Abstract
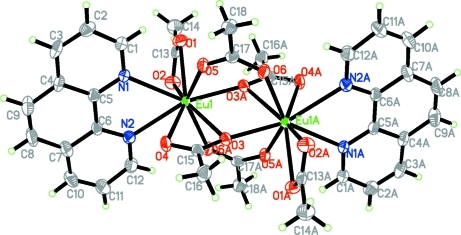
In the title centrosymmetric dinuclear EuIII complex, [Eu2(CH3COO)6(C12H8N2)2], each EuIII cation is coordinated by seven O atoms from five acetate anions and two N atoms from one phenanthroline ligand in a distorted tricapped trigonal-prismatic geometry. Four acetate anions bridge two EuIII cations to form the dinuclear complex, with an Eu⋯Eu distance of 3.9409 (8) Å. Weak intermolecular C—H⋯O hydrogen bonding is present in the crystal structure.
Related literature
For related lanthanide complexes with 1,10-phenanthroline and acetate ligands, see: Hu et al. (2006 ▶); Panagiotopoulos et al. (1995 ▶).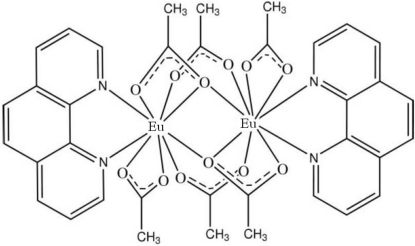
Experimental
Crystal data
[Eu2(C2H3O2)6(C12H8N2)2]
M r = 1018.61
Triclinic,
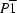
a = 8.7671 (19) Å
b = 8.9265 (19) Å
c = 12.992 (3) Å
α = 103.631 (2)°
β = 109.254 (2)°
γ = 98.300 (3)°
V = 905.1 (3) Å3
Z = 1
Mo Kα radiation
μ = 3.50 mm−1
T = 298 K
0.20 × 0.19 × 0.18 mm
Data collection
Bruker SMART CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2001 ▶) T min = 0.541, T max = 0.571
5010 measured reflections
3474 independent reflections
3062 reflections with I > 2σ(I)
R int = 0.021
Refinement
R[F 2 > 2σ(F 2)] = 0.030
wR(F 2) = 0.062
S = 1.05
3474 reflections
247 parameters
H-atom parameters constrained
Δρmax = 0.80 e Å−3
Δρmin = −0.64 e Å−3
Data collection: SMART (Bruker, 2001 ▶); cell refinement: SAINT (Bruker, 2001 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810015680/xu2753sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810015680/xu2753Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C2—H2⋯O2i | 0.93 | 2.57 | 3.287 (6) | 135 |
| C12—H8⋯O6ii | 0.93 | 2.44 | 3.078 (6) | 126 |
| C16—H10C⋯O1iii | 0.96 | 2.45 | 3.390 (6) | 165 |
Symmetry codes: (i) 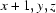 ; (ii)
; (ii) 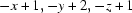 ; (iii)
; (iii) 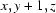 .
.
Acknowledgments
This work was supported financially by Guangdong Provincial Science and Technology Bureau (grant No. 2008B010600009) and the NSFC (grant Nos. 20971047 and U0734005).
supplementary crystallographic information
Comment
Dinuclear lanthanide complexes with 1,10-phenanthroline and acetate ligands had previously been reported (Panagiotopoulos et al., 1995; Hu et al., 2006). In this title complex, each Eu atom is coordinated by two N atoms from one chelating phenanthroline ligand and seven oxygen atoms from acetate ions, to form a distorted tricapped trigonal prism, giving a dimeric structure with an inversion center (Fig.1). The result of the dinuclear centrosymmetric molecule with the Eu···Eu distance of 3.9409 (8) Å was that acetate ions exhibit three different coordination modes: common bidentate chelating mode, bidentate bridging mode and tridentate bridging mode. The Eu1—O bond distances vary from 2.359 (3) Å to 2.586 (3) Å and the Eu1—N bond length are 2.594 (3) Å and 2.649 (4) Å. The C—O distances of CH3COO- are within the range of 1.257 (5)Å to 1.273 (5) Å. This complex exhibits a three-dimensional structure via C—H···O hydrogen-bonds (Table 1).
