Abstract
IMGT, the international ImMunoGeneTics database, freely available at http://imgt.cines.fr:8104, was created in 1989 at the Université Montpellier II, CNRS, Montpellier, France, and is a high quality integrated information system specialising in immunoglobulins, T cell receptors and major histocompatibility complex molecules of human and other vertebrates. IMGT provides researchers and clinicians with a common access to all nucleotide, protein, genetic and structural immunogenetics data. This information is of high value for medical and veterinary research, biotechnology related to antibody and T cell receptor engineering, genome diversity and evolution studies of the immune response.
INTRODUCTION
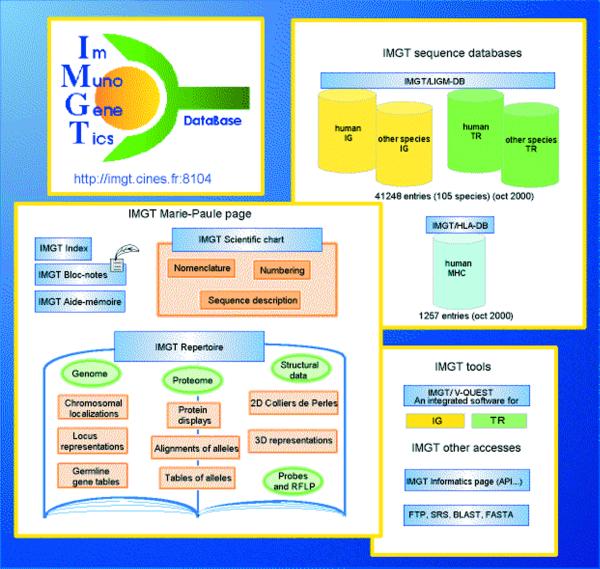
IMGT, the international ImMunoGeneTics database (http://imgt.cines.fr:8104) (1,2), created in 1989 at the Université Montpellier II, CNRS, Montpellier, France, is a high quality integrated information system specialising in Immunoglobulins (Ig), T cell Receptors (TcR) and Major Histocompatibility Complex (MHC) molecules of human and other vertebrates. IMGT provides common access to expertly annotated data on the genome, proteome, genetics and structure of the Ig, TcR and MHC. Due to its high quality and easy data distribution, IMGT has important implications in medical research (repertoire in autoimmune diseases, AIDS, leukemias, lymphomas, myelomas), therapeutic approaches, biotechnology related to antibody engineering, veterinary research, genome diversity and genome evolution studies of the immune response. IMGT consists of databases (‘IMGT sequence databases’), Web resources (‘IMGT Marie-Paule page’) and interactive tools (Fig. 1).
Figure 1.
Overview of the IMGT information system.
IMGT SEQUENCE DATABASES
The IMGT sequence databases comprise at present (i) IMGT/LIGM-DB, a comprehensive database of 41 248 Ig and TcR nucleotide sequences from human and from 104 other vertebrate species, with translation for fully annotated sequences, created by LIGM (Laboratoire d’ImmunoGénétique Moléculaire, Montpellier, France) (1–3) and (ii) IMGT/HLA-DB, a database of the 1257 human MHC allele sequences, developed by ICRF (Imperial Cancer Research Fund, Oxford, UK) and ANRI (Anthony Nolan Research Institute, London, UK) (4).
IMGT MARIE-PAULE PAGE
The IMGT Marie-Paule page comprises Web resources recorded in HTML pages (2780 documents, 3698 internal and external hyperlinks).
The IMGT Scientific chart provides the controlled vocabulary and the annotation rules and concepts defined by IMGT for the identification, description and classification of the Ig and TcR data of all vertebrate species (5). The concepts of classification have been used to set up a unique nomenclature of Ig and TcR genes (6,7), which has been adopted by the HUGO nomenclature committee. The complete list of the IMGT human Ig and TcR gene names has been entered in GDB and LocusLink (1999). A uniform numbering system for Ig and TcR sequences of all species has been established to facilitate sequence comparison and cross-referencing between experiments from different laboratories whatever the antigen receptor (Ig or TcR), chain type or species (8). IMGT has developed a formal specification of the terms to be used in the domain of immunogenetics and bioinformatics to ensure accuracy, consistency and coherence in IMGT. This has been the basis of the IMGT-ONTOLOGY (5), the first ontology in the domain, which allows the management of the immunogenetics knowledge for all vertebrate species. Control of coherence in IMGT combines data integrity control and biological data evaluation (9,10).
The IMGT Repertoire, the global Web resource in ImMunoGeneTics for the immunoglobulins and T cell receptors of human and other vertebrates, based on the ‘IMGT Scientific chart’, provides an easy-to-use interface to carefully expertised data on the genome, proteome, polymorphism and structure of the Ig and TcR (2). Genome data include chromosomal localisations, locus representations and germline gene tables. Proteome and polymorphism data are represented by protein displays, alignments of alleles and tables of alleles. These data are regularly published in the IMGT Locus in Focus section of Experimental and Clinical Immunogenetics (IMGT Index>IMGT Locus in Focus at http://imgt.cines.fr:8104). Structural data comprise 2D graphical representations designated as Colliers de Perles (1) and 3D representations of Ig and TcR variable regions (2,3). This visualisation permits rapid correlation between protein sequences and 3D data retrieved from the Protein Data Bank (PDB). A new section, currently being developed, contains data on probes used for the analysis of Ig and TcR gene rearrangements and expressions, and RFLP (Restriction Fragment Length Polymorphism) studies.
