Abstract
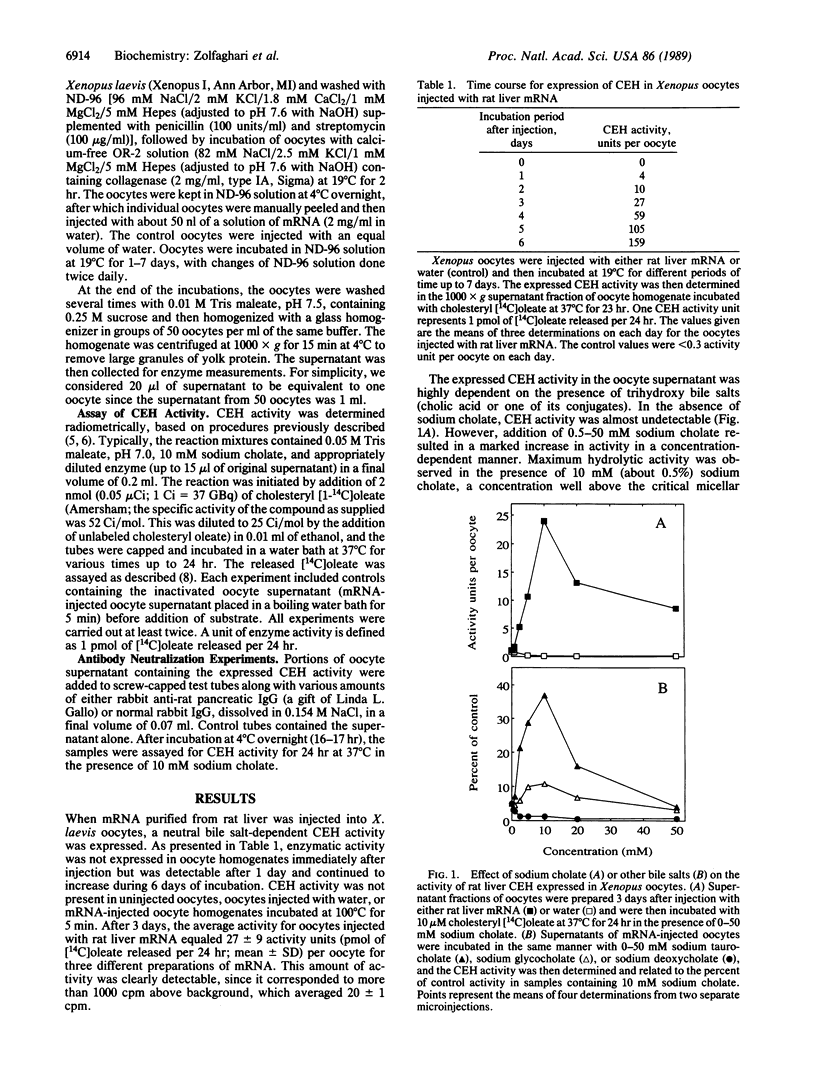
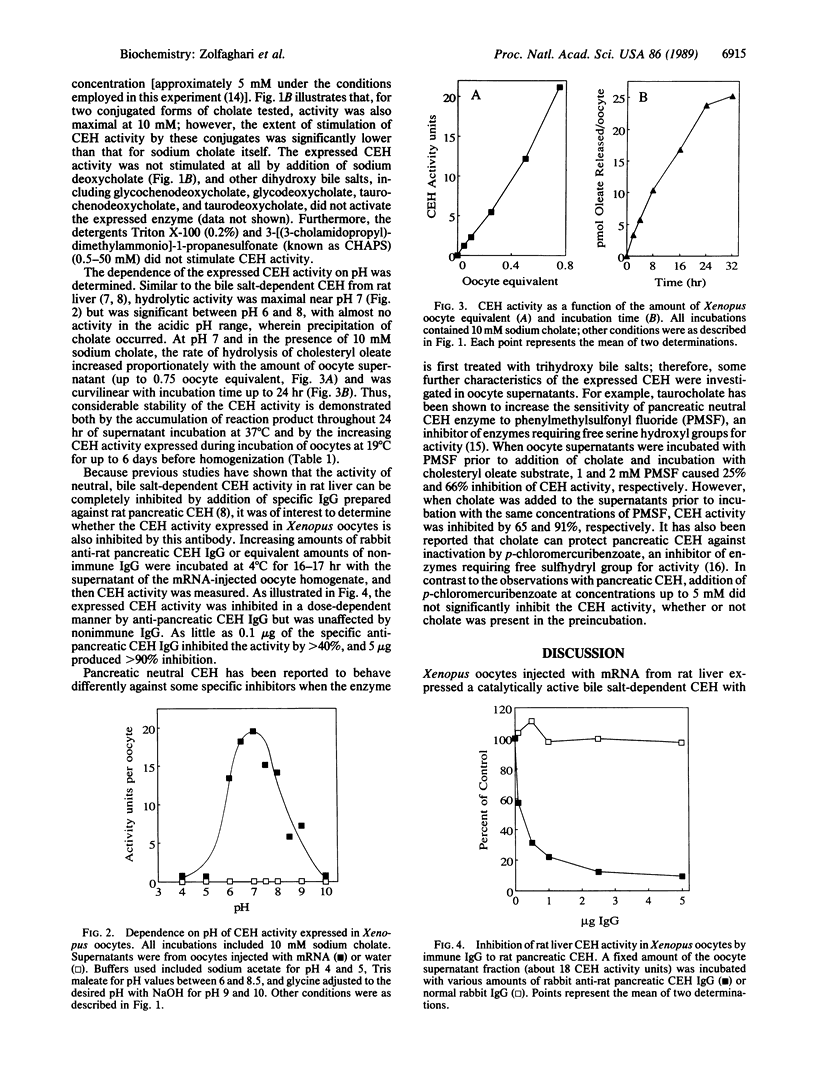
A catalytically active bile salt-dependent cholesteryl ester hydrolase (CEH) was expressed when Xenopus oocytes were injected with rat liver mRNA. The expressed CEH activity was highly dependent on the presence of trihydroxy bile salts (cholate or one of its conjugates); maximum hydrolytic activity was observed in the presence of 10 mM sodium cholate. The expressed CEH was not activated by dihydroxy bile salts (deoxycholate and its conjugates). In the presence of 10 mM sodium cholate, the CEH activity was maximal near pH 7 but was significant between pH 6 and 8. Monospecific immune IgG raised against rat pancreatic CEH completely inhibited the CEH expressed in Xenopus oocytes. Phenylmethylsulfonyl fluoride, a serine enzyme inhibitor, was inhibitory to the expressed CEH activity, whereas p-chloromercuribenzoate (up to 5 mM), a potent thiol-blocking agent, did not significantly inhibit the expressed activity. These experiments clearly demonstrate that the liver contains an mRNA encoding a bile salt-dependent CEH activity and suggest that the uptake of pancreatic enzyme is not necessarily the source of liver CEH as has been speculated.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blaner W. S., Prystowsky J. H., Smith J. E., Goodman D. S. Rat liver retinyl palmitate hydrolase activity. Relationship to cholesteryl oleate and triolein hydrolase activities. Biochim Biophys Acta. 1984 Jul 26;794(3):419–427. doi: 10.1016/0005-2760(84)90008-0. [DOI] [PubMed] [Google Scholar]
- Carey M. C., Small D. M. Micelle formation by bile salts. Physical-chemical and thermodynamic considerations. Arch Intern Med. 1972 Oct;130(4):506–527. [PubMed] [Google Scholar]
