Abstract
In the title compound, C15H11Cl2NO2, the dihedral angle between the two benzene rings is 74.83 (5)°. The N-bound and terminal benzene rings are inclined at dihedral angles of 4.09 (10) and 78.38 (9)°, respectively, to the mean plane through the acetamide group. Intramolecular C—H⋯O and N—H⋯O hydrogen bonds both generate S(6) rings.
Related literature
For the acetylation reaction, see: Greene et al. (1999 ▶); Gupta et al. (2008 ▶). For solvent-free synthesis, see: Roopan et al. (2008 ▶, 2009 ▶). For reactions of acetic anhydride and acetyl chloride, see: Orita et al. (2000 ▶); Procopiou et al. (1998 ▶). For hydrogen-bond motifs, see: Bernstein et al. (1995 ▶).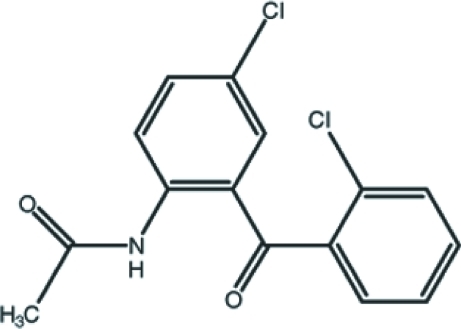
Experimental
Crystal data
C15H11Cl2NO2
M r = 308.15
Monoclinic,
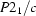
a = 11.1371 (11) Å
b = 5.0661 (6) Å
c = 25.594 (3) Å
β = 100.672 (9)°
V = 1419.1 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.46 mm−1
T = 293 K
0.28 × 0.24 × 0.18 mm
Data collection
Oxford Xcalibur Eos (Nova) CCD detector diffractometer
14628 measured reflections
2633 independent reflections
1537 reflections with I > 2σ(I)
R int = 0.080
Refinement
R[F 2 > 2σ(F 2)] = 0.048
wR(F 2) = 0.107
S = 0.98
2633 reflections
182 parameters
H-atom parameters constrained
Δρmax = 0.18 e Å−3
Δρmin = −0.21 e Å−3
Data collection: CrysAlis PRO CCD (Oxford Diffraction, 2009 ▶); cell refinement: CrysAlis PRO CCD; data reduction: CrysAlis PRO RED (Oxford Diffraction, 2009 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶) and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810018696/tk2672sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810018696/tk2672Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O1 | 0.86 | 1.96 | 2.660 (3) | 138 |
| C5—H5⋯O2 | 0.93 | 2.22 | 2.839 (4) | 124 |
Acknowledgments
We thank the Department of Science and Technology, India, for use of the CCD facility set up under the IRHPA–DST program at IISc, Bangalore. We also thank Professor T. N. Guru Row, IISc, for useful crystallographic discussions. FNK thanks the DST for Fast Track Proposal funding.
supplementary crystallographic information
Comment
The acetylation of phenol, alcohol and amine are important chemical reactions in organic synthesis (Greene et al., 1999, Gupta et al., 2008). Mainly, acylation of amines is used for the protection of an amino functionality in a multi-step synthetic process. Acetic anhydride and acetyl chloride are generally used in the presence of acidic or basic catalysts in an organic medium (Orita et al., 2000; Procopiou et al., 1998). One of the major factors for a green chemical processes are solvent-free reactions. In continuation of our our interest in this area (Roopan et al., 2008, 2009), we herein report the solvent-free acetylation of an amine, leading to the title compound, (I).
