoring programs are likely limited in many instances. As such, short-term studies, with limited spatial replication, which seek general information about host abundance and pathogen dynamics over time, should be interpreted with caution. We observed substantial fluctuations in deer mouse MNA, MNI, and ESP over 14 years, a feature characteristic of highly fecund animals (Krebs 1966, 1996, Singleton 1989, Carver et al. 2008). However, SNV dynamics did not always reflect deer mouse abundance on the same grid and were not always similar between grids. These findings support those of Douglass et al. (2001), who found large spatial and temporal variation in MNA (near 0 to over 120 per 100-trap grid) and MNI (0 to over 60) among 18 grids in Montana. Despite the variable nature of SNV over time, prevalence among deer mouse populations tends to peak in spring, across disparate geographic locations and variable habitat types (Douglass et al. 2001, Mills et al. 1999b and references therein). While it was not the specific focus of this study, understanding processes driving population fluctuations of deer mice (e.g., survival and dispersal) would likely contribute to a better understanding of the dynamics of infection (Douglass et al. 2007, Madhav et al. 2007, Lonner et al. 2008, Luis et al. 2010). Our results indicate that to capture natural variation in deer mouse abundance and dynamics of SNV, surveys need to be spatially replicated and of long duration (Douglass et al. 2001). We intentionally based our less frequent sampling, particularly semiannual and annual sampling, on prior knowledge of deer mouse abundance and SNV dynamics at our grids. We acknowledge that someone establishing a new sampling regime would not have the benefit of this prior knowledge, likely resulting in the detection of fewer annual minima and maxima of MNA, MNI, and ESP. Investigations, with limited resources, that aim to understand spatial variation in host populations and risk of zoonotic pathogen exposure (without a priori knowledge of host–pathogen dynamics) should first seek to understand annual variation in infection and then target spatial surveys during seasons when infection is at a maxima. In such cases, investigators should keep in mind that estimates of high infection prevalence are likely to be inflated. As a corollary, estimates of high infection prevalence could be improved by increasing sampling frequency during periods (epizootics) where ESP exceeds 10%.
In this study, underestimation of deer mouse abundance is important in understanding SNV infection dynamics, because this inflates estimates of ESP. In general, determining the sampling interval to adequately estimate host abundance is likely to depend on the developmental rate of juveniles and the duration and magnitude of natural population fluctuations (Krebs and Myers 1974). For example, there is a 1–2-month lag between the birth of pups and first detection of deer mice by trapping (King et al. 1963, Kirkland and Layne 1989). We have trapped deer mice monthly and this appears to have been a reasonable frequency to capture abundance and follow temporal fluctuations in their abundance, a conclusion also supported by Parmenter et al. (1999). Naturally, the ability to document fluctuations in population abundance and to investigate processes that underlie fluctuations (e.g., survival and recruitment) diminishes as the frequency of sampling decreases (Krebs and Myers 1974). Similar to our findings, that sampling less frequently than monthly leads to underestimates of MNA, attempts to model temporal fluctuations in deer mouse population abundance, using a range of predictors (e.g., temperature, precipitation, survival, and maturation), have also required a monthly sampling to acquire any level of accuracy (Yaffee et al. 2008, Luis et al. 2010). Further, accurate estimation of population demographic parameters [i.e., survival, recruitment (Krebs and Myers 1974), and dispersal (Lonner et al. 2008, Waltee et al. 2009)], all necessary to understand disease dynamics (Keeling and Rohani 2008), require frequent sampling. Understanding the dynamics of SNV among deer mice would greatly benefit from investigations, based on frequent sampling, which link environmental determinants of deer mouse abundance (Luis et al. 2010) and infection.