Experimental
A stoichiometric amount of acetic acid and a quantitative amount of 1,10-phenanthroline (0.5 mmol) were mixed and then dissolved in 95% ethanol solution (20 ml). The pH value of the solution was adjusted to 6.5 by adding 1.0 M NaOH solution, and then added dropwise to the ethanol solution (20 ml) of Eu(NO3)3.6H2O (0.5 mmol). The solution mixture was stirred continuously for 2 h at room temperature and then filtered. Single crystals were obtained by evaporation after one week.
Refinement
H atoms were positioned in calculated positions, with C—H = 0.93 (aromatic) and 0.96 Å (methyl), and refined in riding mode with Uiso(H) = 1.5Ueq(C) for methyl and 1.2Ueq(C) for the others.
Figures
Fig. 1.
Displacement ellipsoid plot (40% probability level) of the title compound [symmetry code: (A) -x+1, -y+2, -z+1].
Crystal data
| [Eu2(C2H3O2)6(C12H8N2)2] | Z = 1 |
| Mr = 1018.61 | F(000) = 500 |
| Triclinic, P1 | Dx = 1.869 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.7671 (19) Å | Cell parameters from 2079 reflections |
| b = 8.9265 (19) Å | θ = 0.7–25.2° |
| c = 12.992 (3) Å | µ = 3.50 mm−1 |
| α = 103.631 (2)° | T = 298 K |
| β = 109.254 (2)° | Block, colorless |
| γ = 98.300 (3)° | 0.20 × 0.19 × 0.18 mm |
| V = 905.1 (3) Å3 |
Data collection
| Bruker SMART CCD diffractometer | 3474 independent reflections |
| Radiation source: fine-focus sealed tube | 3062 reflections with I > 2σ(I) |
| graphite | Rint = 0.021 |
| ω scans | θmax = 26.0°, θmin = 1.7° |
| Absorption correction: multi-scan (SADABS; Bruker, 2001) | h = −10→10 |
| Tmin = 0.541, Tmax = 0.571 | k = −8→10 |
| 5010 measured reflections | l = −14→15 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.030 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.062 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.025P)2] where P = (Fo2 + 2Fc2)/3 |
| 3474 reflections | (Δ/σ)max < 0.001 |
| 247 parameters | Δρmax = 0.80 e Å−3 |
| 0 restraints | Δρmin = −0.64 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Eu1 | 0.47430 (3) | 0.84412 (3) | 0.356229 (17) | 0.02341 (8) | |
| O3 | 0.5788 (4) | 1.1409 (4) | 0.4756 (2) | 0.0312 (7) | |
| O4 | 0.6208 (4) | 1.0735 (4) | 0.3157 (2) | 0.0336 (7) | |
| O2 | 0.2565 (4) | 0.5955 (4) | 0.2687 (3) | 0.0419 (8) | |
| O1 | 0.5000 (4) | 0.5829 (4) | 0.3834 (3) | 0.0373 (8) | |
| O5 | 0.7449 (4) | 0.8969 (4) | 0.4988 (2) | 0.0310 (7) | |
| N1 | 0.6375 (4) | 0.7194 (4) | 0.2384 (3) | 0.0277 (8) | |
| C1 | 0.7671 (6) | 0.6658 (5) | 0.2863 (4) | 0.0362 (11) | |
| H1 | 0.7915 | 0.6637 | 0.3612 | 0.043* | |