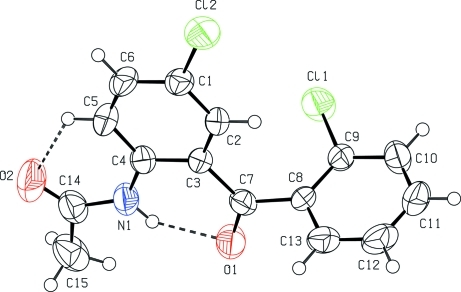
Compound (I), Fig. 1, has two chloro-phenyl groups (Cl2/C1–C6 and Cl1/C8–C13) which make a dihedral angle of 74.83 (5)° with each other. The chloro-phenyl groups are inclined at dihedral angles of 4.09 (10) and 78.38 (9) °, respectively, with the mean plane through the acetamide group (N1/O2/C14/C15). The torsion angles O1—C7—C8—C9, O1—C7—C8—C13, C2—C3—C7—O1 and C4—C3—C7—O1 are 109.0 (3), -68.5 (4), 172.8 (3) and -5.8 (4)°, respectively.
Two intramolecular, i.e. N1—H1···O1 and C5—H5···O2, hydrogen bonds form six-membered rings, producing S(6) ring motifs (Table 1, Fig. 1, Bernstein et al., 1995). In the crystal structure, there are no classical intermolecular hydrogen bonds.
Experimental
2-Amino-5-chloro-phenyl(2-chloro-phenyl)methanone (1 mmol) was stirred with acetyl-chloride (1 mmol) at room temperature for 1 h. The reaction was monitored by TLC. After the completion of the reaction, the contents were cooled and poured onto cold water with stirring. The solid which separated was separated by filtration and dried in air. The dried compound was dissolved in dichloromethane and subjected to slow evaporation to yield single crystals.
Refinement
All the H atoms were discernible in the difference Fourier maps. However, H atoms were located geometrically with N—H = 0.86 and C—H = 0.93-0.96 Å and refined in the riding model approximation, with Uiso(H) = 1.2 or 1.5Ueq(C, N).
Figures
Fig. 1.
The molecular structure of (I) with the atom numbering scheme. Displacement ellipsoids for non-H atoms are drawn at the 50% probability level.
Crystal data
| C15H11Cl2NO2 | F(000) = 632 |
| Mr = 308.15 | Dx = 1.442 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 1298 reflections |
| a = 11.1371 (11) Å | θ = 2.0–20.9° |
| b = 5.0661 (6) Å | µ = 0.46 mm−1 |
| c = 25.594 (3) Å | T = 293 K |
| β = 100.672 (9)° | Block, colourless |
| V = 1419.1 (3) Å3 | 0.28 × 0.24 × 0.18 mm |
| Z = 4 |
Data collection
| Oxford Xcalibur Eos (Nova) CCD detector diffractometer | 1537 reflections with I > 2σ(I) |
| Radiation source: Enhance (Mo) X-ray Source | Rint = 0.080 |
| graphite | θmax = 25.5°, θmin = 2.7° |
| ω scans | h = −13→13 |
| 14628 measured reflections | k = −6→6 |
| 2633 independent reflections | l = −30→30 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.048 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.107 | H-atom parameters constrained |
| S = 0.98 | w = 1/[σ2(Fo2) + (0.0469P)2] where P = (Fo2 + 2Fc2)/3 |
