Abstract
Upon the completion of the Saccharomyces cerevisiae genomic sequence in 1996 [Goffeau,A. et al. (1997) Nature, 387, 5], several creative and ambitious projects have been initiated to explore the functions of gene products or gene expression on a genome-wide scale. To help researchers take advantage of these projects, the Saccharomyces Genome Database (SGD) has created two new tools, Function Junction and Expression Connection. Together, the tools form a central resource for querying multiple large-scale analysis projects for data about individual genes. Function Junction provides information from diverse projects that shed light on the role a gene product plays in the cell, while Expression Connection delivers information produced by the ever-increasing number of microarray projects. WWW access to SGD is available at genome-www.stanford.edu/Saccharomyces/.
FUNCTION JUNCTION
The Function Junction resource allows a Saccharomyces Genome Database (SGD) (1–3) user to simultaneously retrieve genome-wide functional analysis information from a variety of online sources for a selected gene name. The various sites surveyed by Function Junction use a wide assortment of techniques to assay or predict information about a gene product. SGD users can query all sites, or specify an interesting subset. Function Junction, which can be accessed by entering a gene or ORF name at genome-www.stanford.edu/cgi-bin/SGD/functionJunction, currently searches the following sites:
• Portal PathCalling provides data from Uetz et al. (4), investigation of protein–protein interactions and is accessible at: portal.curagen.com/extpc/com.curagen.portal.servlet.Yeast.
• UCLA Function Assignment provides the results of several different gene function prediction methods, including phylogenetic profiling, protein interactions and shared expression patterns (5–7) and can be accessed at www.doe-mbi.ucla.edu/people/marcotte/yeast.html.
• Worm–Yeast Comparison reports potential homologs identified by the Chervitz et al. sequence comparison (8) and can be found at genome-www.stanford.edu/Saccharomyces/worm/.
• Serial Analysis of Gene Expression (SAGE) provides data from the Velculescu et al. study of gene expression (9) and is available at an SGD website, genome-www.stanford.edu/Saccharomyces/SAGE/AdvancedQuery.html.
• Triples Database reports the results of transposon mutagenesis studies (10) providing data about phenotype, expression and localization and is available at ygac.med.yale.edu/triples/triples.htm.
EXPRESSION CONNECTION
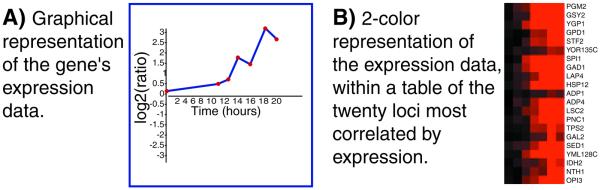
Expression Connection is similar to Function Junction in concept, but it queries only microarray expression data (see Fig. 1 for an example). Using software developed at SGD, downloaded data from several microarray studies are queried to display the expression pattern of a gene and its most similarly expressed neighbors. For serial experiments, a graph showing the expression pattern over the course of the experiment is available. The datasets currently queried by Expression Connection include results of a diauxic shift timecourse (11), a sporulation time series (12), two studies of cell-cycle synchronization (13,14), a study of evolution under glucose limitation (15), a histone depletion timecourse (16), an analysis of gene expression perturbation caused by mutations in the yeast transcriptional machinery (17), an α-factor concentration series and an α-factor treatment timecourse (18). To use Expression Connection, enter a gene or ORF name at the following URL: genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl.
Figure 1.
Expression Connection. For any gene, users may display several sets of microarray data. (A) For each set of serial data, a graphic representation of the gene’s expression pattern is rendered. (B) Two color representations of the expression data are presented, showing the gene’s expression pattern along with the 20 loci whose expression patterns are most similar. The two color representations are accompanied by annotation of gene names, molecular function and biological processes.
CONCLUSIONS
Together, Function Junction and Expression Connection will serve as a central resource on the World Wide Web for quickly retrieving information generated by comprehensive analyses of the yeast genome. As more large-scale datasets are published, SGD will include them on the list of sites queried by these software packages. In this manner, SGD will enable online researchers to benefit from the accumulation of genome-wide studies of Saccharomyces cerevisiae.
Acknowledgments
ACKNOWLEDGEMENT
S.G.D. is supported by a P41, National Resources, grant from the National Genome Research Institute at the US National Institutes of Health.