Detecting annual extremes in host or vector abundance, particularly maxima, is an important facet in predicting infection prevalence of many diseases, including SNV, reflecting the density- or frequency-dependent nature of transmission (Mills et al. 1999b, Madhav et al. 2007, Keeling and Rohani 2008, Carver et al. 2009 and references therein). Similarly, detecting annual extremes, particularly maxima, in infection is important to allocate public-health-related resources. Not surprisingly, a reduction in the frequency of sampling resulted in a consistent reduction in the detection of annual maxima of MNA, MNI, and ESP, and minima of MNA. It should also be acknowledged that annual maxima in deer mouse abundance and infection are observed in winter with reasonable frequency. For example, Calisher et al. (2005) documented a 5-year high in the trap success of deer mice in the winter of 1999. Over the 14 years of our study, we were able to access trapping grids each month, even when environmental conditions, such as deep snow, made site access and trapping challenging. Although trapping in winter is not always possible due to inaccessibility of trapping locations (e.g., Douglass et al. 2001, Pearce-Duvet et al. 2006), investigators should recognize that the detection of annual highs in host abundance and infection is reduced when sampling is less frequent than monthly, and when sampling is not undertaken during winter. Nevertheless, this study suggests that annual highs in deer mouse MNA are most frequently detected between August and October and ESP between April and August, likely reflecting a delayed transmission relationship between deer mouse abundance and infection (Madhav et al. 2007). Our results indicate that missed maxima in host abundance and infection prevalence (such as would likely be the case with semiannual or annual sampling frequencies) would result in inflated predictions and underestimation of true infection prevalence, respectively. This information is of direct relevance to the timing and allocation of management and public health resources.
Conclusions
SNV was first detected as an emerging pathogen following an epidemic of HPS in the Four Corners region of the United States in 1993 (Nichol et al. 1993), and in recent years the number of recognized emergence events of other zoonotic and wildlife infectious diseases has increased (Jones et al. 2008). For example, Nipah virus (reviewed by Mackenzie et al. 2001), chytridiomycosis in amphibians (Berger et al. 1998, Bell et al. 2004), and bat white-nose syndrome (Blehert et al. 2009). Increasingly, ecologists and wildlife biologists are challenged with considering the dynamics of pathogens in addition to the sampling of wildlife. This study suggests that the sampling effort necessary to identify basic demographic patterns in deer mouse populations and dynamics of SNV over time differs. Findings in this study are applicable to implementation of sampling strategies for other hosts and their pathogens. Further, sampling frequency of zoonotic pathogens and their hosts has important implications for the allocation of public health resources and intervention programs. Future research aimed at measuring the data lost by a reduction in sampling frequency would likely benefit from comparing monthly abundance and infection data to models using estimates of survivability and recruitment to predict monthly deer mouse abundance and infection.
Acknowledgments
We thank the private ranch owner at Cascade for allowing us access to his property. Numerous individuals provided valuable assistance in the field, including K. Coffin, R. Van Horn, C. Rognli, T. Wilson, W. Semmens, K. Hughes, A. Skypala, D. Waltee, B. Lonner, J. Wilson, A. Leary, J. Bertoglio, A. Alvarado, K. Richardson, and F. Arneson. K. Wagner provided database support, encouragement, and general advice. S. Zantos provided valuable laboratory assistance. Financial support was provided by NIH Grant P20 RR16455-06-07,08 from the INBRE-BRIN program of the National Center for Research Resources and the U.S. Centers for Disease Control and Prevention, Atlanta, GA, through cooperative agreement. This work followed all relevant environmental and institutional regulations in the collection of data presented here. The findings and conclusions presented here are those of the authors and do not necessarily represent the views of the funding agencies.
Disclosure Statement