| C2 | 0.8702 (6) | 0.6116 (6) | 0.2307 (4) | 0.0423 (12) | |
| H2 | 0.9609 | 0.5753 | 0.2678 | 0.051* | |
| C3 | 0.8341 (6) | 0.6134 (6) | 0.1215 (4) | 0.0450 (13) | |
| H3 | 0.9007 | 0.5782 | 0.0829 | 0.054* | |
| C4 | 0.6972 (6) | 0.6680 (6) | 0.0664 (4) | 0.0368 (11) | |
| C7 | 0.4176 (6) | 0.7809 (6) | −0.0387 (4) | 0.0421 (13) | |
| C6 | 0.4559 (6) | 0.7750 (5) | 0.0747 (3) | 0.0294 (10) | |
| N2 | 0.3612 (5) | 0.8179 (4) | 0.1353 (3) | 0.0323 (9) | |
| C12 | 0.2280 (6) | 0.8637 (6) | 0.0849 (4) | 0.0437 (13) | |
| H8 | 0.1601 | 0.8895 | 0.1249 | 0.052* | |
| C11 | 0.1825 (7) | 0.8757 (7) | −0.0271 (4) | 0.0565 (16) | |
| H7 | 0.0883 | 0.9110 | −0.0592 | 0.068* | |
| C10 | 0.2789 (7) | 0.8348 (7) | −0.0871 (4) | 0.0524 (15) | |
| H6 | 0.2516 | 0.8431 | −0.1607 | 0.063* | |
| C5 | 0.6006 (6) | 0.7203 (5) | 0.1290 (4) | 0.0299 (10) | |
| C15 | 0.6463 (5) | 1.1743 (5) | 0.4079 (4) | 0.0284 (10) | |
| C13 | 0.3479 (6) | 0.5207 (5) | 0.3230 (4) | 0.0327 (11) | |
| C8 | 0.5223 (7) | 0.7281 (7) | −0.0976 (4) | 0.0534 (15) | |
| H5 | 0.4986 | 0.7328 | −0.1718 | 0.064* | |
| O6 | 0.7627 (4) | 1.0478 (4) | 0.6690 (2) | 0.0336 (7) | |
| C14 | 0.2753 (7) | 0.3534 (6) | 0.3168 (5) | 0.0506 (14) | |
| H9A | 0.1575 | 0.3255 | 0.2737 | 0.076* | |
| H9B | 0.2959 | 0.3470 | 0.3927 | 0.076* | |
| H9C | 0.3264 | 0.2814 | 0.2800 | 0.076* | |
| C16 | 0.7559 (6) | 1.3374 (6) | 0.4417 (4) | 0.0422 (12) | |
| H10A | 0.8097 | 1.3412 | 0.3886 | 0.063* | |
| H10B | 0.8385 | 1.3612 | 0.5172 | 0.063* | |
| H10C | 0.6894 | 1.4140 | 0.4409 | 0.063* | |
| C9 | 0.6513 (7) | 0.6730 (7) | −0.0494 (4) | 0.0539 (15) | |
| H4 | 0.7136 | 0.6367 | −0.0914 | 0.065* | |
| C18 | 0.9975 (5) | 0.9554 (6) | 0.6560 (4) | 0.0395 (12) | |
| H11A | 1.0003 | 0.8457 | 0.6445 | 0.059* | |
| H11B | 1.0414 | 1.0106 | 0.7365 | 0.059* | |
| H11C | 1.0636 | 1.0025 | 0.6206 | 0.059* | |
| C17 | 0.8220 (5) | 0.9669 (5) | 0.6038 (4) | 0.0277 (10) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Eu1 | 0.02392 (12) | 0.02795 (13) | 0.02150 (12) | 0.00926 (9) | 0.01093 (9) | 0.00800 (9) |
| O3 | 0.0368 (18) | 0.0376 (18) | 0.0297 (16) | 0.0146 (15) | 0.0192 (14) | 0.0159 (15) |
| O4 | 0.0410 (19) | 0.0386 (19) | 0.0242 (16) | 0.0079 (15) | 0.0181 (14) | 0.0077 (15) |
| O2 | 0.039 (2) | 0.038 (2) | 0.045 (2) | 0.0085 (16) | 0.0110 (16) | 0.0142 (17) |
| O1 | 0.038 (2) | 0.0360 (19) | 0.0442 (19) | 0.0151 (15) | 0.0170 (16) | 0.0167 (16) |
| O5 | 0.0288 (17) | 0.0393 (19) | 0.0243 (16) | 0.0108 (14) | 0.0111 (13) | 0.0056 (14) |
