Abstract
The title compound, C16H15N3O2S, was synthesized by the reaction of 2-amino-5,6,7,8-tetrahydro-4H-cyclohepta[b]thiophene-3-carbonitrile and o-fluoronitrobenzene. The thiophene and nitrophenyl rings and amino and carbonitrile groups are coplanar with a maximum deviation of 0.046 (2) Å and a dihedral angle of 0.92 (6)° between the rings. The cyclohepta ring adopts a chair conformation. Intramolecular N—H⋯O and C—H⋯S interactions occur. In the crystal, the molecules form layers that are linked by π–π stacking interactions between the thiophene and benzene rings [centroid–centroid distances = 3.7089 (12) and 3.6170 (12) Å].
Related literature
For background to 2-substituted thiophenes, see: Campaigne (1984 ▶); Kleemann et al. (2006 ▶). For the biological activity of 2-amino thiophene derivatives, see: Chakrabarti et al. (1982 ▶); Calligaro et al. (1997 ▶); Nikolakopoulos et al. (2006 ▶). For the synthesis of 2-amino thiophenes, see: Gewald (1965 ▶); Gewald et al. (1966 ▶); Sridhar et al. (2007 ▶). For related structures, see: Stephenson et al. (1995 ▶); Yu (2002 ▶); Chen et al. (2005 ▶). For bond-length data, see: Allen et al. (1987 ▶). For puckering parameters, see: Cremer & Pople (1975 ▶).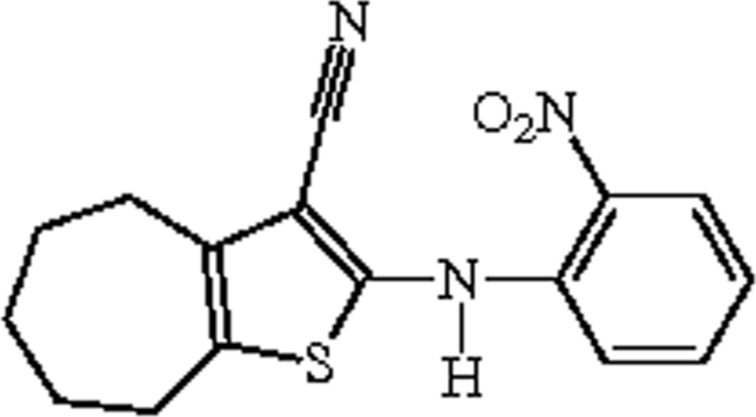
Experimental
Crystal data
C16H15N3O2S
M r = 313.37
Monoclinic,
a = 7.0273 (3) Å
b = 14.4569 (6) Å
c = 14.8867 (7) Å
β = 97.571 (2)°
V = 1499.18 (11) Å3
Z = 4
Mo Kα radiation
μ = 0.23 mm−1
T = 295 K
0.35 × 0.32 × 0.27 mm
Data collection
Nonius KappaCCD diffractometer
9360 measured reflections
3147 independent reflections
2452 reflections with I > 2σ(I)
R int = 0.046
Refinement
R[F 2 > 2σ(F 2)] = 0.052
wR(F 2) = 0.162
S = 1.07
3147 reflections
200 parameters
H-atom parameters constrained
Δρmax = 0.29 e Å−3
Δρmin = −0.31 e Å−3
Data collection: COLLECT (Nonius, 1997 ▶); cell refinement: SCALEPACK (Otwinowski & Minor, 1997 ▶); data reduction: DENZO (Otwinowski & Minor, 1997 ▶) and SCALEPACK; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810017149/dn2562sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810017149/dn2562Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O1 | 0.86 | 1.89 | 2.593 (2) | 138 |
| C15—H15⋯S1 | 0.93 | 2.44 | 3.171 (2) | 135 |
Acknowledgments
This work has received partial support from CNPq, CAPES, FACEPE and FINEP.
supplementary crystallographic information
Comment
The various uses of 2-substituted thiophenes have been well documented (Campaigne, 1984; Kleemann et al., 2006). Amongst these appplications, are neuroleptics activities, as found for a series of thienobenzodiazepines (Chakrabarti et al., 1982; Calligaro et al., 1997; Nikolakopoulos et al., 2006). In this work, we report the structure of the title compound prepared by the reaction of 2-amino-5,6,7, 8-tetrahydro-4H-ciclohepta[b]thiophene-3- carbonitrile and o-fluoro- nitrobenzene.
