Abstract
In the crystal structure of imatinibium dipicrate [systematic name: 1-methyl-4-(4-{4-methyl-3-[4-(3-pyridyl)pyrimidin-2-ylamino]anilinocarbonyl}benzyl)piperazine-1,4-diium dipicrate], C29H33N7O2+·2C6H2N3O7 −, the imatinibium cation is protonated at both of the pyrimidine N atoms. Each of the two picrate anions interacts with the diprotonated cation through bifurcated N—H⋯O hydrogen bonds forming R 1 2(6) ring motifs. Also, an R 2 2(24) graph set is formed between the benzamidium –NH– group and the 4-pyridyl N atom interacting through N—H⋯N hydrogen-bond interactions. Additional weak C—H⋯Cg π-ring and π–π intermolecular interactions are observed which also influence crystal packing.
Related literature
For related structures, see: Bindya et al. (2007 ▶); Harrison, Bindya et al. (2007 ▶); Harrison, Sreevidya et al. (2007 ▶); Jasinski et al. (2009a
▶,b
▶); Swamy et al. (2007 ▶); Szumma et al. (2000 ▶); Yathirajan et al. (2007a
▶,b
▶). For a rationally developed anticancer drug, see: Capdeville et al. (2002 ▶). For its use in chronic myeloid leukaemia, see: Moen et al. (2007 ▶). For puckering parameters, see: Cremer & Pople (1975 ▶).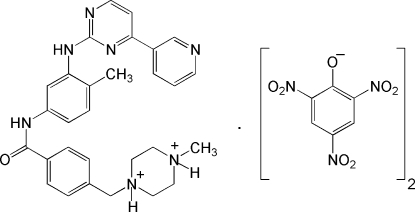
Experimental
Crystal data
C29H33N7O2+·2C6H2N3O7 −
M r = 951.84
Triclinic,
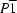
a = 8.560 (1) Å
b = 10.734 (1) Å
c = 23.060 (1) Å
α = 96.74 (3)°
β = 92.69 (2)°
γ = 101.46 (7)°
V = 2056.9 (6) Å3
Z = 2
Cu Kα radiation
μ = 1.02 mm−1
T = 110 K
0.45 × 0.39 × 0.24 mm
Data collection
Oxford Diffraction Xcalibur diffractometer with a Ruby (Gemini Cu) detector
Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2007 ▶) T min = 0.596, T max = 0.782
15890 measured reflections
8082 independent reflections
6946 reflections with I > 2σ(I)
R int = 0.023
Refinement
R[F 2 > 2σ(F 2)] = 0.059
wR(F 2) = 0.158
S = 1.06
8082 reflections
640 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.50 e Å−3
Δρmin = −0.27 e Å−3
Data collection: CrysAlis PRO (Oxford Diffraction, 2007 ▶); cell refinement: CrysAlis RED (Oxford Diffraction, 2007 ▶); data reduction: CrysAlis RED; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶) and Mercury (Macrae et al., 2006 ▶); software used to prepare material for publication: SHELXTL, enCIFer (Allen et al., 2004 ▶) and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810000577/bt5129sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810000577/bt5129Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O1B | 0.85 (3) | 1.85 (3) | 2.658 (3) | 157 (3) |
| N1—H1⋯O62B | 0.85 (3) | 2.35 (3) | 2.890 (3) | 122 (2) |
| N2—H2⋯O1A | 0.89 (4) | 1.85 (4) | 2.678 (3) | 154 (3) |
| N2—H2⋯O62A | 0.89 (4) | 2.41 (4) | 3.009 (3) | 125 (3) |
| N14—H14⋯N31i | 0.85 (3) | 2.23 (3) | 3.069 (3) | 171 (3) |
| C5—H5B⋯O41Aii | 0.98 | 2.48 | 3.258 (4) | 136 |
| C4—H4B⋯O42Biii | 0.99 | 2.33 | 3.199 (3) | 146 |
| C3—H3A⋯O61Biv | 0.99 | 2.57 | 3.199 (3) | 121 |
| C3—H3B⋯O1B | 0.99 | 2.34 | 3.072 (3) | 130 |
| C12—H12A⋯O42Biii | 0.95 | 2.63 | 3.423 (3) | 142 |
| C19—H19A⋯O61Bv | 0.98 | 2.50 | 3.435 (4) | 159 |
| C19—H19A⋯N6Bv | 0.98 | 2.65 | 3.541 (4) | 152 |
Symmetry codes: (i) 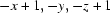 ; (ii)
; (ii) 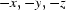 ; (iii)
; (iii) 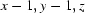 ; (iv)
; (iv) 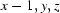 ; (v)
; (v) 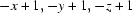 .
