Abstract
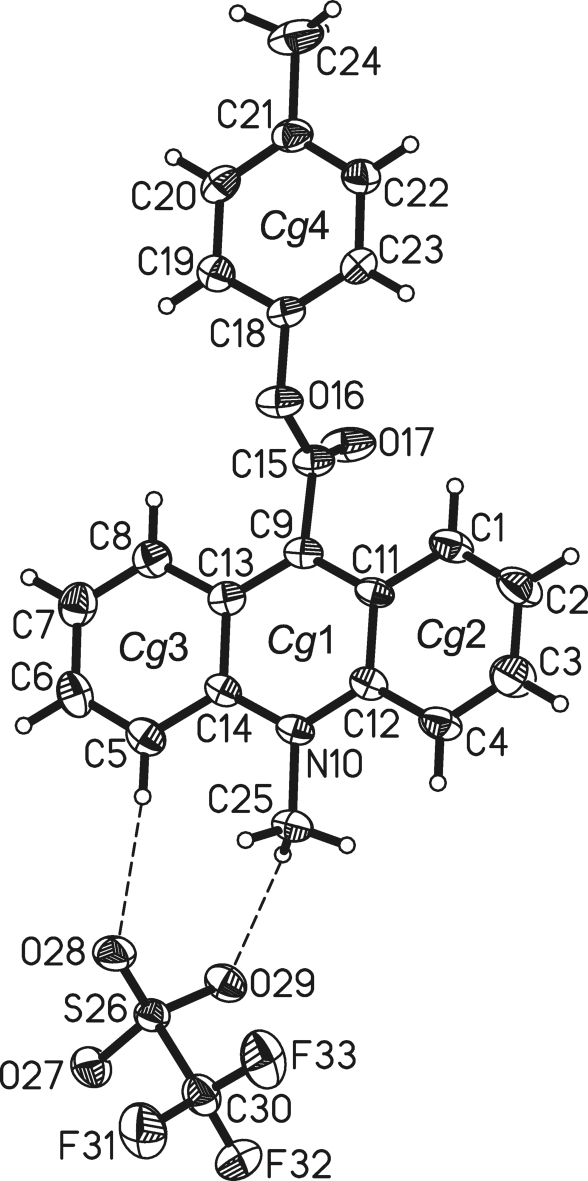
In the crystal structure of the title compound, C22H18NO2 +·CF3SO3 −, adjacent cations are linked through C—H⋯π and π–π interactions, and the cations and anions are connected by C—H⋯O and C—F⋯π interactions. The acridine and benzene ring systems are oriented at a dihedral angle of 3.0 (1)°. The carboxyl group is twisted at an angle of 83.1 (1)° relative to the acridine skeleton. The mean planes of adjacent acridine units are parallel or inclined at an angle of 75.2 (1)° in the crystal structure.
Related literature
For background to the chemiluminogenic properties of 9-phenoxycarbonyl-10-methylacridinium trifluoromethanesulfonates, see: Brown et al. (2009 ▶); Rak et al. (1999 ▶); Roda et al. (2003 ▶); Zomer & Jacquemijns (2001 ▶). For related structures, see: Sikorski et al. (2006 ▶); Trzybiński et al. (2010 ▶). For intermolecular interactions, see: Bianchi et al. (2004 ▶); Dorn et al. (2005 ▶); Hunter et al. (2001 ▶); Novoa et al. (2006 ▶); Takahashi et al. (2001 ▶). For the synthesis, see: Sato (1996 ▶); Sikorski et al. (2006 ▶); Trzybiński et al. (2010 ▶).
Experimental
Crystal data
C22H18NO2 +·CF3O3S−
M r = 477.45
Monoclinic,

a = 13.2686 (6) Å
b = 8.4788 (4) Å
c = 20.4078 (10) Å
β = 106.749 (5)°
V = 2198.51 (19) Å3
Z = 4
Mo Kα radiation
μ = 0.21 mm−1
T = 295 K
0.50 × 0.40 × 0.10 mm
Data collection
Oxford Diffraction Gemini R Ultra Ruby CCD diffractometer
Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2008 ▶) T min = 0.869, T max = 1.000
12172 measured reflections
3892 independent reflections
2096 reflections with I > 2σ(I)
R int = 0.061
Refinement
R[F 2 > 2σ(F 2)] = 0.057
wR(F 2) = 0.137
S = 0.95
3892 reflections
299 parameters
H-atom parameters constrained
Δρmax = 0.23 e Å−3
Δρmin = −0.19 e Å−3
Data collection: CrysAlis CCD (Oxford Diffraction, 2008 ▶); cell refinement: CrysAlis RED; data reduction: CrysAlis RED; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: SHELXL97 and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810016302/ng2760sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810016302/ng2760Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg4 is the centroid of the C18–C23 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C3—H3⋯O27i | 0.93 | 2.57 | 3.314 (5) | 137 |
| C4—H4⋯O29i | 0.93 | 2.44 | 3.319 (4) | 159 |
| C5—H5⋯O28 | 0.93 | 2.44 | 3.364 (5) | 171 |
| C6—H6⋯O28ii | 0.93 | 2.56 | 3.342 (5) | 142 |
| C23—H23⋯O27iii | 0.93 | 2.53 | 3.448 (4) | 169 |
| C25—H25A⋯O29 | 0.96 | 2.56 | 3.415 (5) | 149 |
| C25—H25B⋯Cg4iv | 0.96 | 2.62 | 3.487 (4) | 151 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv) 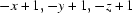 .