| 2633 reflections | (Δ/σ)max < 0.001 |
| 182 parameters | Δρmax = 0.18 e Å−3 |
| 0 restraints | Δρmin = −0.21 e Å−3 |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell esds are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement on F2 for ALL reflections except those flagged by the user for potential systematic errors. Weighted R-factors wR and all goodnesses of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The observed criterion of F2 > 2sigma(F2) is used only for calculating -R-factor-obs etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 1.01228 (8) | 0.21348 (16) | 0.05190 (3) | 0.0676 (3) | |
| Cl2 | 0.68968 (8) | 0.97146 (17) | 0.00452 (3) | 0.0706 (3) | |
| O1 | 0.94298 (18) | 0.2513 (5) | 0.19297 (8) | 0.0777 (9) | |
| O2 | 0.5030 (2) | 0.0779 (5) | 0.17216 (10) | 0.0936 (11) | |
| N1 | 0.7034 (2) | 0.1634 (5) | 0.17250 (9) | 0.0533 (9) | |
| C1 | 0.6927 (2) | 0.7372 (6) | 0.05413 (10) | 0.0477 (10) | |
| C2 | 0.8029 (2) | 0.6603 (5) | 0.08384 (10) | 0.0429 (9) | |
| C3 | 0.8082 (2) | 0.4689 (5) | 0.12330 (10) | 0.0396 (9) | |
| C4 | 0.6978 (2) | 0.3551 (5) | 0.13289 (10) | 0.0431 (10) | |
| C5 | 0.5876 (3) | 0.4396 (6) | 0.10239 (12) | 0.0580 (11) | |
| C6 | 0.5855 (3) | 0.6267 (6) | 0.06384 (12) | 0.0566 (11) | |
| C7 | 0.9300 (2) | 0.3982 (6) | 0.15419 (11) | 0.0459 (10) | |
| C8 | 1.0428 (2) | 0.5175 (5) | 0.13994 (10) | 0.0399 (9) | |
| C9 | 1.0892 (2) | 0.4451 (5) | 0.09588 (10) | 0.0435 (10) | |
| C10 | 1.1978 (3) | 0.5487 (6) | 0.08559 (12) | 0.0565 (11) | |
| C11 | 1.2597 (3) | 0.7314 (7) | 0.11949 (14) | 0.0661 (13) | |
| C12 | 1.2151 (3) | 0.8101 (7) | 0.16279 (13) | 0.0686 (12) | |
| C13 | 1.1075 (3) | 0.7045 (6) | 0.17374 (11) | 0.0586 (11) | |
| C14 | 0.6092 (3) | 0.0330 (6) | 0.18928 (13) | 0.0607 (12) | |
| C15 | 0.6499 (3) | −0.1678 (7) | 0.23219 (13) | 0.0775 (16) | |
| H1 | 0.77560 | 0.12130 | 0.18860 | 0.0640* | |
| H2 | 0.87480 | 0.73680 | 0.07750 | 0.0510* | |
| H5 | 0.51450 | 0.36760 | 0.10840 | 0.0700* | |
| H6 | 0.51110 | 0.68050 | 0.04390 | 0.0680* | |
| H10 | 1.22820 | 0.49410 | 0.05590 | 0.0680* | |
| H11 | 1.33250 | 0.80190 | 0.11290 | 0.0790* | |
| H12 | 1.25710 | 0.93660 | 0.18540 | 0.0820* | |
| H13 | 1.07860 | 0.75900 | 0.20380 | 0.0700* | |
| H15A | 0.63180 | −0.10450 | 0.26520 | 0.1160* | |
| H15B | 0.73630 | −0.19610 | 0.23590 | 0.1160* | |
| H15C | 0.60750 | −0.33100 | 0.22290 | 0.1160* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0736 (6) | 0.0686 (6) | 0.0632 (5) | −0.0187 (5) | 0.0197 (4) | −0.0213 (4) |