.
Table 2. π-Ring hydrogen-bond geometry (Å, °) for (I).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C33—H33A⋯Cg5vi | 0.95 | 2.90 | 3.545 (8) | 127 |
Symmetry code: (vi) x + 1, y, z. Cg5 is the centroid of the C15–C21 ring.
Table 3. π–π stacking geometry (Å) for (I).
| Cg2⋯Cg7v | 3.740 (4) |
| Cg3⋯Cg3v | 3.496 (7) |
| Cg6⋯Cg6vii | 3.396 (0) |
Symmetry codes: (v) −x + 1, −y + 1, −z + 1; (vii) −x + 2, −y + 2, −z. Cg2, Cg3, Cg6 and Cg7 are the centroids of the C25–C27/N28/C23/N4, C32–C34/C29//C30/N31, C1A–C6A and C1B–C6B rings, respectively.
Acknowledgments
QNMHA thanks the University of Mysore for use of its research facilities. RJB acknowledges the NSF MRI program (grant No. CHE-0619278) for funds to purchase an X-ray diffractometer.
supplementary crystallographic information
Comment
Imatinib, marketed as a cancer drug by Novartix, [Gleevec(USA), Glivec(Europe/Australia)] (systematic name: 4-(4-methyl-piperazin-1-yl-methyl-N-[4-methyl-3-(4-pyridin-3-ylpyrimidin-2-yl-amino)-phenyl]-benzamide) is a synthetic tyrosine kinase inhibitor used in treating chronic myelogenous leukemia (CML), gastrointestinal stromal tumours (GISTs) and a number of other malignancies. It is a 2-phenylaminopyrimidine derivative and is the first member of a new class of agents that act by inhibiting particular tyrpsine kinase enzymes, instead of non-specifically inhibiting rapidly dividing cells. Reviews on the use of imatinib in chronic myeloid leukaemia (Moen et al., 2007) and on the rationally developed targeted anticancer drug have been published (Capdeville et al., 2002). Picrates form charge-transfer complexes with organic compounds, function as acceptors in the formation of π- stacking complexes with aromatic biomolecules and as an acidic ligand forming salts with polar biomolecules. In this context, the crystal and molecular structures of related compounds include amitriptylinium picrate (Bindya et al., 2007), mepazinium picrate (Yathirajan et al., 2007a), trifluperazinium dipicrate (Yathirajan et al., 2007b), imipraminium picrate (Harrison, Bindya et al., 2007), nevirapiniumpicrate (Harrison, Sreevidya et al., 2007), desipraminium picrate (Swamy et al., 2007) and propiverinium picrate (Jasinski et al., 2009a) have been reported. In view of the importance of imatinib and to study the hydrogen bonding patterns in the title compound, (I), C29H33N7O2+ (C6H2N3O7-)2, a dipicrate salt of Imatinib, a crystal structure is reported.