- DEYKIN D., GOODMAN D. S. The hydrolysis of long-chain fatty acid esters of cholesterol with rat liver enzymes. J Biol Chem. 1962 Dec;237:3649–3656. [PubMed] [Google Scholar]
- Gallo L. L., Cheriathundam E., Vahouny G. V., Treadwell C. R. Immunological comparison of cholesterol esterases. Arch Biochem Biophys. 1978 Nov;191(1):42–48. doi: 10.1016/0003-9861(78)90065-6. [DOI] [PubMed] [Google Scholar]
- Gallo L. L., Chiang Y., Vahouny G. V., Treadwell C. R. Localization and origin at rat intestinal cholesterol esterase determined by immunocytochemistry. J Lipid Res. 1980 Jul;21(5):537–545. [PubMed] [Google Scholar]
- Gallo L. L., Newbill T., Hyun J., Vahouny G. V., Treadwell C. R. Role of pancreatic cholesterol esterase in the uptake and esterification of cholesterol by isolated intestinal cells. Proc Soc Exp Biol Med. 1977 Nov;156(2):277–281. doi: 10.3181/00379727-156-39921. [DOI] [PubMed] [Google Scholar]
- Goller H. J., Sgoutas D. S. Further studies on the fatty acid specificity of rat liver sterol-ester hydrolase. Biochemistry. 1970 Nov 24;9(24):4801–4806. doi: 10.1021/bi00826a026. [DOI] [PubMed] [Google Scholar]
- Goller H. J., Sgoutas D. S., Ismail I. A., Gunstone F. D. Dependence of sterol ester hydrolase activity on the position of ethylenic bond in cholesteryl cis-octadecenoates. Biochemistry. 1970 Jul 21;9(15):3072–3076. doi: 10.1021/bi00817a021. [DOI] [PubMed] [Google Scholar]
- Han J. H., Stratowa C., Rutter W. J. Isolation of full-length putative rat lysophospholipase cDNA using improved methods for mRNA isolation and cDNA cloning. Biochemistry. 1987 Mar 24;26(6):1617–1625. doi: 10.1021/bi00380a020. [DOI] [PubMed] [Google Scholar]
- Harrison E. H. Bile salt-dependent, neutral cholesteryl ester hydrolase of rat liver: possible relationship with pancreatic cholesteryl ester hydrolase. Biochim Biophys Acta. 1988 Nov 4;963(1):28–34. doi: 10.1016/0005-2760(88)90334-7. [DOI] [PubMed] [Google Scholar]