| N1 | 0.030 (2) | 0.028 (2) | 0.0260 (19) | 0.0091 (16) | 0.0132 (16) | 0.0064 (16) |
| C1 | 0.044 (3) | 0.038 (3) | 0.032 (3) | 0.017 (2) | 0.019 (2) | 0.011 (2) |
| C2 | 0.034 (3) | 0.047 (3) | 0.050 (3) | 0.018 (2) | 0.018 (2) | 0.013 (3) |
| C3 | 0.046 (3) | 0.047 (3) | 0.048 (3) | 0.016 (3) | 0.031 (3) | 0.004 (3) |
| C4 | 0.043 (3) | 0.037 (3) | 0.032 (3) | 0.011 (2) | 0.021 (2) | 0.003 (2) |
| C7 | 0.050 (3) | 0.048 (3) | 0.025 (2) | 0.007 (3) | 0.014 (2) | 0.010 (2) |
| C6 | 0.036 (3) | 0.027 (2) | 0.022 (2) | 0.004 (2) | 0.011 (2) | 0.0032 (19) |
| N2 | 0.034 (2) | 0.035 (2) | 0.027 (2) | 0.0104 (18) | 0.0096 (17) | 0.0088 (18) |
| C12 | 0.039 (3) | 0.061 (4) | 0.031 (3) | 0.024 (3) | 0.011 (2) | 0.012 (3) |
| C11 | 0.064 (4) | 0.074 (4) | 0.033 (3) | 0.033 (3) | 0.009 (3) | 0.021 (3) |
| C10 | 0.069 (4) | 0.060 (4) | 0.027 (3) | 0.018 (3) | 0.013 (3) | 0.016 (3) |
| C5 | 0.037 (3) | 0.025 (2) | 0.028 (2) | 0.005 (2) | 0.015 (2) | 0.005 (2) |
| C15 | 0.027 (2) | 0.034 (3) | 0.031 (2) | 0.012 (2) | 0.014 (2) | 0.015 (2) |
| C13 | 0.043 (3) | 0.033 (3) | 0.029 (2) | 0.012 (2) | 0.024 (2) | 0.006 (2) |
| C8 | 0.067 (4) | 0.071 (4) | 0.029 (3) | 0.018 (3) | 0.027 (3) | 0.015 (3) |
| O6 | 0.0333 (18) | 0.0416 (19) | 0.0257 (16) | 0.0171 (15) | 0.0109 (14) | 0.0052 (15) |
| C14 | 0.064 (4) | 0.033 (3) | 0.057 (3) | 0.004 (3) | 0.027 (3) | 0.016 (3) |
| C16 | 0.049 (3) | 0.036 (3) | 0.046 (3) | 0.007 (2) | 0.027 (3) | 0.009 (2) |
| C9 | 0.063 (4) | 0.066 (4) | 0.037 (3) | 0.017 (3) | 0.030 (3) | 0.008 (3) |
| C18 | 0.030 (3) | 0.044 (3) | 0.041 (3) | 0.013 (2) | 0.009 (2) | 0.009 (2) |
| C17 | 0.027 (2) | 0.028 (2) | 0.034 (3) | 0.0086 (19) | 0.014 (2) | 0.014 (2) |
Geometric parameters (Å, °)
| Eu1—O3i | 2.358 (3) | C7—C10 | 1.384 (7) |
| Eu1—O6i | 2.374 (3) | C7—C6 | 1.415 (6) |
| Eu1—O5 | 2.377 (3) | C7—C8 | 1.438 (7) |
| Eu1—O2 | 2.453 (3) | C6—N2 | 1.355 (5) |
| Eu1—O1 | 2.470 (3) | C6—C5 | 1.448 (6) |
| Eu1—O4 | 2.513 (3) | N2—C12 | 1.311 (6) |
| Eu1—O3 | 2.586 (3) | C12—C11 | 1.411 (7) |
| Eu1—N1 | 2.594 (3) | C12—H8 | 0.9300 |
| Eu1—N2 | 2.649 (4) | C11—C10 | 1.358 (7) |
| Eu1—C13 | 2.815 (5) | C11—H7 | 0.9300 |
| Eu1—C15 | 2.920 (4) | C10—H6 | 0.9300 |
| Eu1—Eu1i | 3.9409 (8) | C15—C16 | 1.500 (6) |
| O3—C15 | 1.276 (5) | C13—C14 | 1.508 (6) |
| O3—Eu1i | 2.358 (3) | C8—C9 | 1.324 (8) |
| O4—C15 | 1.245 (5) | C8—H5 | 0.9300 |
| O2—C13 | 1.262 (6) | O6—C17 | 1.273 (5) |
| O1—C13 | 1.262 (5) | O6—Eu1i | 2.374 (3) |
| O5—C17 | 1.256 (5) | C14—H9A | 0.9600 |
| N1—C1 | 1.319 (6) | C14—H9B | 0.9600 |