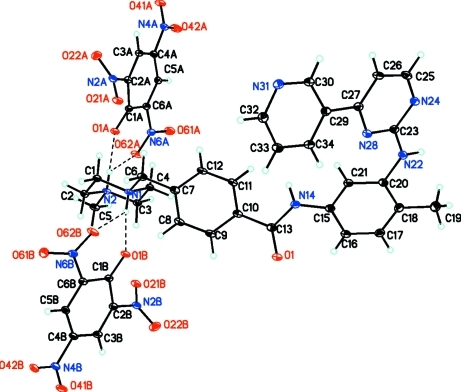
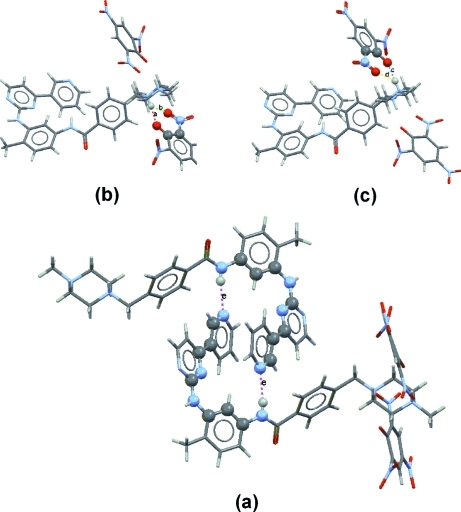
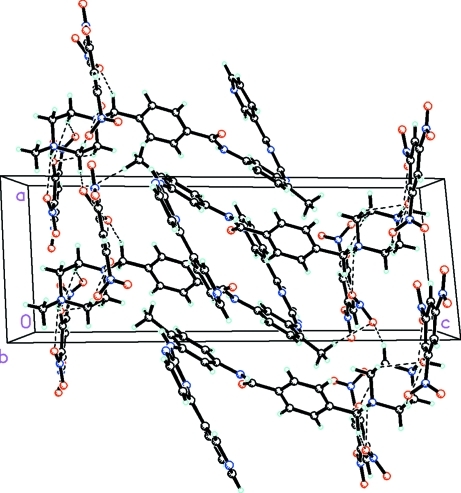
The imatinibium cation contains a doubly charged methyl piperazine group bonded at the 4 position of a p-methyl benzamide group and a 2-phenylaminopyrimidine(pyridine) derivative bonded to the amino end. The 6-membered methyl piperazine group adopts a slightly distorted chair conformation (Cremer & Pople, 1975) with puckering parameters Q, θ and φ of 0.572 (5) Å, 176.1 (5)° and 168.174 (3)°, respectively (Fig. 1). For an ideal chair θ has a value of 0 or 180°. An R22(24) graph-set motif is formed between the benzamidium –NH– group and the 4-pyridiyl N atom interacting through a N–H···N hydrogen bond interaction (Fig.2a). The dihedral angle between the mean plane of the benzyl ring in the benzamide group and the mean planes of the piperazine, amino phenyl, pyrimidine and pyridine groups are 81.1 (7)°, 50.8 (5)°, 57.1 (7)° and 46.1 (4)°, respectively. The mean planes of the pyrimidine and pyridine rings are twisted by 11.1 (9)°. The dihedral angles between mean planes of the aminobenzyl group and the pyrimidine and pyridine groups are 30.7 (3)° and 32.3 (2)°, while the dihedral angles between the mean planes of the piperizine group and the aminobenzyl, pyrimidine and pyridine groups are 48.3 (9)°, 59.2 (1)° and 69.3 (9)°, respectively. The two picrate anions, labeled A and B, each interact with the diprotonated cation through bifurcated N–H···O hydrogen bonds forming an R21(6) ring-motif creating an ···ab··· and ···cd··· array of hydrogen bonding patterns (Fig.2 b,c). The mean plane of the two o-NO2 groups in the two picrate anions are twisted by 16.1 (9)° and 39.1 (9)° in the A-ring and 27.1 (5)° and 47.4 (1)° in the B-ring with respect to the mean planes of the 6-membered benzene rings. The difference in the twist angles of the mean planes of the two o-NO2 groups in each picrate anion can be attributed to an intermolecular "side" hydrogen bond interaction (Szumma et al., 2000) between the N1 and N2 atoms of the cation piperizine group with a two-centered hydrogen bond to the singly bonded oxygen atom (O1A & O1B) and to one oxygen atom of an adjacent o-NO2 group (O62A & O62B), respectively, [N1—H1···O1B & N1—H1···O62B and N2—H2···O1A & N2—H2···O62A, see Table 1, Fig.1]. The difference in angles between the mean planes of the o-O61A—N6A—O62A (16.1 (9)°) and o-O21a—N2a—O22A (39.1 (9)°) groups with the mean plane of the benzene ring in picrate A (23°) and those of the o-O21B—N2B—O22B (47.4 (1)°) and o-O61B—N6B—O62B (27.1 (5)°) with the mean plane of the benzene ringin picrate B (16.2 (3)°) are a direct result of the N2—H2···O62A and N1—H1···O62B hydrogen bonds. The p-NO2 groups in both picrate anions are essentially in the plane of the ring (torsion angles C5A—C4A—N4A—O41A = 179.9 (2)°; C5B—C4B—N4B—O41B = 176.1 (2)°). Crystal packing is also influenced by N—H···N hydrogen bond interactions between the benzamide and pyridine groups (N14—H14···N31), intermediate C—H···O hydrogen bond interactions (C5—H5B···O41A, C4—H4···H42B & C3—H3A···O61B) between the piperizine group and o-NO2 & p-NO2 groups of picrates A & B and weak C—H···O hydrogen bond interactions involving the benzamide, phenyl, o-NO2 andp-NO2 groups (C12—H12A···O42B, C19—H19···O61B & C19—H19A···N6B;Table 1) which produces a two-dimensional network arranged along the (101)plane of the unit cell (Fig.3). In addition there are weak C—H···π (Table 2) and weak π-π intermolecular interactions (Table 3) similar to that observed in 3-(2-Chloroethyl)-2-methyl-4H-pyrido[1,2-a] pyrimidinium-4-one picrate (Jasinski et al., 2009b).