.
Table 2. C–F⋯π interactions (Å,°).
Cg1 and Cg2 are the centroids of the C9/N10/C11–C14 and C1–C4/C11/C12 rings, respectively.
| X | I | J | I⋯J | X⋯J | X–I⋯J |
|---|---|---|---|---|---|
| C30 | F31 | Cg2i | 3.420 (3) | 4.044 (4) | 108.9 (2) |
| C30 | F32 | Cg1i | 3.441 (3) | 4.032 (4) | 107.1 (2) |
| C30 | F32 | Cg2i | 3.788 (4) | 4.044 (4) | 91.5 (2) |
| C30 | F33 | Cg1i | 3.669 (3) | 4.032 (4) | 96.2 (2) |
Symmetry code: (i)  .
.
Table 3. π–π interactions (Å,°).
Cg3 and Cg4 are the centroids of the C5–C8/C13/C14 and C18–C23 rings, respectively. CgI⋯CgJ is the distance between ring centroids. The dihedral angle is that between the planes of the rings I and J. CgI_Perp is the perpendicular distance of CgI from ring J. CgI_Offset is the distance between CgI and perpendicular projection of CgJ on ring I.
| I | J | CgI⋯CgJ | Dihedral angle | CgI_Perp | CgI_Offset |
|---|---|---|---|---|---|
| 3 | 4v | 3.913 (2) | 4.80 (17) | 3.472 (2) | 1.805 (2) |
| 4 | 3v | 3.913 (2) | 4.80 (17) | 3.565 (2) | 1.613 (2) |
Symmetry code: (v)  .
.
Acknowledgments
This study was financed by the State Funds for Scientific Research (grant No. N204 123 32/3143, contract No. 3143/H03/2007/32, of the Polish Ministry of Research and Higher Education) for the period 2007–2010.
supplementary crystallographic information
Comment
9-(Phenoxycarbonyl)-10-methylacridinium salts appear to be convenient chemiluminescent indicators or the chemiluminogenic fragments of chemiluminescent labels (Zomer & Jacquemijns, 2001), which are widely applied in assays of biologically and environmentally important entities such as antigens, antibodies, enzymes or DNA fragments (Roda et al., 2003; Brown et al., 2009). Oxidation of the cations of these salts with hydrogen peroxide in alkaline media is accompanied by the removal of the phenoxycarbonyl fragment and the conversion of the remaining part of the molecule to electronically excited, light emitting 10-methyl-9-acridinone (Rak et al., 1999; Zomer & Jacquemijns, 2001). The efficiency of chemiluminescence – crucial for analytical applications – is affected by the constitution of the phenyl fragment (Zomer & Jacquemijns, 2001). This prompted us to undertake investigations on derivatives substituted in this fragment. Here we present the structure of 9-(4-methylphenoxy)carbonyl-10-methylacridinium trifluoromethanesulfonate, a structural isomer of the 2-methyl substituted salt, whose structure has already been refined (Sikorski et al., 2006).
In the cation of the title compound (Fig. 1), the bond lengths and angles characterizing the geometry of the acridinium moiety are typical of acridine-based derivatives (Sikorski et al., 2006; Trzybiński et al., 2010). With respective average deviations from planarity of 0.0386 (3) Å and 0.0017 (3) Å, the acridine and benzene ring systems are oriented at a dihedral angle of 3.0 (1)°. The carboxyl group is twisted at an angle of 83.1 (1)° relative to the acridine skeleton. The mean planes of the adjacent acridine moieties are parallel (remain at an angle of 0.0 (1)°) or inclined at an angle of 75.2 (1)° in the lattice.
In the crystal structure, the adjacent cations are linked through C–H···π (Table 1, Fig. 2) and π–π (involving acridine and phenyl moieties) (Table 3, Fig. 2) interactions, and cations and anions are connected by multidirectional C–H···O (Table 1, Figs. 1 and 2) and C–F···π (Table 2, Fig. 2) interactions. The C–H···O interactions are of the hydrogen bond type (Bianchi et al. 2004; Novoa et al., 2006). The C–H···π interactions should be of an attractive nature (Takahashi et al., 2001), like the C–F···π (Dorn et al., 2005) and π–π (Hunter et al., 2001) interactions. The crystal structure is stabilized by a network of these short-range specific interactions and by long-range electrostatic interactions between ions.