As indicated in the literature, compounds that presents o-nitrophenyl group linked to 2-amino-thiophene ring have great potential to produce crystalline structures, as found in 5-methyl-2-[(2-nitrophenyl)-amino]- thiophene-3-carbonitrile (ROY), that presents seven polymorphs coexisting at room temperature (Stephenson et al., 1995; Yu, 2002; Chen et al., 2005).
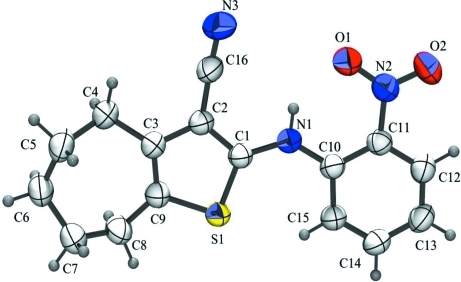
In the title compound,the least squares plane passing through all atoms of thiophene and nitrophenyl rings, and amino and carbonitrile groups, show planarity with maximum deviation of [0.046 (2) Å] for atom N3 (Fig.1). Bond lengths and angles are in good agreement with the expected values reported in the literature (Allen et al., 1987). The cyclohepta ring adopts a chair conformation and the calculated puckering parameters are: q2 = 0.175 (9) Å, q3 = 0.619 (9) Å, QT = 0.643 (9) Å, θ = 15.8 (9)° , φ2 = 56.1 (8)° and φ3 = 78.4 (8)° (Cremer & Pople, 1975).
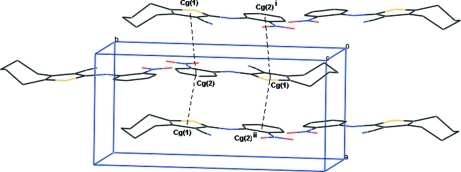
In the molecule there are, intramolecular N–H··· O, C—H··· O and C—H··· S interactions that are responsible for the roughly planar arrangement (Table 1). In the packing molecules form layers that extends along a direction parallel to the (100) plane, and are linked by π-π stacking interactions between thiophene [Cg1] and benzene [Cg2] rings(Table 2, Fig. 2).
Experimental
To a stirred suspension mixture of dry THF (20 ml) and NaH (0,105 mol) at 0 °C under nitrogen was added dropwise a solution of 2-amino-5,6,7,8- tetrahydro-4H-ciclohepta[b]thiophene-3-carbonitrile (0,07 mol), o-fluoro-nitrobenzene (0,07 mol) in 80 ml of dry THF. The reaction mixture was stirred under room temperature for 22 h. The resulting mixture was adjusted to pH = 5 with hydroclorid acid 2 N and them extracted with CHCl3. The extracted was washed with aqueous Na2CO3, water, dried (CaCl2) and evaporated under reduced pression. The dark red solid obtained was purified by recrystallization from absolute ethanol, affording the title compound as red crystals (21.4 g, (98%), m.p. 100-102 °C)(Gewald, 1965; Gewald et al., 1966; Sridhar et al., 2007). Crystals suitable for single-crystal X-ray diffraction were grown by slow evaporation at room temperature of a solution of the pure title compound in ethanol/dichloromethane (1:1).
NMR 1H (200 MHz, CDCl3) δ : 1.60-1.70 (m, 4H), 1.80-1.90 (m, 2H), 2,70-2,80 (m, 4H), 6.90 (dt, 1H, J = 8.6, 1.6 Hz), 7.20 (dd, 1H, J = 8.6, 0.8 Hz), 7.5 (dt, 1H, J = 8.6, 8.2 Hz), 8.2 (dd, 1H, J = 8.4, 1.6 Hz), 9.6 (s, 1H).
NMR 13C (200 MHz, CDCl3) δ : 27.2-31.9 (5 CH2), 107.7, 113.9, 116.1, 119.5, 126.6, 133.9, 135.7, 136.1, 139.0, 141.4, 145.3.