Experimental
The title compound was synthesized by mixing an aqueous solution (10 ml) of picric acid (0.92 g, 2 mmol) and N-(4-methyl-3-(4-(pyridin-3-yl)pyrimidin-2-ylamino)phenyl)-4-((4-methyl piperazin-1-yl)methyl)benzamide (1.18 g, 2 mmol) in methanolic aqueous solution (10 ml) and the resulting solution was stirred well at 313 K. The formation of a yellow precipitate of the charge transfer complex was noticed almost instantaneously. The formed complex was filtered off, washed with distilled water and dried in vacuo over CaCl2. The purity of the synthesized compound was improved by a successive recrystallization process with methanol (yield: 76.2%). The crystals for X-ray studies were grown from slow evaporation of a methanol solution. The melting range was found to be 490–493 K.
Refinement
All of the H atoms were placed in their calculated positions and then refined using the riding model with N—H = 0.85–0.89, C—H = 0.95–0.99 Å, and with Uiso(H) = 1.15–1.51Ueq(C,N).
Figures
Fig. 1.
Molecular structure of the title compound, C29H33N7O2+ (C6H2N3O7-)2, showing the cation-dianion unit that comprises the asymmetric unit, the atom labeling scheme and 30% probability displacement ellipsoids. Picrate anions A & B are labeled accordingly.
Fig. 2.
Diagrams of the (a) R22(24) ···ee···, (b) R21(6) ···ab··· and (c) R21(6) ···cd··· graph-set motifs in the cation (a) and anions (b, c) of the title compound, (I).
Fig. 3.
Packing diagram of the title compound, (I), viewed down the b axis. Dashed lines indicate intermolecular N—H···O, N—H···N & C—H···O hydrogen bond interactions which produces a two-dimensional network arranged along the (101) plane of the unit cell.