Experimental
The compound was synthesized as described elsewhere (Sikorski et al., 2006; Trzybiński et al., 2010), i.e., 9-(chlorocarbonyl)acridine obtained by treating acridine-9-carboxylic acid with a tenfold molar excess of thionyl chloride was esterified with 4-methylphenol in anhydrous dichloromethane in the presence of N,N-diethylethanamine and a catalytic amount of N,N-dimethyl-4-pyridinamine (room temperature, 15h) (Sato, 1996). The product – 4-methylphenyl acridine-9-carboxylate, purified chromatographically (SiO2, cyclohexane/ethyl acetate, 3/2 v/v) – was quaternarized with a fivefold molar excess of methyl trifluoromethanesulfonate dissolved in anhydrous dichloromethane. The crude 9-(4-methylphenoxycarbonyl)-10-methylacridinium trifluoromethanesulfonate was dissolved in a small amount of ethanol, filtered and precipitated with 25 v/v excess of diethyl ether. Light-orange crystals suitable for X-ray investigations were grown from absolute ethanol solution (m.p. 445 - 447 K).
Refinement
H atoms were positioned geometrically, with C—H = 0.93 Å and 0.96 Å for the aromatic and methyl H atoms, respectively, and constrained to ride on their parent atoms with Uiso(H) = xUeq(C), where x = 1.2 for the aromatic and x = 1.5 for the methyl H atoms.
Figures
Fig. 1.
The molecular structure of the title compound showing the atom-labeling scheme. Displacement ellipsoids are drawn at the 25% probability level and H atoms are shown as small spheres of arbitrary radius. Cg1, Cg2, Cg3 and Cg4 denote the ring centroids. The C–H···O hydrogen bonds are represented by dashed lines.
Fig. 2.
The arrangement of the ions in the crystal structure. The C–H···O interactions are represented by dashed lines, the C–H···π, C–F···π and π–π contacts by dotted lines. H atoms not involved in interactions have been omitted. [Symmetry codes: (i) –x + 3/2, y + 1/2, –z + 1/2; (ii) –x + 2, –y + 1, –z + 1; (iii) x – 1, y, z; (iv) –x + 1, –y + 1, –z + 1; (v) –x + 1, –y, –z + 1.]
Crystal data
| C22H18NO2+·CF3O3S− | F(000) = 984 |
| Mr = 477.45 | Dx = 1.442 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 3355 reflections |
| a = 13.2686 (6) Å | θ = 3.1–29.2° |
| b = 8.4788 (4) Å | µ = 0.21 mm−1 |
| c = 20.4078 (10) Å | T = 295 K |
| β = 106.749 (5)° | Plate, light-orange |
| V = 2198.51 (19) Å3 | 0.50 × 0.40 × 0.10 mm |
| Z = 4 |
Data collection
| Oxford Diffraction Gemini R Ultra Ruby CCD diffractometer | 3892 independent reflections |
| Radiation source: enhanced (Mo) X-Ray Source | 2096 reflections with I > 2σ(I) |
| graphite | Rint = 0.061 |
| Detector resolution: 10.4002 pixels mm-1 | θmax = 25.1°, θmin = 3.1° |
| ω scans | h = −15→15 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2008) | k = −10→10 |
| Tmin = 0.869, Tmax = 1.000 | l = −24→24 |
| 12172 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.057 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.137 | H-atom parameters constrained |