| Cl2 | 0.0681 (6) | 0.0796 (6) | 0.0622 (5) | 0.0248 (5) | 0.0069 (4) | 0.0221 (5) |
| O1 | 0.0495 (13) | 0.117 (2) | 0.0670 (14) | 0.0078 (13) | 0.0115 (11) | 0.0456 (14) |
| O2 | 0.0531 (15) | 0.121 (2) | 0.109 (2) | −0.0246 (16) | 0.0209 (14) | 0.0138 (17) |
| N1 | 0.0417 (14) | 0.0620 (17) | 0.0590 (16) | −0.0031 (13) | 0.0164 (12) | 0.0079 (14) |
| C1 | 0.0450 (18) | 0.054 (2) | 0.0428 (16) | 0.0096 (15) | 0.0050 (14) | −0.0018 (14) |
| C2 | 0.0343 (15) | 0.0506 (18) | 0.0446 (16) | 0.0030 (13) | 0.0098 (13) | −0.0003 (15) |
| C3 | 0.0334 (15) | 0.0486 (18) | 0.0367 (15) | 0.0029 (13) | 0.0061 (12) | 0.0003 (14) |
| C4 | 0.0394 (16) | 0.0466 (18) | 0.0446 (16) | 0.0048 (14) | 0.0110 (13) | −0.0019 (15) |
| C5 | 0.0325 (16) | 0.077 (2) | 0.064 (2) | 0.0005 (16) | 0.0074 (15) | −0.0017 (19) |
| C6 | 0.0357 (17) | 0.075 (2) | 0.0552 (19) | 0.0166 (16) | −0.0017 (14) | −0.0033 (18) |
| C7 | 0.0460 (17) | 0.056 (2) | 0.0368 (16) | 0.0067 (15) | 0.0108 (13) | 0.0062 (15) |
| C8 | 0.0304 (14) | 0.0515 (19) | 0.0357 (15) | 0.0058 (14) | 0.0004 (12) | 0.0049 (14) |
| C9 | 0.0392 (16) | 0.0481 (18) | 0.0427 (16) | −0.0041 (14) | 0.0066 (13) | −0.0055 (14) |
| C10 | 0.0502 (19) | 0.064 (2) | 0.060 (2) | −0.0023 (17) | 0.0223 (16) | −0.0031 (17) |
| C11 | 0.0431 (18) | 0.081 (3) | 0.072 (2) | −0.0194 (19) | 0.0052 (17) | 0.001 (2) |
| C12 | 0.066 (2) | 0.071 (2) | 0.061 (2) | −0.022 (2) | −0.0085 (18) | −0.0118 (19) |
| C13 | 0.060 (2) | 0.071 (2) | 0.0425 (17) | −0.0004 (18) | 0.0032 (15) | −0.0111 (17) |
| C14 | 0.062 (2) | 0.066 (2) | 0.059 (2) | −0.0170 (19) | 0.0242 (18) | −0.0106 (18) |
| C15 | 0.098 (3) | 0.071 (3) | 0.071 (2) | −0.026 (2) | 0.035 (2) | 0.003 (2) |
Geometric parameters (Å, °)
| Cl1—C9 | 1.738 (3) | C8—C9 | 1.374 (3) |
| Cl2—C1 | 1.734 (3) | C9—C10 | 1.388 (4) |
| O1—C7 | 1.228 (4) | C10—C11 | 1.365 (5) |
| O2—C14 | 1.205 (4) | C11—C12 | 1.356 (5) |
| N1—C4 | 1.397 (3) | C12—C13 | 1.388 (5) |
| N1—C14 | 1.374 (4) | C14—C15 | 1.504 (5) |
| N1—H1 | 0.8600 | C2—H2 | 0.9300 |
| C1—C6 | 1.382 (4) | C5—H5 | 0.9300 |
| C1—C2 | 1.375 (3) | C6—H6 | 0.9300 |
| C2—C3 | 1.393 (4) | C10—H10 | 0.9300 |
| C3—C4 | 1.420 (3) | C11—H11 | 0.9300 |
| C3—C7 | 1.482 (3) | C12—H12 | 0.9300 |
| C4—C5 | 1.394 (4) | C13—H13 | 0.9300 |
| C5—C6 | 1.365 (4) | C15—H15A | 0.9600 |
| C7—C8 | 1.499 (3) | C15—H15B | 0.9600 |
| C8—C13 | 1.391 (4) | C15—H15C | 0.9600 |
