Abstract
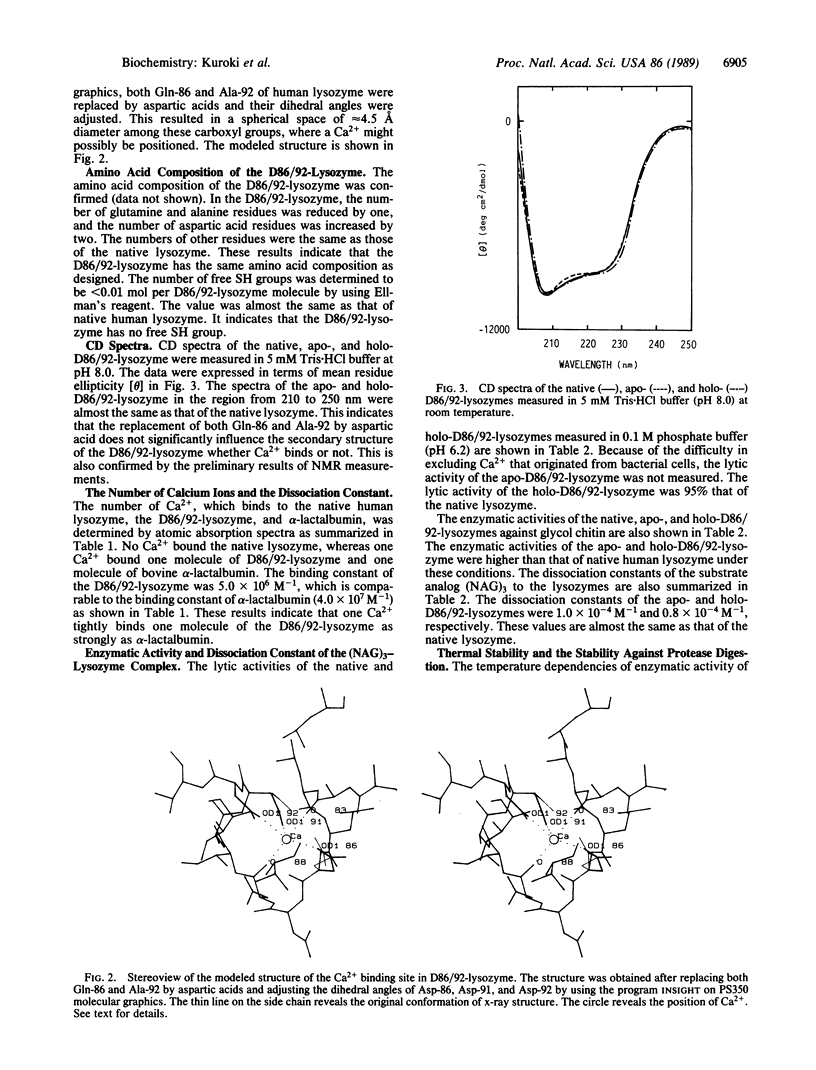
A Ca2+ binding site like an EF-hand motif was designed and created in human lysozyme by replacing both Gln-86 and Ala-92 with aspartic acids by site-directed mutagenesis. The mutant human lysozyme (D86/92-lysozyme) was expressed and secreted by yeast. One Ca2+ was found to bind one molecule of the purified protein with the binding constant 5.0 x 10(6) M-1. The enzymatic activity of holo-D86/92-lysozyme against glycol chitin at 40 degrees C was 2-fold higher than that of the native lysozyme. Maximal activity of the holo-D86/92-lysozyme was observed at 80 degrees C, where its relative activity normalized to the value at 40 degrees C was 6-fold and 17-fold higher than those of the native and apoenzymes, respectively. The activities of the native lysozyme and apo-D86/92-lysozyme were maximum at 65 degrees C-70 degrees C. Moreover, D86/92-lysozyme was more stable against protease digestion than the native lysozyme. These results indicate that the creation of the calcium binding site like an EF-hand motif in the human lysozyme enhances its structural stability.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T., Sun D. P., Wilson K., Wozniak J. A., Cook S. P., Matthews B. W. Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme. Nature. 1987 Nov 5;330(6143):41–46. doi: 10.1038/330041a0. [DOI] [PubMed] [Google Scholar]
- Artymiuk P. J., Blake C. C. Refinement of human lysozyme at 1.5 A resolution analysis of non-bonded and hydrogen-bond interactions. J Mol Biol. 1981 Nov 15;152(4):737–762. doi: 10.1016/0022-2836(81)90125-x. [DOI] [PubMed] [Google Scholar]
- Blake C. C., Mair G. A., North A. C., Phillips D. C., Sarma V. R. On the conformation of the hen egg-white lysozyme molecule. Proc R Soc Lond B Biol Sci. 1967 Apr 18;167(1009):365–377. doi: 10.1098/rspb.1967.0034. [DOI] [PubMed] [Google Scholar]
- Brew K., Castellino F. J., Vanaman T. C., Hill R. L. The complete amino acid sequence of bovine alpha-lactalbumin. J Biol Chem. 1970 Sep 10;245(17):4570–4582. [PubMed] [Google Scholar]
- Dahlquist F. W., Jao L., Raftery M. On the binding of chitin oligosaccharides to lysozyme. Proc Natl Acad Sci U S A. 1966 Jul;56(1):26–30. doi: 10.1073/pnas.56.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELLMAN G. L. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959 May;82(1):70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- Grütter M. G., Weaver L. H., Matthews B. W. Goose lysozyme structure: an evolutionary link between hen and bacteriophage lysozymes? Nature. 1983 Jun 30;303(5920):828–831. doi: 10.1038/303828a0. [DOI] [PubMed] [Google Scholar]