No competing financial interests exist.
References
- Bell BD. Carver S. Mitchell NJ. Pledger S. The dramatic decline of a New Zealand endemic: how and why did populations of Archey's frog Leiopelma archeyi crash over 1996–2001? Biol Conserv. 2004;120:193–203. [Google Scholar]
- Berger L. Speare R. Daszak P. Green DE, et al. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc Natl Acad Sci USA. 1998;95:9031–9036. doi: 10.1073/pnas.95.15.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs JR. Bennett KD. Mullen MA. Haarmann TK, et al. Relationship of ecological variables to Sin Nombre virus antibody seroprevalence in populations of deer mice. J Mammal. 2000a;81:676–682. [Google Scholar]
- Biggs JR. Bennett KD. Torrez-Martinez N. Hjelle BL. Sin nombre virus antibody prevalence in rodents of north-central New Mexico. Southwest Nat. 2000b;45:61–66. [Google Scholar]
- Blehert DS. Hicks AC. Behr M. Meteyer CU, et al. Bat white-nose syndrome: an emerging fungal pathogen? Science. 2009;323:227. doi: 10.1126/science.1163874. [DOI] [PubMed] [Google Scholar]
- Botten J. Mirowsky K. Kusewitt D. Bharadwaj M, et al. Experimental infection model for Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus) Proc Natl Acad Sci USA. 2000;97:10578–10583. doi: 10.1073/pnas.180197197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botten J. Mirowsky K. Ye CY. Gottlieb K, et al. Shedding and intracage transmission of Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus) model. J Virol. 2002;76:7587–7594. doi: 10.1128/JVI.76.15.7587-7594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher CH. Root JJ. Mills JN. Rowe JE, et al. Epizootiology of Sin Nombre and El Moro Canyon hantaviruses, southeastern Colorado, 1995–2000. J Wildl Dis. 2005;41:1–11. doi: 10.7589/0090-3558-41.1.1. [DOI] [PubMed] [Google Scholar]
- Carver S. Bestall A. Jardine A. Ostfeld RS. The influence of hosts on the ecology of arboviral transmission: potential mechanisms influencing dengue, Murray Valley encephalitis and Ross River virus in Australia. Vector Borne Zoonot Dis. 2009;9:51–64. doi: 10.1089/vbz.2008.0040. [DOI] [PubMed] [Google Scholar]
- Carver S. Sakalidis V. Weinstein P. House mouse abundance and Ross River virus notifications in Victoria, Australia. Int J Infect Dis. 2008;12:528–533. doi: 10.1016/j.ijid.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Cavanagh RD. Lambin X. Ergon T. Bennett M, et al. Disease dynamics in cyclic populations of field voles (Microtus agrestis): cowpox virus and vole tuberculosis (Mycobacterium microti) Proc Roy Soc Lond B Biol. 2004;271:859–867. doi: 10.1098/rspb.2003.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass RJ. Calisher CH. Wagoner KD. Milis JN. Sin Nombre virus infection of deer mice in Montana: characteristics of newly infected mice, incidence, and temporal pattern of infection. J Wildl Dis. 2007;43:12–22. doi: 10.7589/0090-3558-43.1.12. [DOI] [PubMed] [Google Scholar]
- Douglass RJ. VanHorn R. Coffin KW. Zanto SN. Hantavirus in Montana deer mouse populations: preliminary results. J Wildl Dis. 1996;32:527–530. doi: 10.7589/0090-3558-32.3.527. [DOI] [PubMed] [Google Scholar]
- Douglass RJ. Wilson T. Semmens WJ. Zanto SN, et al. Longitudinal studies of Sin Nombre virus in deer mouse-dominated ecosystems of Montana. Am J Trop Med Hyg. 2001;65:33–41. doi: 10.4269/ajtmh.2001.65.33. [DOI] [PubMed] [Google Scholar]
- Escutenaire S. Chalon P. Verhagen R. Heyman P, et al. Spatial and temporal dynamics of Puumala hantavirus infection in red bank vole (Clethrionomys glareolus) populations in Belgium. Virus Res. 2000;67:91–107. doi: 10.1016/s0168-1702(00)00136-2. [DOI] [PubMed] [Google Scholar]
- Flowerdew JR. Shore RF. Poulton SMC. Sparks TH. Live trapping to monitor small mammals in Britain. Mammal Rev. 2004;34:31–50. [Google Scholar]
- Gortazar C. Ferroglio E. Hofle U. Frolich K, et al. Diseases shared between wildlife and livestock: a European perspective. Eur J Wildl Res. 2007;53:241–256. [Google Scholar]
- Jones KE. Patel NG. Levy MA. Storeygard A, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling MJ. Rohani P. Modeling Infectious Diseases in Humans and Animals. Princeton, NJ: Princeton University Press; 2008. [Google Scholar]
- King JA. Deshaies JC. Webster R. Age of weaning in two subspecies of deer mice. Science. 1963;139:483–484. doi: 10.1126/science.139.3554.483. [DOI] [PubMed] [Google Scholar]