| Cl1···C2 | 3.455 (3) | C13···O1x | 3.407 (4) |
| Cl1···C3 | 3.423 (3) | C7···H1 | 2.5000 |
| Cl1···Cl1i | 3.3974 (12) | C8···H2 | 2.4900 |
| Cl2···C11ii | 3.650 (4) | C9···H2 | 2.7700 |
| Cl1···H2iii | 3.0000 | C12···H15Axi | 3.0900 |
| Cl2···H6iv | 2.9400 | C14···H5 | 2.7300 |
| Cl2···H10v | 3.0500 | C15···H12xii | 2.9500 |
| O1···N1 | 2.660 (3) | H1···O1 | 1.9600 |
| O1···C13iii | 3.407 (4) | H1···C7 | 2.5000 |
| O2···C5 | 2.839 (4) | H1···H15B | 2.1100 |
| O2···C11vi | 3.300 (4) | H2···Cl1x | 3.0000 |
| O1···H13vii | 2.7000 | H2···C8 | 2.4900 |
| O1···H1 | 1.9600 | H2···C9 | 2.7700 |
| O1···H13iii | 2.9000 | H5···O2 | 2.2200 |
| O2···H5 | 2.2200 | H5···C14 | 2.7300 |
| O2···H11vi | 2.6100 | H6···Cl2iv | 2.9400 |
| O2···H12vi | 2.9100 | H10···Cl2v | 3.0500 |
| O2···H15Aviii | 2.8800 | H11···O2ix | 2.6100 |
| N1···O1 | 2.660 (3) | H12···O2ix | 2.9100 |
| C2···Cl1 | 3.455 (3) | H12···C15xiii | 2.9500 |
| C2···C9 | 3.330 (3) | H13···O1x | 2.9000 |
| C3···Cl1 | 3.423 (3) | H13···O1xi | 2.7000 |
| C5···O2 | 2.839 (4) | H15A···O2xiv | 2.8800 |
| C9···C2 | 3.330 (3) | H15A···C12vii | 3.0900 |
| C11···O2ix | 3.300 (4) | H15B···H1 | 2.1100 |
| C11···Cl2ii | 3.650 (4) | ||
| C4—N1—C14 | 128.8 (2) | C11—C12—C13 | 120.8 (3) |
| C14—N1—H1 | 116.00 | C8—C13—C12 | 120.3 (3) |
| C4—N1—H1 | 116.00 | N1—C14—C15 | 114.2 (3) |
| Cl2—C1—C6 | 120.6 (2) | O2—C14—N1 | 123.4 (3) |
| C2—C1—C6 | 119.9 (3) | O2—C14—C15 | 122.4 (3) |
| Cl2—C1—C2 | 119.55 (19) | C1—C2—H2 | 120.00 |
| C1—C2—C3 | 120.8 (2) | C3—C2—H2 | 120.00 |
| C2—C3—C7 | 117.8 (2) | C4—C5—H5 | 120.00 |
| C2—C3—C4 | 119.1 (2) | C6—C5—H5 | 120.00 |
| C4—C3—C7 | 123.1 (2) | C1—C6—H6 | 120.00 |
| N1—C4—C5 | 122.4 (2) | C5—C6—H6 | 120.00 |
| C3—C4—C5 | 118.6 (2) | C9—C10—H10 | 120.00 |
| N1—C4—C3 | 119.0 (2) | C11—C10—H10 | 120.00 |
| C4—C5—C6 | 120.9 (3) | C10—C11—H11 | 120.00 |
| C1—C6—C5 | 120.8 (3) | C12—C11—H11 | 120.00 |
| O1—C7—C3 | 122.5 (2) | C11—C12—H12 | 120.00 |
| C3—C7—C8 | 119.9 (2) | C13—C12—H12 | 120.00 |
| O1—C7—C8 | 117.6 (2) | C8—C13—H13 | 120.00 |
| C7—C8—C13 | 119.0 (2) | C12—C13—H13 | 120.00 |
| C9—C8—C13 | 117.5 (2) | C14—C15—H15A | 109.00 |
| C7—C8—C9 | 123.5 (2) | C14—C15—H15B | 110.00 |
| C8—C9—C10 | 121.9 (2) | C14—C15—H15C | 109.00 |
| Cl1—C9—C8 | 119.76 (18) | H15A—C15—H15B | 109.00 |
| Cl1—C9—C10 | 118.4 (2) | H15A—C15—H15C | 109.00 |
| C9—C10—C11 | 119.4 (3) | H15B—C15—H15C | 110.00 |
| C10—C11—C12 | 120.0 (3) | ||
| C14—N1—C4—C3 | 178.3 (3) | N1—C4—C5—C6 | −179.7 (3) |