Refinement
All H atoms attached to C atoms and N atom were fixed geometrically and treated as riding with C—H = 0.93 Å (aromatic) or 0.97 Å (methylene) and N—H = 0.86 Å with Uiso(H) = 1.2Ueq(C or N).The maximum and minimum residual electron density peaks were located 1.10 and 0.78 Å, from the H4A and S1 atoms respectively.
Figures
Fig. 1.
Molecular view of C16H15N3O2S, showing the atom labelling scheme. Ellipsoids are drawn at the 50% probability level. H atoms are represented as small spheres of arbitrary radii.
Fig. 2.
View showing π- π stacking interactions between the molecules. For the sake of clarity, H atoms have been omitted.
Crystal data
| C16H15N3O2S | F(000) = 656 |
| Mr = 313.37 | Dx = 1.388 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 6392 reflections |
| a = 7.0273 (3) Å | θ = 2.9–26.7° |
| b = 14.4569 (6) Å | µ = 0.23 mm−1 |
| c = 14.8867 (7) Å | T = 295 K |
| β = 97.571 (2)° | Prism, colorless |
| V = 1499.18 (11) Å3 | 0.35 × 0.32 × 0.27 mm |
| Z = 4 |
Data collection
| Nonius KappaCCD diffractometer | 2452 reflections with I > 2σ(I) |
| Radiation source: Enraf Nonius FR590 | Rint = 0.046 |
| horizonally mounted graphite crystal | θmax = 26.6°, θmin = 3.1° |
| Detector resolution: 9 pixels mm-1 | h = −8→8 |
| CCD rotation images,thick slices scans | k = −15→18 |
| 9360 measured reflections | l = −18→18 |
| 3147 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.052 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.162 | H-atom parameters constrained |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.1026P)2 + 0.1476P] where P = (Fo2 + 2Fc2)/3 |
| 3147 reflections | (Δ/σ)max = 0.001 |
| 200 parameters | Δρmax = 0.29 e Å−3 |
| 0 restraints | Δρmin = −0.30 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.29875 (7) | 0.33887 (3) | 0.05524 (3) | 0.0529 (2) | |
| O1 | 0.1864 (3) | 0.64817 (11) | −0.14472 (10) | 0.0763 (5) | |
| O2 | 0.1729 (3) | 0.78484 (12) | −0.09126 (12) | 0.0887 (6) | |
| N1 | 0.2433 (2) | 0.50661 (11) | −0.03762 (10) | 0.0531 (4) | |
| H1 | 0.2277 | 0.5305 | −0.0910 | 0.064* | |
| N2 | 0.1906 (2) | 0.70191 (13) | −0.07994 (12) | 0.0594 (4) | |
| N3 | 0.2128 (4) | 0.44432 (17) | −0.26828 (13) | 0.0871 (7) | |
| C1 | 0.2620 (2) | 0.41167 (13) | −0.03697 (12) | 0.0474 (4) | |
| C2 | 0.2544 (3) | 0.36042 (14) | −0.11578 (12) | 0.0501 (4) | |
| C3 | 0.2772 (3) | 0.26261 (14) | −0.10156 (13) | 0.0527 (5) | |
| C4 | 0.2731 (4) | 0.19525 (16) | −0.17935 (14) | 0.0673 (6) | |
| H4A | 0.4041 | 0.1775 | −0.1850 | 0.081* | |
| H4B | 0.2221 | 0.2269 | −0.2348 | 0.081* | |
| C5 | 0.1554 (3) | 0.10721 (16) | −0.17147 (15) | 0.0688 (6) | |
| H5A | 0.0271 | 0.1251 | −0.1609 | 0.083* | |