| S = 0.95 | w = 1/[σ2(Fo2) + (0.0678P)2] where P = (Fo2 + 2Fc2)/3 |
| 3892 reflections | (Δ/σ)max < 0.001 |
| 299 parameters | Δρmax = 0.23 e Å−3 |
| 0 restraints | Δρmin = −0.19 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.3367 (3) | 0.4502 (4) | 0.3326 (2) | 0.0699 (10) | |
| H1 | 0.2898 | 0.4002 | 0.3519 | 0.084* | |
| C2 | 0.2997 (3) | 0.5409 (5) | 0.2766 (2) | 0.0918 (13) | |
| H2 | 0.2276 | 0.5502 | 0.2563 | 0.110* | |
| C3 | 0.3705 (3) | 0.6211 (5) | 0.2491 (2) | 0.0957 (14) | |
| H3 | 0.3441 | 0.6854 | 0.2111 | 0.115* | |
| C4 | 0.4760 (3) | 0.6083 (4) | 0.2759 (2) | 0.0691 (10) | |
| H4 | 0.5208 | 0.6634 | 0.2565 | 0.083* | |
| C5 | 0.7739 (3) | 0.3924 (4) | 0.44967 (19) | 0.0631 (9) | |
| H5 | 0.8203 | 0.4433 | 0.4302 | 0.076* | |
| C6 | 0.8115 (3) | 0.3064 (5) | 0.5083 (2) | 0.0735 (10) | |
| H6 | 0.8838 | 0.3013 | 0.5287 | 0.088* | |
| C7 | 0.7440 (3) | 0.2253 (5) | 0.5386 (2) | 0.0766 (11) | |
| H7 | 0.7713 | 0.1667 | 0.5783 | 0.092* | |
| C8 | 0.6389 (3) | 0.2334 (4) | 0.50934 (18) | 0.0639 (10) | |
| H8 | 0.5944 | 0.1786 | 0.5291 | 0.077* | |
| C9 | 0.4876 (3) | 0.3337 (4) | 0.41922 (16) | 0.0479 (8) | |
| N10 | 0.62352 (19) | 0.4910 (3) | 0.36094 (13) | 0.0486 (7) | |
| C11 | 0.4461 (2) | 0.4303 (4) | 0.36217 (16) | 0.0498 (8) | |
| C12 | 0.5177 (2) | 0.5109 (4) | 0.33342 (16) | 0.0495 (8) | |
| C13 | 0.5948 (3) | 0.3229 (4) | 0.44951 (16) | 0.0507 (8) | |
| C14 | 0.6651 (2) | 0.4037 (4) | 0.41908 (16) | 0.0473 (8) | |
| C15 | 0.4137 (3) | 0.2312 (4) | 0.44360 (17) | 0.0548 (9) | |
| O16 | 0.38896 (18) | 0.2932 (3) | 0.49667 (12) | 0.0648 (7) | |
| O17 | 0.3828 (2) | 0.1080 (3) | 0.41764 (13) | 0.0858 (9) | |
| C18 | 0.3185 (3) | 0.2065 (4) | 0.52390 (16) | 0.0500 (8) | |
| C19 | 0.3597 (3) | 0.1129 (4) | 0.57992 (17) | 0.0609 (9) | |
| H19 | 0.4321 | 0.0997 | 0.5978 | 0.073* | |
| C20 | 0.2903 (3) | 0.0381 (4) | 0.60930 (17) | 0.0652 (10) | |
| H20 | 0.3170 | −0.0262 | 0.6473 | 0.078* | |
| C21 | 0.1839 (3) | 0.0565 (4) | 0.58395 (19) | 0.0647 (10) | |
| C22 | 0.1461 (3) | 0.1515 (5) | 0.52795 (19) | 0.0671 (10) | |
| H22 | 0.0739 | 0.1655 | 0.5102 | 0.081* | |
| C23 | 0.2132 (3) | 0.2273 (4) | 0.49707 (17) | 0.0593 (9) | |
| H23 | 0.1866 | 0.2908 | 0.4589 | 0.071* | |
| C24 | 0.1088 (4) | −0.0241 (5) | 0.6176 (2) | 0.1041 (15) | |
| H24A | 0.0428 | 0.0309 | 0.6055 | 0.156* | |
| H24B | 0.1387 | −0.0225 | 0.6664 | 0.156* | |
| H24C | 0.0978 | −0.1313 | 0.6021 | 0.156* | |
| C25 | 0.6972 (3) | 0.5623 (4) | 0.32692 (17) | 0.0655 (10) | |
| H25A | 0.7583 | 0.4964 | 0.3343 | 0.098* | |
| H25B | 0.7180 | 0.6650 | 0.3458 | 0.098* | |
| H25C | 0.6632 | 0.5715 | 0.2787 | 0.098* | |
| S26 | 0.98783 (7) | 0.47984 (11) | 0.33273 (4) | 0.0560 (3) | |
| O27 | 1.09157 (18) | 0.4193 (3) | 0.34606 (12) | 0.0801 (8) | |
| O28 | 0.96339 (18) | 0.5485 (3) | 0.39004 (12) | 0.0844 (8) | |
| O29 | 0.90554 (18) | 0.3856 (3) | 0.28984 (12) | 0.0688 (7) | |
| C30 | 0.9911 (3) | 0.6484 (5) | 0.2789 (2) | 0.0737 (11) | |