| C14—N1—C4—C5 | −1.6 (4) | C3—C4—C5—C6 | 0.4 (4) |
| C4—N1—C14—O2 | −3.1 (5) | C4—C5—C6—C1 | −0.1 (5) |
| C4—N1—C14—C15 | 178.4 (3) | O1—C7—C8—C9 | 109.0 (3) |
| Cl2—C1—C2—C3 | −178.8 (2) | O1—C7—C8—C13 | −68.5 (4) |
| C6—C1—C2—C3 | 0.9 (4) | C3—C7—C8—C9 | −73.8 (4) |
| Cl2—C1—C6—C5 | 179.1 (2) | C3—C7—C8—C13 | 108.7 (3) |
| C2—C1—C6—C5 | −0.6 (4) | C7—C8—C9—Cl1 | 2.8 (4) |
| C1—C2—C3—C4 | −0.6 (4) | C7—C8—C9—C10 | −176.2 (3) |
| C1—C2—C3—C7 | −179.3 (2) | C13—C8—C9—Cl1 | −179.7 (2) |
| C2—C3—C4—N1 | −180.0 (2) | C13—C8—C9—C10 | 1.4 (4) |
| C2—C3—C4—C5 | −0.1 (4) | C7—C8—C13—C12 | 177.2 (3) |
| C7—C3—C4—N1 | −1.3 (4) | C9—C8—C13—C12 | −0.4 (4) |
| C7—C3—C4—C5 | 178.5 (3) | Cl1—C9—C10—C11 | 179.8 (2) |
| C2—C3—C7—O1 | 172.8 (3) | C8—C9—C10—C11 | −1.2 (4) |
| C2—C3—C7—C8 | −4.3 (4) | C9—C10—C11—C12 | 0.0 (5) |
| C4—C3—C7—O1 | −5.8 (4) | C10—C11—C12—C13 | 0.9 (5) |
| C4—C3—C7—C8 | 177.1 (2) | C11—C12—C13—C8 | −0.7 (5) |
Symmetry codes: (i) −x+2, −y, −z; (ii) −x+2, −y+2, −z; (iii) x, y−1, z; (iv) −x+1, −y+2, −z; (v) −x+2, −y+1, −z; (vi) x−1, y−1, z; (vii) −x+2, y−1/2, −z+1/2; (viii) −x+1, y+1/2, −z+1/2; (ix) x+1, y+1, z; (x) x, y+1, z; (xi) −x+2, y+1/2, −z+1/2; (xii) −x+2, y−3/2, −z+1/2; (xiii) −x+2, y+3/2, −z+1/2; (xiv) −x+1, y−1/2, −z+1/2.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1 | 0.86 | 1.96 | 2.660 (3) | 138 |
| C5—H5···O2 | 0.93 | 2.22 | 2.839 (4) | 124 |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: TK2672).
References
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl.34, 1555–1573.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Greene, T. W. & Wuts, P. G. M. (1999). Protective Groups in Organic Synthesis, 3rd ed. New York: Wiley & Sons.
- Gupta, R., Kumar, V., Gupta, M., Paul, S. & Gupta, R. (2008). Indian J. Chem. Sect. B, 47, 1739–1743.
- Orita, A., Tanahashi, C., Kakuda, A. & Otera, J. (2000). Angew. Chem. Int. Ed.39, 2877–2879. [DOI] [PubMed]
- Oxford Diffraction (2009). CrysAlis PRO CCD and CrysAlis PRO RED Oxford Diffraction Ltd, Yarnton, England.
- Procopiou, P. A., Baugh, S. P. D., Flack, S. S. & Inglis, G. G. A. (1998). J. Org. Chem.63, 2342–2347.
- Roopan, S. M. & Khan, F. N. (2009). Arkivoc, xiii, 161–169.
- Roopan, S. M., Maiyalagan, T. & Khan, F. N. (2008). Can. J. Chem 86, 1019–1025.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810018696/tk2672sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810018696/tk2672Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report