Crystal data
| C29H33N7O2+·2C6H2N3O7− | Z = 2 |
| Mr = 951.84 | F(000) = 988 |
| Triclinic, P1 | Dx = 1.537 Mg m−3 |
| Hall symbol: -P 1 | Cu Kα radiation, λ = 1.54178 Å |
| a = 8.560 (1) Å | Cell parameters from 9389 reflections |
| b = 10.734 (1) Å | θ = 4.2–74.0° |
| c = 23.060 (1) Å | µ = 1.02 mm−1 |
| α = 96.74 (3)° | T = 110 K |
| β = 92.69 (2)° | Chunk, pale yellow |
| γ = 101.46 (7)° | 0.45 × 0.39 × 0.24 mm |
| V = 2056.9 (6) Å3 |
Data collection
| Oxford Diffraction Xcalibur diffractometer with a Ruby (Gemini Cu) detector | 8082 independent reflections |
| Radiation source: Enhance (Cu) X-ray Source | 6946 reflections with I > 2σ(I) |
| graphite | Rint = 0.023 |
| Detector resolution: 10.5081 pixels mm-1 | θmax = 74.1°, θmin = 4.2° |
| ω scans | h = −10→8 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2007) | k = −13→13 |
| Tmin = 0.596, Tmax = 0.782 | l = −28→28 |
| 15890 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.059 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.158 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0615P)2 + 3.3059P] where P = (Fo2 + 2Fc2)/3 |
| 8082 reflections | (Δ/σ)max = 0.001 |
| 640 parameters | Δρmax = 0.50 e Å−3 |
| 0 restraints | Δρmin = −0.27 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.3153 (2) | 0.58462 (17) | 0.49702 (9) | 0.0356 (4) | |
| N1 | 0.4213 (2) | 0.4360 (2) | 0.19545 (10) | 0.0242 (4) | |
| H1 | 0.465 (4) | 0.514 (3) | 0.2058 (13) | 0.028 (8)* | |
| N2 | 0.2325 (3) | 0.4773 (2) | 0.09313 (10) | 0.0259 (5) | |
| H2 | 0.182 (4) | 0.397 (4) | 0.0809 (15) | 0.044 (9)* | |
| C1 | 0.4665 (3) | 0.4017 (2) | 0.13475 (12) | 0.0274 (5) | |
| H1A | 0.4187 | 0.3106 | 0.1211 | 0.033* | |
| H1B | 0.5841 | 0.4127 | 0.1349 | 0.033* | |
| C2 | 0.4094 (3) | 0.4851 (2) | 0.09331 (12) | 0.0291 (5) | |
| H2A | 0.4655 | 0.5752 | 0.1049 | 0.035* | |
| H2B | 0.4371 | 0.4581 | 0.0532 | 0.035* | |
| C3 | 0.1853 (3) | 0.5056 (2) | 0.15391 (11) | 0.0264 (5) | |
| H3A | 0.0675 | 0.4925 | 0.1534 | 0.032* | |
| H3B | 0.2307 | 0.5965 | 0.1689 | 0.032* | |
| C4 | 0.2433 (3) | 0.4209 (2) | 0.19414 (11) | 0.0255 (5) | |
| H4A | 0.2118 | 0.4431 | 0.2342 | 0.031* | |
| H4B | 0.1922 | 0.3303 | 0.1807 | 0.031* | |
| C5 | 0.1813 (4) | 0.5660 (3) | 0.05447 (13) | 0.0342 (6) | |
| H5A | 0.0661 | 0.5612 | 0.0558 | 0.051* | |
| H5B | 0.2058 | 0.5413 | 0.0142 | 0.051* | |
| H5C | 0.2386 | 0.6539 | 0.0680 | 0.051* | |
| C6 | 0.4812 (3) | 0.3553 (2) | 0.23719 (12) | 0.0294 (6) | |
| H6A | 0.4361 | 0.2637 | 0.2235 | 0.035* | |
| H6B | 0.5990 | 0.3686 | 0.2370 | 0.035* | |
| C7 | 0.4370 (3) | 0.3868 (2) | 0.29871 (12) | 0.0284 (5) | |
| C8 | 0.5091 (3) | 0.5012 (2) | 0.33399 (12) | 0.0305 (6) | |
| H8A | 0.5904 | 0.5608 | 0.3193 | 0.037* | |
| C9 | 0.4627 (3) | 0.5279 (2) | 0.38995 (12) | 0.0304 (6) | |