| F31 | 1.0614 (2) | 0.7536 (3) | 0.31098 (15) | 0.1247 (10) | |
| F32 | 1.0156 (2) | 0.6073 (4) | 0.22295 (13) | 0.1123 (9) | |
| F33 | 0.8988 (2) | 0.7204 (3) | 0.25876 (13) | 0.1117 (9) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.057 (2) | 0.061 (2) | 0.098 (3) | −0.003 (2) | 0.033 (2) | 0.014 (2) |
| C2 | 0.050 (2) | 0.096 (3) | 0.131 (4) | 0.012 (2) | 0.029 (2) | 0.033 (3) |
| C3 | 0.078 (3) | 0.090 (3) | 0.117 (3) | 0.012 (3) | 0.027 (3) | 0.046 (3) |
| C4 | 0.056 (2) | 0.060 (2) | 0.098 (3) | −0.0012 (19) | 0.032 (2) | 0.019 (2) |
| C5 | 0.060 (2) | 0.052 (2) | 0.079 (3) | −0.0077 (19) | 0.022 (2) | −0.011 (2) |
| C6 | 0.061 (2) | 0.063 (3) | 0.085 (3) | −0.001 (2) | 0.003 (2) | −0.010 (2) |
| C7 | 0.086 (3) | 0.063 (3) | 0.073 (3) | −0.001 (2) | 0.011 (2) | 0.004 (2) |
| C8 | 0.071 (3) | 0.054 (2) | 0.067 (2) | −0.008 (2) | 0.020 (2) | −0.003 (2) |
| C9 | 0.058 (2) | 0.0330 (18) | 0.060 (2) | −0.0060 (16) | 0.0285 (18) | −0.0113 (17) |
| N10 | 0.0474 (17) | 0.0375 (15) | 0.0676 (17) | −0.0061 (13) | 0.0274 (14) | −0.0070 (14) |
| C11 | 0.048 (2) | 0.0370 (18) | 0.071 (2) | −0.0068 (16) | 0.0289 (18) | −0.0049 (18) |
| C12 | 0.052 (2) | 0.0331 (19) | 0.067 (2) | −0.0010 (16) | 0.0229 (17) | −0.0037 (17) |
| C13 | 0.063 (2) | 0.0320 (18) | 0.060 (2) | −0.0040 (17) | 0.0225 (18) | −0.0059 (16) |
| C14 | 0.046 (2) | 0.0351 (18) | 0.063 (2) | −0.0052 (15) | 0.0203 (17) | −0.0105 (17) |
| C15 | 0.068 (2) | 0.040 (2) | 0.066 (2) | −0.0062 (18) | 0.0334 (19) | −0.0037 (18) |
| O16 | 0.0833 (17) | 0.0462 (14) | 0.0804 (16) | −0.0143 (13) | 0.0485 (14) | −0.0130 (12) |
| O17 | 0.122 (2) | 0.0581 (17) | 0.1013 (19) | −0.0380 (16) | 0.0701 (18) | −0.0252 (16) |
| C18 | 0.066 (2) | 0.0374 (19) | 0.054 (2) | −0.0074 (18) | 0.0285 (19) | −0.0066 (17) |
| C19 | 0.061 (2) | 0.057 (2) | 0.066 (2) | −0.0032 (19) | 0.0198 (19) | −0.008 (2) |
| C20 | 0.091 (3) | 0.054 (2) | 0.0529 (19) | −0.004 (2) | 0.024 (2) | 0.0038 (18) |
| C21 | 0.074 (3) | 0.068 (3) | 0.060 (2) | −0.014 (2) | 0.032 (2) | −0.014 (2) |
| C22 | 0.061 (2) | 0.078 (3) | 0.068 (2) | −0.001 (2) | 0.027 (2) | −0.004 (2) |
| C23 | 0.072 (3) | 0.055 (2) | 0.053 (2) | 0.002 (2) | 0.0214 (19) | 0.0023 (17) |
| C24 | 0.141 (4) | 0.097 (3) | 0.099 (3) | −0.038 (3) | 0.074 (3) | −0.008 (3) |
| C25 | 0.058 (2) | 0.068 (2) | 0.079 (2) | −0.0098 (19) | 0.0331 (19) | −0.0052 (19) |
| S26 | 0.0511 (5) | 0.0592 (6) | 0.0605 (5) | 0.0019 (5) | 0.0208 (4) | 0.0024 (5) |
| O27 | 0.0581 (15) | 0.0891 (19) | 0.0933 (18) | 0.0225 (14) | 0.0221 (13) | 0.0194 (15) |
| O28 | 0.0716 (16) | 0.118 (2) | 0.0732 (16) | −0.0188 (16) | 0.0366 (14) | −0.0244 (15) |
| O29 | 0.0627 (15) | 0.0584 (15) | 0.0878 (17) | −0.0099 (12) | 0.0258 (13) | −0.0128 (13) |
| C30 | 0.062 (3) | 0.066 (3) | 0.087 (3) | −0.012 (2) | 0.011 (2) | 0.000 (2) |
| F31 | 0.122 (2) | 0.0878 (19) | 0.140 (2) | −0.0465 (17) | 0.0005 (17) | 0.0175 (16) |
| F32 | 0.114 (2) | 0.147 (2) | 0.0809 (15) | −0.0172 (17) | 0.0356 (14) | 0.0272 (16) |