- Kirkland GL. Layne JN. Advances in the study of Peromyscus (Rodentia) Lubbock: Texas Tech University Press; 1989. [Google Scholar]
- Krebs CJ. Demographic changes in fluctuating populations of Microtus californicus. Ecol Monogr. 1966;36:239–273. [Google Scholar]
- Krebs CJ. Population cycles revisited. J Mammal. 1996;77:8–24. [Google Scholar]
- Krebs CJ. Myers JM. Population cycles in small mammals. Adv Ecol Res. 1974;8:267–399. [Google Scholar]
- Larsen KW. Adams IT. Haughland DL. Small mammal communities in a pyrogenic habitat mosaic. Int J Wildland Fire. 2007;16:728–740. [Google Scholar]
- Lonner BN. Douglass RJ. Kuenzi AJ. Hughes K. Seroprevalence against Sin Nombre virus in resident and dispersing deer mice. Vector Borne Zoonot Dis. 2008;8:433–442. doi: 10.1089/vbz.2007.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis AD. Douglass RJ. Mills JN. Bjornstad ON. The effect of seasonality, density and climate on the population dynamics of Montana deer mice, important reservoir hosts for Sin Nombre hantavirus. J Anim Ecol. 2010;79:462–470. doi: 10.1111/j.1365-2656.2009.01646.x. [DOI] [PubMed] [Google Scholar]
- Mackenzie JS. Chua KB. Daniels PW. Eaton BT, et al. Emerging viral diseases of Southeast Asia and the Western Pacific. Emerg Infect Dis. 2001;7:497–504. doi: 10.3201/eid0707.017703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhav NK. Wagoner KD. Douglass RJ. Mills JN. Delayed density-dependent prevalence of Sin Nombre virus antibody in Montana deer mice (Peromyscus maniculatus) and implications for human disease risk. Vector Borne Zoonot Dis. 2007;7:353–364. doi: 10.1089/vbz.2006.0605. [DOI] [PubMed] [Google Scholar]
- Mills JN. Ksiazek TG. Peters CJ. Childs JE. Long-term studies of hantavirus reservoir populations in the southwestern United States: a synthesis. Emerg Infect Dis. 1999a;5:135–142. doi: 10.3201/eid0501.990116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JN. Yates TL. Childs JE. Parmenter RR, et al. Guidelines for working with rodents potentially infected with hantavirus. J Mammal. 1995;76:716–722. [Google Scholar]
- Mills JN. Yates TL. Ksiazek TG. Peters CJ, et al. Long-term studies of hantavirus reservoir populations in the southwestern United States: rationale, potential, and methods. Emerg Infect Dis. 1999b;5:95–101. doi: 10.3201/eid0501.990111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichol ST. Spiropoulou CF. Morzunov S. Rollin PE, et al. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- Olsson G. Ahlm C. Elgh F. Verlemyr A, et al. Hantavirus antibody occurrence in bank voles (Clethrionomys glareolus) during a vole population cycle. J Wildl Dis. 2003;39:299–305. doi: 10.7589/0090-3558-39.2.299. [DOI] [PubMed] [Google Scholar]
- Parmenter CA. Yates TL. Parmenter RR. Dunnum JL. Statistical sensitivity for detection of spatial and temporal patterns in rodent population densities. Emerg Infect Dis. 1999;5:118–125. doi: 10.3201/eid0501.990114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce-Duvet JMC. Jeor SCS. Boone JD. Dearing MD. Changes in Sin Nombre virus antibody prevalence in deer mice across seasons: the interaction between habitat, sex, and infection in deer mice. J Wildl Dis. 2006;42:819–824. doi: 10.7589/0090-3558-42.4.819. [DOI] [PubMed] [Google Scholar]
- Saitoh T. Bjornstad ON. Stenseth NC. Density dependence in voles and mice: a comparative study. Ecology. 1999;80:638–650. [Google Scholar]
- Singleton G. Population dynamics of an outbreak of house mouse (Mus domesticus) in the Mallee wheatlands of Australia—hypothesis of plague formation. J Zool. 1989;219:495–515. [Google Scholar]
- Strann KB. Yoccoz NG. Ims RA. Is the heart of Fennoscandian rodent cycle still beating? A 14-year study of small mammals and Tengmalm's owls in northern Norway. Ecography. 2002;25:81–87. [Google Scholar]
- Waltee D. Lonner BN. Kuenzi AJ. Douglass RJ. Seasonal dispersal patterns of sylvan deer mice (Peromyscus maniculatus) within Montana rangelands. J Wildl Dis. 2009;45:998–1007. doi: 10.7589/0090-3558-45.4.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffee RA. Nikolopoulos K. Reilly DP. Crone SF, et al. An experiment in epidemiological forecasting: comparison of forecast accuracies among different methods of forecasting deer mouse population densities in Montana. www.forecasters.org/submissions/ISF2008RobertAYaffeeCDCprojectarticle.pdf Proceedings of the 28th International Symposium on Forecasting, Nice, France. 2008 [Google Scholar]