- Hyun J., Kothari H., Herm E., Mortenson J., Treadwell C. R., Vahouny G. V. Purification and properties of pancreatic juice cholesterol esterase. J Biol Chem. 1969 Apr 10;244(7):1937–1945. [PubMed] [Google Scholar]
- Hyun J., Treadwell C. R., Vahouny G. V. Pancreatic juice cholesterol esterase. Studies on molecular weight and bile salt-induced polymerization. Arch Biochem Biophys. 1972 Sep;152(1):233–242. doi: 10.1016/0003-9861(72)90211-1. [DOI] [PubMed] [Google Scholar]
- Jacobson A. Purification and fractionation of poly(A)+ RNA. Methods Enzymol. 1987;152:254–261. doi: 10.1016/0076-6879(87)52028-6. [DOI] [PubMed] [Google Scholar]
- Julius D., MacDermott A. B., Axel R., Jessell T. M. Molecular characterization of a functional cDNA encoding the serotonin 1c receptor. Science. 1988 Jul 29;241(4865):558–564. doi: 10.1126/science.3399891. [DOI] [PubMed] [Google Scholar]
- Prystowsky J. H., Smith J. E., Goodman D. S. Retinyl palmitate hydrolase activity in normal rat liver. J Biol Chem. 1981 May 10;256(9):4498–4503. [PubMed] [Google Scholar]
- Ruggieri S., Roblin R., Black P. H. Lipids of whole cells and plasma membrane fractions from Balb/c3T3, SV3T3, and concanavalin A-selected revertant cells. J Lipid Res. 1979 Aug;20(6):760–771. [PubMed] [Google Scholar]
- Vahouny G. W., Weersing S., Treadwell C. R. Function of specific bile acids in cholesterol esterase activity in vitro. Biochim Biophys Acta. 1965 Jun 1;98(3):607–616. doi: 10.1016/0005-2760(65)90158-x. [DOI] [PubMed] [Google Scholar]
- White M. M., Mayne K. M., Lester H. A., Davidson N. Mouse-Torpedo hybrid acetylcholine receptors: functional homology does not equal sequence homology. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4852–4856. doi: 10.1073/pnas.82.14.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]