| N1—C5 | 1.351 (5) | C14—H9C | 0.9600 |
| C1—C2 | 1.402 (6) | C16—H10A | 0.9600 |
| C1—H1 | 0.9300 | C16—H10B | 0.9600 |
| C2—C3 | 1.352 (7) | C16—H10C | 0.9600 |
| C2—H2 | 0.9300 | C9—H4 | 0.9300 |
| C3—C4 | 1.399 (7) | C18—C17 | 1.492 (6) |
| C3—H3 | 0.9300 | C18—H11A | 0.9600 |
| C4—C5 | 1.410 (6) | C18—H11B | 0.9600 |
| C4—C9 | 1.437 (7) | C18—H11C | 0.9600 |
| O3i—Eu1—O6i | 75.03 (10) | C5—N1—Eu1 | 120.7 (3) |
| O3i—Eu1—O5 | 76.96 (10) | N1—C1—C2 | 123.7 (4) |
| O6i—Eu1—O5 | 137.07 (10) | N1—C1—H1 | 118.2 |
| O3i—Eu1—O2 | 86.29 (10) | C2—C1—H1 | 118.2 |
| O6i—Eu1—O2 | 81.08 (11) | C3—C2—C1 | 118.2 (5) |
| O5—Eu1—O2 | 128.67 (11) | C3—C2—H2 | 120.9 |
| O3i—Eu1—O1 | 77.36 (10) | C1—C2—H2 | 120.9 |
| O6i—Eu1—O1 | 127.36 (11) | C2—C3—C4 | 120.5 (4) |
| O5—Eu1—O1 | 75.84 (10) | C2—C3—H3 | 119.8 |
| O2—Eu1—O1 | 53.10 (11) | C4—C3—H3 | 119.8 |
| O3i—Eu1—O4 | 125.07 (10) | C3—C4—C5 | 117.4 (4) |
| O6i—Eu1—O4 | 90.28 (11) | C3—C4—C9 | 123.4 (5) |
| O5—Eu1—O4 | 79.96 (10) | C5—C4—C9 | 119.2 (5) |
| O2—Eu1—O4 | 144.17 (10) | C10—C7—C6 | 117.6 (5) |
| O1—Eu1—O4 | 141.79 (10) | C10—C7—C8 | 123.9 (5) |
| O3i—Eu1—O3 | 74.40 (11) | C6—C7—C8 | 118.5 (5) |
| O6i—Eu1—O3 | 72.72 (10) | N2—C6—C7 | 122.5 (4) |
| O5—Eu1—O3 | 68.79 (10) | N2—C6—C5 | 118.0 (4) |
| O2—Eu1—O3 | 150.59 (10) | C7—C6—C5 | 119.5 (4) |
| O1—Eu1—O3 | 138.62 (10) | C12—N2—C6 | 117.9 (4) |
| O4—Eu1—O3 | 50.80 (9) | C12—N2—Eu1 | 122.8 (3) |
| O3i—Eu1—N1 | 143.33 (11) | C6—N2—Eu1 | 118.7 (3) |
| O6i—Eu1—N1 | 139.90 (10) | N2—C12—C11 | 123.2 (5) |
| O5—Eu1—N1 | 77.84 (10) | N2—C12—H8 | 118.4 |
| O2—Eu1—N1 | 89.07 (11) | C11—C12—H8 | 118.4 |
| O1—Eu1—N1 | 70.92 (11) | C10—C11—C12 | 118.8 (5) |
| O4—Eu1—N1 | 75.40 (10) | C10—C11—H7 | 120.6 |
| O3—Eu1—N1 | 119.64 (10) | C12—C11—H7 | 120.6 |
| O3i—Eu1—N2 | 149.00 (11) | C11—C10—C7 | 120.0 (5) |
| O6i—Eu1—N2 | 77.11 (11) | C11—C10—H6 | 120.0 |
| O5—Eu1—N2 | 133.80 (10) | C7—C10—H6 | 120.0 |
| O2—Eu1—N2 | 76.19 (11) | N1—C5—C4 | 122.1 (4) |
| O1—Eu1—N2 | 110.07 (11) | N1—C5—C6 | 118.6 (4) |
| O4—Eu1—N2 | 67.99 (10) | C4—C5—C6 | 119.3 (4) |
| O3—Eu1—N2 | 109.76 (10) | O4—C15—O3 | 120.4 (4) |
| N1—Eu1—N2 | 62.79 (11) | O4—C15—C16 | 121.0 (4) |
| O3i—Eu1—C13 | 79.18 (11) | O3—C15—C16 | 118.5 (4) |
| O6i—Eu1—C13 | 103.79 (13) | O4—C15—Eu1 | 58.8 (2) |
| O5—Eu1—C13 | 102.11 (13) | O3—C15—Eu1 | 62.3 (2) |
| O2—Eu1—C13 | 26.59 (12) | C16—C15—Eu1 | 172.3 (3) |
| O1—Eu1—C13 | 26.61 (12) | O2—C13—O1 | 121.4 (4) |
| O4—Eu1—C13 | 154.90 (11) | O2—C13—C14 | 119.9 (4) |
| O3—Eu1—C13 | 153.35 (11) | O1—C13—C14 | 118.8 (5) |