| H5B | 0.1438 | 0.0750 | −0.2291 | 0.083* | |
| C6 | 0.2345 (4) | 0.04019 (15) | −0.09784 (16) | 0.0707 (6) | |
| H6A | 0.3688 | 0.0293 | −0.1031 | 0.085* | |
| H6B | 0.1677 | −0.0183 | −0.1086 | 0.085* | |
| C7 | 0.2190 (4) | 0.07060 (16) | −0.00136 (16) | 0.0710 (6) | |
| H7A | 0.0872 | 0.0888 | 0.0021 | 0.085* | |
| H7B | 0.2471 | 0.0179 | 0.0385 | 0.085* | |
| C8 | 0.3487 (4) | 0.14916 (15) | 0.03367 (16) | 0.0669 (6) | |
| H8A | 0.4798 | 0.1324 | 0.0270 | 0.080* | |
| H8B | 0.3418 | 0.1564 | 0.0979 | 0.080* | |
| C9 | 0.3044 (3) | 0.24118 (14) | −0.01205 (13) | 0.0551 (5) | |
| C10 | 0.2447 (2) | 0.57016 (13) | 0.03089 (12) | 0.0473 (4) | |
| C11 | 0.2192 (3) | 0.66561 (13) | 0.01206 (13) | 0.0491 (4) | |
| C12 | 0.2185 (3) | 0.73041 (15) | 0.08074 (14) | 0.0587 (5) | |
| H12 | 0.2012 | 0.7927 | 0.0663 | 0.070* | |
| C13 | 0.2430 (3) | 0.70360 (16) | 0.16918 (15) | 0.0635 (5) | |
| H13 | 0.2427 | 0.7472 | 0.2151 | 0.076* | |
| C14 | 0.2683 (3) | 0.61106 (16) | 0.18993 (14) | 0.0636 (5) | |
| H14 | 0.2845 | 0.5924 | 0.2503 | 0.076* | |
| C15 | 0.2700 (3) | 0.54584 (15) | 0.12264 (13) | 0.0594 (5) | |
| H15 | 0.2884 | 0.4840 | 0.1386 | 0.071* | |
| C16 | 0.2285 (3) | 0.40474 (15) | −0.20159 (13) | 0.0594 (5) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0672 (3) | 0.0504 (3) | 0.0413 (3) | −0.00605 (19) | 0.0074 (2) | 0.00132 (18) |
| O1 | 0.1190 (14) | 0.0599 (10) | 0.0478 (8) | −0.0002 (8) | 0.0034 (8) | 0.0025 (7) |
| O2 | 0.1394 (16) | 0.0503 (10) | 0.0753 (11) | 0.0042 (9) | 0.0103 (10) | 0.0123 (8) |
| N1 | 0.0707 (10) | 0.0480 (9) | 0.0407 (8) | −0.0010 (7) | 0.0077 (7) | 0.0000 (7) |
| N2 | 0.0684 (10) | 0.0538 (10) | 0.0558 (10) | −0.0019 (7) | 0.0072 (8) | 0.0042 (8) |
| N3 | 0.1274 (18) | 0.0861 (15) | 0.0481 (11) | 0.0209 (13) | 0.0123 (10) | 0.0112 (10) |
| C1 | 0.0508 (9) | 0.0485 (10) | 0.0427 (9) | −0.0040 (7) | 0.0061 (7) | 0.0005 (7) |
| C2 | 0.0563 (10) | 0.0523 (10) | 0.0422 (9) | −0.0028 (8) | 0.0078 (7) | 0.0000 (8) |
| C3 | 0.0604 (10) | 0.0505 (10) | 0.0475 (10) | −0.0033 (8) | 0.0084 (8) | −0.0032 (8) |
| C4 | 0.0929 (15) | 0.0587 (12) | 0.0513 (11) | −0.0070 (11) | 0.0131 (10) | −0.0077 (10) |
| C5 | 0.0836 (14) | 0.0569 (12) | 0.0647 (13) | 0.0012 (10) | 0.0048 (11) | −0.0140 (10) |
| C6 | 0.0909 (16) | 0.0494 (12) | 0.0705 (14) | −0.0006 (11) | 0.0057 (11) | −0.0047 (10) |
| C7 | 0.0915 (15) | 0.0502 (12) | 0.0715 (14) | −0.0034 (10) | 0.0116 (11) | 0.0040 (11) |
| C8 | 0.0859 (14) | 0.0559 (13) | 0.0566 (12) | −0.0038 (10) | 0.0010 (10) | 0.0049 (9) |
| C9 | 0.0650 (11) | 0.0505 (11) | 0.0497 (10) | −0.0063 (8) | 0.0069 (8) | −0.0009 (8) |
| C10 | 0.0481 (9) | 0.0511 (10) | 0.0432 (9) | −0.0042 (7) | 0.0081 (7) | −0.0011 (8) |