The IMGT Index and the IMGT Aide-mémoire represent useful biological resources for students and researchers. The IMGT Bloc-notes provides numerous hyperlinks to the Web servers specialising in immunology, genetics, molecular biology and bioinformatics (11).
IMGT TOOLS AND OTHER ACCESSES
IMGT/V-QUEST (V-QUEry and STandardization) is an integrated software for Ig and TcR. This tool analyses the input Ig or TcR variable nucleotide sequence and provides the nucleotide alignment by comparison with the IMGT reference directory, the protein translation of the input sequence, and its 2D Colliers de Perles representation. IMGT/LIGM-DB can be searched by BLAST or FASTA on different servers.
Since July 1995, IMGT has been available on the Web at http://imgt.cines.fr:8104. IMGT provides biologists with an easy to use and friendly interface. From January 1996 to October 2000, the IMGT WWW server at Montpellier was accessed by more than 95 000 sites. IMGT has an exceptional response with more than 6000 requests a week. IMGT data are also distributed by EBI (distribution of CD-ROM, network fileserver: netserv@ebi.ac.uk, and anonymous FTP server), and from many SRS (Sequence Retrieval System) sites. To facilitate the integration of IMGT data into applications developed by other laboratories, we have built an API (Application Programming Interface) to access the database and its software tools (10).
ELECTRONIC AND MAILING ADDRESSES
IMGT home page: http://imgt.cines.fr:8104 (IMGT contact lefranc@ligm.igh.cnrs.fr).
IMGT page at EBI: http://www.ebi.ac.uk/imgt, ftp://ftp.ebi.ac.uk/pub/databases/imgt
IMGT/LIGM-DB: http://imgt.cines.fr:8104 (contacts lefranc@ligm.igh.cnrs.fr, giudi@ligm.igh.cnrs.fr), ftp://imgt.cines.fr/IMGT (contact denys.chaume@igh.cnrs.fr).
IMGT/HLA-DB: http://www.ebi.ac.uk/imgt/hla/ (contacts jrobinso@ebi.ac.uk, julia@icrf.icnet.uk, marsh@icrf.icnet.uk).
CITING IMGT
Authors who make use of the information provided by IMGT should cite this article as a general reference for the access to and content of IMGT, and quote the IMGT home page URL, http://imgt.cines.fr:8104.
Acknowledgments
ACKNOWLEDGEMENTS
I am deeply grateful to the IMGT team for its expertise and motivation. IMGT is funded by the European Union’s 5th PCRDT programme QLG2-2000-01287, the CNRS (Centre National de la Recherche Scientifique), the Ministère de l’Education Nationale and the Ministère de la Recherche. Subventions have been received from ARC (Association pour la Recherche sur le Cancer) and the Région Languedoc-Roussillon.
References
- 1.Lefranc M.-P., Giudicelli,V., Ginestoux,C., Bodmer,J., Müller,W., Bontrop,R., Lemaitre,M., Malik,A., Barbié,V. and Chaume,D. (1999) IMGT, the international ImMunoGeneTics database. Nucleic Acids Res., 27, 209–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruiz M., Giudicelli,V., Ginestoux,C., Stoehr,P., Robinson,J., Bodmer,J., Marsh,S.G., Bontrop,R., Lemaitre,M., Lefranc,G., Chaume,D. and Lefranc,M.-P. (2000) IMGT, the international ImMunoGeneTics database. Nucleic Acids Res., 28, 219–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lefranc M.-P. (2000) IMGT ImMunoGeneTics Database. International BIOforum, 4, 98–100. [Google Scholar]
- 4.Robinson J., Malik,A., Parham,P., Bodmer,J.G. and Marsh,S.G.E. (2000) IMGT/HLA Database – a sequence database for the human major histocompatibility complex. Tissue Antigens, 55, 280–287. [DOI] [PubMed] [Google Scholar]
- 5.Giudicelli V. and Lefranc,M.-P. (1999) Ontology for Immunogenetics: IMGT-ONTOLOGY. Bioinformatics, 12, 1047–1054. [DOI] [PubMed] [Google Scholar]
- 6.Lefranc M.-P. (2000) Locus maps and genomic repertoire of the human Ig genes. Immunologist, 8, 80–87. [Google Scholar]
- 7.Lefranc M.-P. (2000) Locus maps and genomic repertoire of the human T-cell receptor genes. Immunologist, 8, 72–79. [Google Scholar]
- 8.Lefranc M.-P. (1999) The IMGT unique numbering for Immunoglobulins, T cell receptors and Ig-like domains. Immunologist, 7, 132–136. [Google Scholar]
- 9.Giudicelli V., Chaume,D., Mennessier,G., Althaus,H.H., Müller,W., Bodmer,J., Malik,A. and Lefranc,M.-P. (1998) IMGT, the international ImMunoGeneTics database: a new Design for Immunogenetics Data Access. In Cesnik,B. et al. (eds), Proceedings of the Ninth World Congress on Medical Informatics, MEDINFO’98. IOS Press, Amsterdam, 351–355. [PubMed]
- 10.Giudicelli V., Chaume,D. and Lefranc,M.-P. (1998) IMGT/LIGM-DB: A systematized approach for ImMunoGeneTics database coherence and data distribution improvement. Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology, ISBM-98, 59–68. [PubMed]
- 11.Lefranc M.-P. (2000) Web sites of Interest to Immunologists. Curr. Protocols Immunol ., A.1J.1–A.1J.33. [DOI] [PubMed] [Google Scholar]