| F33 | 0.0992 (18) | 0.0718 (16) | 0.142 (2) | 0.0200 (15) | −0.0010 (16) | 0.0185 (15) |
Geometric parameters (Å, °)
| C1—C2 | 1.347 (5) | C15—O16 | 1.327 (4) |
| C1—C11 | 1.412 (4) | O16—C18 | 1.421 (3) |
| C1—H1 | 0.9300 | C18—C23 | 1.357 (4) |
| C2—C3 | 1.401 (5) | C18—C19 | 1.370 (4) |
| C2—H2 | 0.9300 | C19—C20 | 1.388 (5) |
| C3—C4 | 1.352 (5) | C19—H19 | 0.9300 |
| C3—H3 | 0.9300 | C20—C21 | 1.366 (5) |
| C4—C12 | 1.411 (5) | C20—H20 | 0.9300 |
| C4—H4 | 0.9300 | C21—C22 | 1.369 (5) |
| C5—C6 | 1.366 (5) | C21—C24 | 1.524 (5) |
| C5—C14 | 1.401 (4) | C22—C23 | 1.388 (4) |
| C5—H5 | 0.9300 | C22—H22 | 0.9300 |
| C6—C7 | 1.406 (5) | C23—H23 | 0.9300 |
| C6—H6 | 0.9300 | C24—H24A | 0.9600 |
| C7—C8 | 1.351 (5) | C24—H24B | 0.9600 |
| C7—H7 | 0.9300 | C24—H24C | 0.9600 |
| C8—C13 | 1.413 (4) | C25—H25A | 0.9600 |
| C8—H8 | 0.9300 | C25—H25B | 0.9600 |
| C9—C13 | 1.381 (4) | C25—H25C | 0.9600 |
| C9—C11 | 1.400 (4) | S26—O27 | 1.420 (2) |
| C9—C15 | 1.499 (4) | S26—O28 | 1.425 (2) |
| N10—C12 | 1.364 (4) | S26—O29 | 1.430 (2) |
| N10—C14 | 1.372 (4) | S26—C30 | 1.811 (4) |
| N10—C25 | 1.481 (4) | C30—F32 | 1.321 (4) |
| C11—C12 | 1.427 (4) | C30—F31 | 1.321 (4) |
| C13—C14 | 1.434 (4) | C30—F33 | 1.324 (4) |
| C15—O17 | 1.189 (4) | ||
| C2—C1—C11 | 120.7 (3) | O16—C15—C9 | 112.4 (3) |
| C2—C1—H1 | 119.7 | C15—O16—C18 | 117.3 (2) |
| C11—C1—H1 | 119.7 | C23—C18—C19 | 121.9 (3) |
| C1—C2—C3 | 119.5 (4) | C23—C18—O16 | 119.4 (3) |
| C1—C2—H2 | 120.2 | C19—C18—O16 | 118.5 (3) |
| C3—C2—H2 | 120.2 | C18—C19—C20 | 118.1 (3) |
| C4—C3—C2 | 122.3 (4) | C18—C19—H19 | 121.0 |
| C4—C3—H3 | 118.9 | C20—C19—H19 | 121.0 |
| C2—C3—H3 | 118.9 | C21—C20—C19 | 121.8 (3) |
| C3—C4—C12 | 119.7 (3) | C21—C20—H20 | 119.1 |
| C3—C4—H4 | 120.1 | C19—C20—H20 | 119.1 |
| C12—C4—H4 | 120.1 | C20—C21—C22 | 118.2 (3) |
| C6—C5—C14 | 119.7 (3) | C20—C21—C24 | 121.1 (4) |
| C6—C5—H5 | 120.1 | C22—C21—C24 | 120.7 (4) |
| C14—C5—H5 | 120.1 | C21—C22—C23 | 121.5 (3) |
| C5—C6—C7 | 121.8 (4) | C21—C22—H22 | 119.2 |
| C5—C6—H6 | 119.1 | C23—C22—H22 | 119.2 |
| C7—C6—H6 | 119.1 | C18—C23—C22 | 118.5 (3) |
| C8—C7—C6 | 119.2 (4) | C18—C23—H23 | 120.7 |
| C8—C7—H7 | 120.4 | C22—C23—H23 | 120.7 |
| C6—C7—H7 | 120.4 | C21—C24—H24A | 109.5 |
| C7—C8—C13 | 121.7 (3) | C21—C24—H24B | 109.5 |
| C7—C8—H8 | 119.1 | H24A—C24—H24B | 109.5 |
| C13—C8—H8 | 119.1 | C21—C24—H24C | 109.5 |
| C13—C9—C11 | 121.2 (3) | H24A—C24—H24C | 109.5 |
| C13—C9—C15 | 120.1 (3) | H24B—C24—H24C | 109.5 |
| C11—C9—C15 | 118.5 (3) | N10—C25—H25A | 109.4 |
| C12—N10—C14 | 122.2 (2) | N10—C25—H25B | 109.5 |
| C12—N10—C25 | 119.8 (3) | H25A—C25—H25B | 109.5 |
| C14—N10—C25 | 118.0 (3) | N10—C25—H25C | 109.6 |
| C9—C11—C1 | 122.4 (3) | H25A—C25—H25C | 109.5 |
| C9—C11—C12 | 118.2 (3) | H25B—C25—H25C | 109.5 |
| C1—C11—C12 | 119.4 (3) | O27—S26—O28 | 115.33 (16) |
| N10—C12—C4 | 121.7 (3) | O27—S26—O29 | 116.23 (16) |
| N10—C12—C11 | 120.0 (3) | O28—S26—O29 | 114.60 (14) |