| N1—Eu1—C13 | 80.57 (12) | O2—C13—Eu1 | 60.5 (2) |
| N2—Eu1—C13 | 94.65 (12) | O1—C13—Eu1 | 61.3 (2) |
| O3i—Eu1—C15 | 99.99 (11) | C14—C13—Eu1 | 173.6 (3) |
| O6i—Eu1—C15 | 82.87 (11) | C9—C8—C7 | 122.1 (5) |
| O5—Eu1—C15 | 70.72 (11) | C9—C8—H5 | 119.0 |
| O2—Eu1—C15 | 160.62 (12) | C7—C8—H5 | 119.0 |
| O1—Eu1—C15 | 146.09 (11) | C17—O6—Eu1i | 136.1 (3) |
| O4—Eu1—C15 | 25.08 (10) | C13—C14—H9A | 109.5 |
| O3—Eu1—C15 | 25.89 (10) | C13—C14—H9B | 109.5 |
| N1—Eu1—C15 | 96.31 (11) | H9A—C14—H9B | 109.5 |
| N2—Eu1—C15 | 89.71 (11) | C13—C14—H9C | 109.5 |
| C13—Eu1—C15 | 172.71 (13) | H9A—C14—H9C | 109.5 |
| O3i—Eu1—Eu1i | 39.20 (7) | H9B—C14—H9C | 109.5 |
| O6i—Eu1—Eu1i | 69.54 (7) | C15—C16—H10A | 109.5 |
| O5—Eu1—Eu1i | 68.14 (7) | C15—C16—H10B | 109.5 |
| O2—Eu1—Eu1i | 122.20 (8) | H10A—C16—H10B | 109.5 |
| O1—Eu1—Eu1i | 111.19 (7) | C15—C16—H10C | 109.5 |
| O4—Eu1—Eu1i | 85.93 (7) | H10A—C16—H10C | 109.5 |
| O3—Eu1—Eu1i | 35.19 (6) | H10B—C16—H10C | 109.5 |
| N1—Eu1—Eu1i | 143.56 (8) | C8—C9—C4 | 121.4 (5) |
| N2—Eu1—Eu1i | 137.30 (8) | C8—C9—H4 | 119.3 |
| C13—Eu1—Eu1i | 118.30 (9) | C4—C9—H4 | 119.3 |
| C15—Eu1—Eu1i | 60.89 (9) | C17—C18—H11A | 109.5 |
| C15—O3—Eu1i | 160.5 (3) | C17—C18—H11B | 109.5 |
| C15—O3—Eu1 | 91.8 (3) | H11A—C18—H11B | 109.5 |
| Eu1i—O3—Eu1 | 105.60 (10) | C17—C18—H11C | 109.5 |
| C15—O4—Eu1 | 96.1 (2) | H11A—C18—H11C | 109.5 |
| C13—O2—Eu1 | 92.9 (3) | H11B—C18—H11C | 109.5 |
| C13—O1—Eu1 | 92.1 (3) | O5—C17—O6 | 125.1 (4) |
| C17—O5—Eu1 | 139.2 (3) | O5—C17—C18 | 117.4 (4) |
| C1—N1—C5 | 118.2 (4) | O6—C17—C18 | 117.5 (4) |
| C1—N1—Eu1 | 120.9 (3) |
Symmetry codes: (i) −x+1, −y+2, −z+1.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C2—H2···O2ii | 0.93 | 2.57 | 3.287 (6) | 135 |
| C12—H8···O6i | 0.93 | 2.44 | 3.078 (6) | 126 |
| C16—H10C···O1iii | 0.96 | 2.45 | 3.390 (6) | 165 |
Symmetry codes: (ii) x+1, y, z; (i) −x+1, −y+2, −z+1; (iii) x, y+1, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: XU2753).
References
- Bruker (2001). SMART, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Hu, X.-L., Qiu, L., Sun, W.-B. & Chen, Z. (2006). Acta Cryst. E62, m3213–m3214.
- Panagiotopoulos, A., Zafiropoulos, T. F., Perlepes, S. P., Bakalbassis, E., Masson-Ramade, I., Kahn, O., Terzis, A. & Raptopoulou, C. P. (1995). Inorg. Chem.34, 4918–4923.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810015680/xu2753sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810015680/xu2753Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report