| C11 | 0.0493 (9) | 0.0508 (11) | 0.0474 (10) | −0.0045 (7) | 0.0071 (7) | 0.0001 (7) |
| C12 | 0.0653 (11) | 0.0520 (11) | 0.0597 (12) | 0.0000 (9) | 0.0120 (9) | −0.0070 (9) |
| C13 | 0.0725 (13) | 0.0632 (13) | 0.0569 (11) | −0.0048 (10) | 0.0169 (9) | −0.0144 (10) |
| C14 | 0.0811 (13) | 0.0665 (13) | 0.0448 (11) | −0.0042 (10) | 0.0147 (9) | −0.0043 (9) |
| C15 | 0.0786 (13) | 0.0532 (11) | 0.0478 (11) | −0.0023 (10) | 0.0134 (9) | 0.0008 (9) |
| C16 | 0.0750 (12) | 0.0587 (12) | 0.0442 (10) | 0.0058 (9) | 0.0066 (8) | −0.0043 (9) |
Geometric parameters (Å, °)
| S1—C1 | 1.7220 (18) | C6—C7 | 1.520 (3) |
| S1—C9 | 1.735 (2) | C6—H6A | 0.9700 |
| O1—N2 | 1.236 (2) | C6—H6B | 0.9700 |
| O2—N2 | 1.215 (2) | C7—C8 | 1.506 (3) |
| N1—C10 | 1.372 (2) | C7—H7A | 0.9700 |
| N1—C1 | 1.379 (2) | C7—H7B | 0.9700 |
| N1—H1 | 0.8600 | C8—C9 | 1.508 (3) |
| N2—C11 | 1.456 (2) | C8—H8A | 0.9700 |
| N3—C16 | 1.139 (3) | C8—H8B | 0.9700 |
| C1—C2 | 1.383 (2) | C10—C15 | 1.399 (3) |
| C2—C16 | 1.419 (3) | C10—C11 | 1.415 (3) |
| C2—C3 | 1.436 (3) | C11—C12 | 1.387 (3) |
| C3—C9 | 1.357 (3) | C12—C13 | 1.361 (3) |
| C3—C4 | 1.510 (3) | C12—H12 | 0.9300 |
| C4—C5 | 1.531 (3) | C13—C14 | 1.379 (3) |
| C4—H4A | 0.9700 | C13—H13 | 0.9300 |
| C4—H4B | 0.9700 | C14—C15 | 1.377 (3) |
| C5—C6 | 1.513 (3) | C14—H14 | 0.9300 |
| C5—H5A | 0.9700 | C15—H15 | 0.9300 |
| C5—H5B | 0.9700 | ||
| C1—S1—C9 | 92.84 (9) | C8—C7—C6 | 115.5 (2) |
| C10—N1—C1 | 132.09 (16) | C8—C7—H7A | 108.4 |
| C10—N1—H1 | 114.0 | C6—C7—H7A | 108.4 |
| C1—N1—H1 | 114.0 | C8—C7—H7B | 108.4 |
| O2—N2—O1 | 121.38 (19) | C6—C7—H7B | 108.4 |
| O2—N2—C11 | 119.03 (18) | H7A—C7—H7B | 107.5 |
| O1—N2—C11 | 119.59 (17) | C7—C8—C9 | 115.43 (19) |
| N1—C1—C2 | 122.32 (17) | C7—C8—H8A | 108.4 |
| N1—C1—S1 | 128.18 (14) | C9—C8—H8A | 108.4 |
| C2—C1—S1 | 109.50 (14) | C7—C8—H8B | 108.4 |
| C1—C2—C16 | 120.52 (18) | C9—C8—H8B | 108.4 |
| C1—C2—C3 | 114.31 (17) | H8A—C8—H8B | 107.5 |
| C16—C2—C3 | 125.17 (18) | C3—C9—C8 | 129.69 (19) |
| C9—C3—C2 | 111.61 (17) | C3—C9—S1 | 111.73 (15) |
| C9—C3—C4 | 126.28 (19) | C8—C9—S1 | 118.46 (15) |
| C2—C3—C4 | 122.11 (18) | N1—C10—C15 | 123.03 (18) |
| C3—C4—C5 | 115.66 (17) | N1—C10—C11 | 121.17 (16) |
| C3—C4—H4A | 108.4 | C15—C10—C11 | 115.79 (17) |
| C5—C4—H4A | 108.4 | C12—C11—C10 | 121.68 (18) |
| C3—C4—H4B | 108.4 | C12—C11—N2 | 115.89 (18) |
| C5—C4—H4B | 108.4 | C10—C11—N2 | 122.43 (16) |
| H4A—C4—H4B | 107.4 | C13—C12—C11 | 120.6 (2) |
| C6—C5—C4 | 115.93 (19) | C13—C12—H12 | 119.7 |
| C6—C5—H5A | 108.3 | C11—C12—H12 | 119.7 |
| C4—C5—H5A | 108.3 | C12—C13—C14 | 119.26 (19) |
| C6—C5—H5B | 108.3 | C12—C13—H13 | 120.4 |