| C4—C12—C11 | 118.3 (3) | O27—S26—C30 | 102.13 (17) |
| C9—C13—C8 | 122.6 (3) | O28—S26—C30 | 103.11 (19) |
| C9—C13—C14 | 119.3 (3) | O29—S26—C30 | 102.54 (16) |
| C8—C13—C14 | 118.2 (3) | F32—C30—F31 | 107.0 (3) |
| N10—C14—C5 | 121.9 (3) | F32—C30—F33 | 106.9 (3) |
| N10—C14—C13 | 118.9 (3) | F31—C30—F33 | 107.5 (3) |
| C5—C14—C13 | 119.3 (3) | F32—C30—S26 | 111.8 (3) |
| O17—C15—O16 | 125.2 (3) | F31—C30—S26 | 111.7 (3) |
| O17—C15—C9 | 122.4 (3) | F33—C30—S26 | 111.7 (3) |
| C11—C1—C2—C3 | −2.5 (6) | C6—C5—C14—C13 | −0.7 (5) |
| C1—C2—C3—C4 | 1.5 (7) | C9—C13—C14—N10 | 0.7 (4) |
| C2—C3—C4—C12 | 0.2 (6) | C8—C13—C14—N10 | 180.0 (3) |
| C14—C5—C6—C7 | 1.3 (5) | C9—C13—C14—C5 | −179.8 (3) |
| C5—C6—C7—C8 | −0.5 (5) | C8—C13—C14—C5 | −0.5 (4) |
| C6—C7—C8—C13 | −0.8 (5) | C13—C9—C15—O17 | −94.0 (4) |
| C13—C9—C11—C1 | −176.4 (3) | C11—C9—C15—O17 | 81.1 (4) |
| C15—C9—C11—C1 | 8.5 (4) | C13—C9—C15—O16 | 85.3 (3) |
| C13—C9—C11—C12 | 3.6 (4) | C11—C9—C15—O16 | −99.6 (3) |
| C15—C9—C11—C12 | −171.5 (3) | O17—C15—O16—C18 | −1.1 (5) |
| C2—C1—C11—C9 | −178.2 (3) | C9—C15—O16—C18 | 179.6 (3) |
| C2—C1—C11—C12 | 1.8 (5) | C15—O16—C18—C23 | −87.3 (4) |
| C14—N10—C12—C4 | 176.3 (3) | C15—O16—C18—C19 | 97.4 (3) |
| C25—N10—C12—C4 | −5.0 (4) | C23—C18—C19—C20 | 0.0 (5) |
| C14—N10—C12—C11 | −4.6 (4) | O16—C18—C19—C20 | 175.2 (3) |
| C25—N10—C12—C11 | 174.0 (3) | C18—C19—C20—C21 | −0.3 (5) |
| C3—C4—C12—N10 | 178.1 (3) | C19—C20—C21—C22 | 0.1 (5) |
| C3—C4—C12—C11 | −0.9 (5) | C19—C20—C21—C24 | −178.9 (3) |
| C9—C11—C12—N10 | 0.9 (4) | C20—C21—C22—C23 | 0.3 (5) |
| C1—C11—C12—N10 | −179.1 (3) | C24—C21—C22—C23 | 179.3 (3) |
| C9—C11—C12—C4 | 180.0 (3) | C19—C18—C23—C22 | 0.4 (5) |
| C1—C11—C12—C4 | 0.0 (4) | O16—C18—C23—C22 | −174.8 (3) |
| C11—C9—C13—C8 | 176.4 (3) | C21—C22—C23—C18 | −0.5 (5) |
| C15—C9—C13—C8 | −8.6 (4) | O27—S26—C30—F32 | 58.5 (3) |
| C11—C9—C13—C14 | −4.4 (4) | O28—S26—C30—F32 | 178.4 (3) |
| C15—C9—C13—C14 | 170.6 (3) | O29—S26—C30—F32 | −62.3 (3) |
| C7—C8—C13—C9 | −179.4 (3) | O27—S26—C30—F31 | −61.3 (3) |
| C7—C8—C13—C14 | 1.3 (5) | O28—S26—C30—F31 | 58.6 (3) |
| C12—N10—C14—C5 | −175.6 (3) | O29—S26—C30—F31 | 177.9 (3) |
| C25—N10—C14—C5 | 5.7 (4) | O27—S26—C30—F33 | 178.2 (3) |
| C12—N10—C14—C13 | 3.8 (4) | O28—S26—C30—F33 | −61.8 (3) |
| C25—N10—C14—C13 | −174.8 (3) | O29—S26—C30—F33 | 57.5 (3) |
| C6—C5—C14—N10 | 178.8 (3) |
Hydrogen-bond geometry (Å, °)
| Cg4 is the centroid of the C18–C23 ring. |
| D—H···A | D—H | H···A | D···A | D—H···A |
| C3—H3···O27i | 0.93 | 2.57 | 3.314 (5) | 137 |
| C4—H4···O29i | 0.93 | 2.44 | 3.319 (4) | 159 |
| C5—H5···O28 | 0.93 | 2.44 | 3.364 (5) | 171 |
| C6—H6···O28ii | 0.93 | 2.56 | 3.342 (5) | 142 |
| C23—H23···O27iii | 0.93 | 2.53 | 3.448 (4) | 169 |
| C25—H25A···O29 | 0.96 | 2.56 | 3.415 (5) | 149 |
| C25—H25B···Cg4iv | 0.96 | 2.62 | 3.487 (4) | 151 |
Symmetry codes: (i) −x+3/2, y+1/2, −z+1/2; (ii) −x+2, −y+1, −z+1; (iii) x−1, y, z; (iv) −x+1, −y+1, −z+1.