References
- 1.Cherry J.M., Adler,C., Ball,C., Chervitz,S.A., Dwight,S.S., Hester,E.T., Jia,Y., Juvik,G., Roe,T., Schroeder,M., Weng,S. and Botstein,D. (1998) SGD: Saccharomyces Genome Database. Nucleic Acids Res., 26, 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chervitz S.A., Hester,E.T., Ball,C.A., Dolinski,K., Dwight,S.S., Harris,M.A., Juvik,G., Malekian,A., Roberts,S., Roe,T., Scafe,C., Schroeder,M., Sherlock,G., Weng,S., Zhu,Y., Cherry,J.M. and Botstein,D. (1999) Using the Saccharomyces Genome Database (SGD) for analysis of protein similarities and structure. Nucleic Acids Res., 27, 74–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball C.A., Dolinski,K., Dwight,S.S., Harris,M.A., Issel-Tarver,L., Kasarskis,A., Scafe,C.R., Sherlock,G., Binkley,G., Jin,H., Kaloper,M., Orr,S.D., Schroeder,M., Weng,S., Zhu,Y., Botstein,D. and Cherry,J.M. (2000) Integrating functional genomic information into the Saccharomyces genome database. Nucleic Acids Res., 28, 77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uetz P., Giot,L., Cagney,G., Mansfield,T.A., Judson,R.S., Knight,J.R., Lockshon,D., Narayan,V., Srinivasan,M., Pochart,P., Qureshi-Emili,A., Li,Y., Godwin,B., Conover,D., Kalbfleisch,T., Vijayadamodar,G., Yang,M., Johnston,M., Fields,S. and Rothberg,J.M. (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- 5.Marcotte E.M., Pellegrini,M., Thompson,M.J., Yeates,T.O. and Eisenberg,D. (1999) A combined algorithm for genome-wide prediction of protein function. Nature, 402, 83–86. [DOI] [PubMed] [Google Scholar]
- 6.Marcotte E.M., Pellegrini,M., Ng,H.L., Rice,D.W., Yeates,T.O. and Eisenberg,D. (1999) Detecting protein function and protein-protein interactions from genome sequences. Science, 285, 751–753. [DOI] [PubMed] [Google Scholar]
- 7.Pellegrini M., Marcotte,E.M., Thompson,M.J., Eisenberg,D. and Yeates,T.O. (1999) Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA, 96, 4285–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chervitz S.A., Aravind,L., Sherlock,G., Ball,C.A., Koonin,E.V., Dwight,S.S., Harris,M.A., Dolinski,K., Mohr,S., Smith,T., Weng,S., Cherry,J.M. and Botstein,D. (1998) Comparison of the complete protein sets of worm and yeast: orthology and divergence. Science, 282, 2022–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Velculescu V.E., Zhang,L., Zhou,W., Vogelstein,J., Basrai,M.A., Bassett,D.E.,Jr, Hieter,P., Vogelstein,B. and Kinzler,K.W. (1997) Characterization of the yeast transcriptome. Cell, 88, 243–251. [DOI] [PubMed] [Google Scholar]
- 10.Ross-Macdonald P., Coelho,P.S., Roemer,T., Agarwal,S., Kumar,A., Jansen,R., Cheung,K.H., Sheehan,A., Symoniatis,D., Umansky,L., Heidtman,M., Nelson,F.K., Iwasaki,H., Hager,K., Gerstein,M., Miller,P., Roeder,G.S. and Snyder,M. (1999) Large-scale analysis of the yeast genome by transposon tagging and gene disruption [see comments]. Nature, 402, 413–418. [DOI] [PubMed] [Google Scholar]
- 11.DeRisi J.L., Iyer,V.R. and Brown,P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 12.Chu S., DeRisi,J., Eisen,M., Mulholland,J., Botstein,D., Brown,P.O. and Herskowitz,I. (1998) The transcriptional program of sporulation in budding yeast [published erratum appears in Science (1998) 20, 1421]. Science, 282, 699–705. [DOI] [PubMed] [Google Scholar]
- 13.Spellman P.T., Sherlock,G., Zhang,M.Q., Iyer,V.R., Anders,K., Eisen,M.B., Brown,P.O., Botstein,D. and Futcher,B. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell, 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho R.J., Campbell,M.J., Winzeler,E.A., Steinmetz,L., Conway,A., Wodicka,L., Wolfsberg,T.G., Gabrielian,A.E., Landsman,D., Lockhart,D.J. and Davis,R.W. (1998) A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell, 2, 65–73. [DOI] [PubMed] [Google Scholar]
- 15.Ferea T.L., Botstein,D., Brown,P.O. and Rosenzweig,R.F. (1999) Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc. Natl Acad. Sci. USA, 96, 9721–9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyrick J.J., Holstege,F.C., Jennings,E.G., Causton,H.C., Shore,D., Grunstein,M., Lander,E.S. and Young,R.A. (1999) Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature, 402, 418–421. [DOI] [PubMed] [Google Scholar]
- 17.Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- 18.Roberts C.J., Nelson,B., Marton,M.J., Stoughton,R., Meyer,M.R., Bennett,H.A., He,Y.D., Dai,H., Walker,W.L., Hughes,T.R., Tyers,M., Boone,C. and Friend,S.H. (2000) Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science, 287, 873–880. [DOI] [PubMed] [Google Scholar]