- Herzberg O., James M. N. Structure of the calcium regulatory muscle protein troponin-C at 2.8 A resolution. Nature. 1985 Feb 21;313(6004):653–659. doi: 10.1038/313653a0. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y., Segawa T., Kuwajima K., Sugai S., Murai N. alpha-Lactalbumin: a calcium metalloprotein. Biochem Biophys Res Commun. 1980 Aug 14;95(3):1098–1104. doi: 10.1016/0006-291x(80)91585-5. [DOI] [PubMed] [Google Scholar]
- Imoto T., Yamada H., Ueda T. Unfolding rates of globular proteins determined by kinetics of proteolysis. J Mol Biol. 1986 Aug 20;190(4):647–649. doi: 10.1016/0022-2836(86)90250-0. [DOI] [PubMed] [Google Scholar]
- Kikuchi M., Yamamoto Y., Taniyama Y., Ishimaru K., Yoshikawa W., Kaisho Y., Ikehara M. Secretion in yeast of human lysozymes with different specific activities created by replacing valine-110 with proline by site-directed mutagenesis. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9411–9415. doi: 10.1073/pnas.85.24.9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretsinger R. H., Nockolds C. E. Carp muscle calcium-binding protein. II. Structure determination and general description. J Biol Chem. 1973 May 10;248(9):3313–3326. [PubMed] [Google Scholar]
- Matthews B. W., Grütter M. G., Anderson W. F., Remington S. J. Common precursor of lysozymes of hen egg-white and bacteriophage T4. Nature. 1981 Mar 26;290(5804):334–335. doi: 10.1038/290334a0. [DOI] [PubMed] [Google Scholar]
- Matthews B. W., Remington S. J., Grütter M. G., Anderson W. F. Relation between hen egg white lysozyme and bacteriophage T4 lysozyme: evolutionary implications. J Mol Biol. 1981 Apr 25;147(4):545–558. doi: 10.1016/0022-2836(81)90399-5. [DOI] [PubMed] [Google Scholar]
- Matthews B. W., Remington S. J. The three dimensional structure of the lysozyme from bacteriophage T4. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4178–4182. doi: 10.1073/pnas.71.10.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson H., Becktel W. J., Matthews B. W. Enhanced protein thermostability from designed mutations that interact with alpha-helix dipoles. Nature. 1988 Dec 15;336(6200):651–656. doi: 10.1038/336651a0. [DOI] [PubMed] [Google Scholar]
- Nitta K., Tsuge H., Shimazaki K., Sugai S. Calcium-binding lysozymes. Biol Chem Hoppe Seyler. 1988 Aug;369(8):671–675. doi: 10.1515/bchm3.1988.369.2.671. [DOI] [PubMed] [Google Scholar]
- Nitta K., Tsuge H., Sugai S., Shimazaki K. The calcium-binding property of equine lysozyme. FEBS Lett. 1987 Nov 2;223(2):405–408. doi: 10.1016/0014-5793(87)80328-9. [DOI] [PubMed] [Google Scholar]
- Pantoliano M. W., Whitlow M., Wood J. F., Rollence M. L., Finzel B. C., Gilliland G. L., Poulos T. L., Bryan P. N. The engineering of binding affinity at metal ion binding sites for the stabilization of proteins: subtilisin as a test case. Biochemistry. 1988 Nov 1;27(22):8311–8317. doi: 10.1021/bi00422a004. [DOI] [PubMed] [Google Scholar]