Table 2 C–F···π interactions (Å,°).
| X | I | J | I···J | X···J | X–I···J |
| C30 | F31 | Cg2i | 3.420 (3) | 4.044 (4) | 108.9 (2) |
| C30 | F32 | Cg1i | 3.441 (3) | 4.032 (4) | 107.1 (2) |
| C30 | F32 | Cg2i | 3.788 (4) | 4.044 (4) | 91.5 (2) |
| C30 | F33 | Cg1i | 3.669 (3) | 4.032 (4) | 96.2 (2) |
Symmetry code: (i) –x + 3/2, y + 1/2, –z + 1/2. Notes: Cg1 and Cg2 are the centroids of the C9/N10/C11–C14 and C1–C4/C11/C12 rings, respectively.
Table 3 π–π interactions (Å,°).
| I | J | CgI···CgJ | Dihedral angle | CgI_Perp | CgI_Offset |
| 3 | 4v | 3.913 (2) | 4.80 (17) | 3.472 (2) | 1.805 (2) |
| 4 | 3v | 3.913 (2) | 4.80 (17) | 3.565 (2) | 1.613 (2) |
Symmetry code: (v) –x + 1, –y, –z + 1.Notes: Cg3 and Cg4 are the centroids of the C5–C8/C13/C14 and C18–C23 rings, respectively. CgI···CgJ is the distance between ring centroids. The dihedral angle is that between the planes of the rings I and J. CgI_Perp is the perpendicular distance of CgI from ring J. CgI_Offset is the distance between CgI and perpendicular projection of CgJ on ring I.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: NG2760).
References
- Bianchi, R., Forni, A. & Pilati, T. (2004). Acta Cryst. B60, 559–568. [DOI] [PubMed]
- Brown, R. C., Li, Z., Rutter, A. J., Mu, X., Weeks, O. H., Smith, K. & Weeks, I. (2009). Org. Biomol. Chem.7, 386–394. [DOI] [PubMed]
- Dorn, T., Janiak, C. & Abu-Shandi, K. (2005). CrystEngComm, 7, 633–641.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Hunter, C. A., Lawson, K. R., Perkins, J. & Urch, C. J. (2001). J. Chem. Soc. Perkin Trans. 2, pp. 651–669.
- Novoa, J. J., Mota, F. & D’Oria, E. (2006). Hydrogen Bonding – New Insights, edited by S. Grabowski, pp. 193–244. The Netherlands: Springer.
- Oxford Diffraction (2008). CrysAlis CCD and CrysAlis RED Oxford Diffraction Ltd, Yarnton, England.
- Rak, J., Skurski, P. & Błażejowski, J. (1999). J. Org. Chem.64, 3002–3008. [DOI] [PubMed]
- Roda, A., Guardigli, M., Michelini, E., Mirasoli, M. & Pasini, P. (2003). Anal. Chem.A75, 462–470. [PubMed]
- Sato, N. (1996). Tetrahedron Lett.37, 8519–8522.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sikorski, A., Krzymiński, K., Białońska, A., Lis, T. & Błażejowski, J. (2006). Acta Cryst. E62, o822–o824.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Takahashi, O., Kohno, Y., Iwasaki, S., Saito, K., Iwaoka, M., Tomada, S., Umezawa, Y., Tsuboyama, S. & Nishio, M. (2001). Bull. Chem. Soc. Jpn, 74, 2421–2430.
- Trzybiński, D., Krzymiński, K., Sikorski, A., Malecha, P. & Błażejowski, J. (2010). Acta Cryst. E66, o826–o827. [DOI] [PMC free article] [PubMed]
- Zomer, G. & Jacquemijns, M. (2001). Chemiluminescence in Analytical Chemistry, edited by A.M. Garcia-Campana & W. R. G. Baeyens, pp. 529–549. New York: Marcel Dekker.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810016302/ng2760sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810016302/ng2760Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report