| C4—C5—H5B | 108.3 | C14—C13—H13 | 120.4 |
| H5A—C5—H5B | 107.4 | C15—C14—C13 | 120.97 (19) |
| C5—C6—C7 | 115.73 (19) | C15—C14—H14 | 119.5 |
| C5—C6—H6A | 108.3 | C13—C14—H14 | 119.5 |
| C7—C6—H6A | 108.3 | C14—C15—C10 | 121.7 (2) |
| C5—C6—H6B | 108.3 | C14—C15—H15 | 119.1 |
| C7—C6—H6B | 108.3 | C10—C15—H15 | 119.1 |
| H6A—C6—H6B | 107.4 | N3—C16—C2 | 176.3 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1 | 0.86 | 1.89 | 2.593 (2) | 138 |
| C12—H12···O2 | 0.93 | 2.33 | 2.657 (3) | 100 |
| C15—H15···S1 | 0.93 | 2.44 | 3.171 (2) | 135 |
Table 2 π–π stacking interactions (Å, °) between the thiophene (Cg1) and benzene (Cg2) rings.
| Cg1···Cg2 | Cg1 to plane 2 | Cg2 to plane 1 | Offset | |
| Cg1···Cg2i | 3.7089 (12) | -3.5039 (9) | -3.5021 (9) | 19.4 |
| Cg1···Cg2ii | 3.6170 (12) | 3.4761 (9) | 3.4878 (9) | 16.0 |
Symmetry codes: (i) -x, 1-y, -z; (ii) 1-x, 1-y, -z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: DN2562).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. J. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19
- Calligaro, D. O., Fairhurst, J., Hotten, T. M., Moore, N. A. & Tupper, D. E. (1997). Bioorg. Med. Chem. Lett.7, 25–30.
- Campaigne, E. (1984). Comprehensive Heterocyclic Chemistry, Vol. 4, edited by A. R. Katritzky & C. W. Rees, pp. 863–934. Oxford: Pergamon.
- Chakrabarti, J. K., Hotten, T. M., Morgan, S. E., Pullar, I. A., Rackham, D. M., Risius, F. C., Wedley, S., Chaney, M. & Jones, N. D. (1982). J. Med. Chem.25, 1133–1140. [DOI] [PubMed]
- Chen, S., Guzei, I. A. & Yu, L. (2005). J. Am. Chem. Soc.127, 9881–9885. [DOI] [PubMed]
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc.97, 1354–1358.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Gewald, K. (1965). Chem. Ber.98, 3571–3577.
- Gewald, K., Schinke, E. & Bottcher, H. (1966). Chem. Ber.99, 99–100.
- Kleemann, A., Engel, J. B., Kutscher, B. & Reichert, D. (2006). Pharmaceutical Substances New York, Stuttgart: Georg Thieme Verlag.
- Nikolakopoulos, G., Figler, H., Linden, J. & Scammells, P. J. (2006). Bioorg. Med. Chem.14, 2358–2365. [DOI] [PubMed]
- Nonius (1997). COLLECT Nonius BV, Delft, The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sridhar, M., Rao, R. M., Baba, N. H. K. & Kumbhare, R. M. (2007). Tetrahedron Lett.48, 3171–3172.
- Stephenson, G. A., Borchardt, T. B., Byrn, S. R., Bowyer, J., Bunnell, C. A., Snorek, S. V. & Yu, L. (1995). J. Pharm. Sci.84, 1385–1386. [DOI] [PubMed]
- Yu, L. (2002). J. Phys. Chem. A, 106, 544–550.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810017149/dn2562sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810017149/dn2562Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report