- Perry L. J., Wetzel R. Disulfide bond engineered into T4 lysozyme: stabilization of the protein toward thermal inactivation. Science. 1984 Nov 2;226(4674):555–557. doi: 10.1126/science.6387910. [DOI] [PubMed] [Google Scholar]
- Qasba P. K., Safaya S. K. Similarity of the nucleotide sequences of rat alpha-lactalbumin and chicken lysozyme genes. Nature. 1984 Mar 22;308(5957):377–380. doi: 10.1038/308377a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. J., Begg G. S., Dorow D. S., Morgan F. J. Complete amino acid sequence of the goose-type lysozyme from the egg white of the black swan. Biochemistry. 1980 Apr 29;19(9):1814–1819. doi: 10.1021/bi00550a013. [DOI] [PubMed] [Google Scholar]
- Stuart D. I., Acharya K. R., Walker N. P., Smith S. G., Lewis M., Phillips D. C. Alpha-lactalbumin possesses a novel calcium binding loop. Nature. 1986 Nov 6;324(6092):84–87. doi: 10.1038/324084a0. [DOI] [PubMed] [Google Scholar]
- Taniyama Y., Yamamoto Y., Nakao M., Kikuchi M., Ikehara M. Role of disulfide bonds in folding and secretion of human lysozyme in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1988 May 16;152(3):962–967. doi: 10.1016/s0006-291x(88)80377-2. [DOI] [PubMed] [Google Scholar]
- To-E A., Ueda Y., Kakimoto S. I., Oshima Y. Isolation and characterization of acid phosphatase mutants in Saccharomyces cerevisiae. J Bacteriol. 1973 Feb;113(2):727–738. doi: 10.1128/jb.113.2.727-738.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanaman T. C., Brew K., Hill R. L. The disulfide bonds of bovine alpha-lactalbumin. J Biol Chem. 1970 Sep 10;245(17):4583–4590. [PubMed] [Google Scholar]
- Wakabayashi T., Huxley H. E., Amos L. A., Klug A. Three-dimensional image reconstruction of actin-tropomyosin complex and actin-tropomyosin-troponin T-troponin I complex. J Mol Biol. 1975 Apr 25;93(4):477–497. doi: 10.1016/0022-2836(75)90241-7. [DOI] [PubMed] [Google Scholar]
- Yamada H., Imoto T. A convenient synthesis of glycolchitin, a substrate of lysozyme. Carbohydr Res. 1981 May 18;92(1):160–162. doi: 10.1016/s0008-6215(00)85993-5. [DOI] [PubMed] [Google Scholar]
- Yoshimura K., Toibana A., Kikuchi K., Kobayashi M., Hayakawa T., Nakahama K., Kikuchi M., Ikehara M. Differences between Saccharomyces cerevisiae and Bacillus subtilis in secretion of human lysozyme. Biochem Biophys Res Commun. 1987 Jun 15;145(2):712–718. doi: 10.1016/0006-291x(87)91023-0. [DOI] [PubMed] [Google Scholar]
- Yutani K., Ogasahara K., Tsujita T., Sugino Y. Dependence of conformational stability on hydrophobicity of the amino acid residue in a series of variant proteins substituted at a unique position of tryptophan synthase alpha subunit. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4441–4444. doi: 10.1073/pnas.84.13.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]