| H9A | 0.5108 | 0.6066 | 0.4130 | 0.036* | |
| C10 | 0.3464 (3) | 0.4409 (2) | 0.41296 (12) | 0.0276 (5) | |
| C11 | 0.2752 (3) | 0.3270 (2) | 0.37767 (12) | 0.0279 (5) | |
| H11A | 0.1957 | 0.2667 | 0.3927 | 0.033* | |
| C12 | 0.3182 (3) | 0.3006 (2) | 0.32147 (12) | 0.0289 (6) | |
| H12A | 0.2668 | 0.2233 | 0.2980 | 0.035* | |
| C13 | 0.2977 (3) | 0.4734 (2) | 0.47391 (12) | 0.0292 (5) | |
| N14 | 0.2326 (3) | 0.3702 (2) | 0.49985 (10) | 0.0290 (5) | |
| H14 | 0.250 (4) | 0.298 (3) | 0.4857 (13) | 0.030 (8)* | |
| C15 | 0.1556 (3) | 0.3663 (2) | 0.55298 (12) | 0.0276 (5) | |
| C16 | 0.0846 (3) | 0.4640 (3) | 0.57780 (12) | 0.0314 (6) | |
| H16A | 0.0945 | 0.5428 | 0.5620 | 0.038* | |
| C17 | −0.0012 (3) | 0.4419 (3) | 0.62653 (13) | 0.0339 (6) | |
| H17A | −0.0498 | 0.5080 | 0.6438 | 0.041* | |
| C18 | −0.0199 (3) | 0.3286 (3) | 0.65133 (12) | 0.0315 (6) | |
| C19 | −0.1298 (4) | 0.3073 (3) | 0.70004 (14) | 0.0437 (7) | |
| H19A | −0.0708 | 0.2871 | 0.7340 | 0.066* | |
| H19B | −0.2194 | 0.2357 | 0.6867 | 0.066* | |
| H19C | −0.1708 | 0.3851 | 0.7111 | 0.066* | |
| C20 | 0.0583 (3) | 0.2345 (2) | 0.62720 (11) | 0.0264 (5) | |
| C21 | 0.1447 (3) | 0.2542 (2) | 0.57814 (12) | 0.0274 (5) | |
| H21A | 0.1971 | 0.1896 | 0.5617 | 0.033* | |
| N22 | 0.0369 (3) | 0.1166 (2) | 0.65091 (10) | 0.0295 (5) | |
| H22 | −0.053 (4) | 0.098 (3) | 0.6678 (15) | 0.040 (9)* | |
| C23 | 0.1200 (3) | 0.0202 (2) | 0.64192 (11) | 0.0264 (5) | |
| N24 | 0.0467 (3) | −0.0938 (2) | 0.65697 (10) | 0.0304 (5) | |
| C25 | 0.1278 (3) | −0.1869 (3) | 0.64831 (13) | 0.0341 (6) | |
| H25A | 0.0816 | −0.2684 | 0.6589 | 0.041* | |
| C26 | 0.2749 (3) | −0.1733 (2) | 0.62498 (13) | 0.0322 (6) | |
| H26A | 0.3286 | −0.2426 | 0.6189 | 0.039* | |
| C27 | 0.3399 (3) | −0.0522 (2) | 0.61095 (11) | 0.0267 (5) | |
| N28 | 0.2636 (3) | 0.0452 (2) | 0.62024 (9) | 0.0270 (5) | |
| C29 | 0.4951 (3) | −0.0229 (2) | 0.58435 (11) | 0.0267 (5) | |
| C30 | 0.5691 (3) | −0.1204 (2) | 0.56171 (12) | 0.0290 (5) | |
| H30A | 0.5167 | −0.2065 | 0.5635 | 0.035* | |
| N31 | 0.7095 (3) | −0.1001 (2) | 0.53756 (10) | 0.0314 (5) | |
| C32 | 0.7788 (3) | 0.0217 (3) | 0.53364 (12) | 0.0303 (6) | |
| H32A | 0.8790 | 0.0380 | 0.5169 | 0.036* | |
| C33 | 0.7121 (3) | 0.1247 (3) | 0.55272 (12) | 0.0308 (6) | |
| H33A | 0.7635 | 0.2094 | 0.5479 | 0.037* | |
| C34 | 0.5695 (3) | 0.1028 (2) | 0.57896 (12) | 0.0291 (5) | |
| H34A | 0.5225 | 0.1724 | 0.5932 | 0.035* | |
| O1A | 0.1518 (2) | 0.22087 (16) | 0.07622 (8) | 0.0291 (4) | |
| O21A | 0.3381 (2) | 0.07732 (17) | 0.12752 (9) | 0.0346 (4) | |
| O22A | 0.3131 (2) | −0.0904 (2) | 0.06313 (10) | 0.0396 (5) | |
| O41A | −0.2360 (2) | −0.33957 (17) | 0.04481 (10) | 0.0395 (5) | |
| O42A | −0.4305 (2) | −0.2407 (2) | 0.03722 (12) | 0.0493 (6) | |
| O61A | −0.3332 (3) | 0.2150 (2) | 0.07480 (12) | 0.0520 (6) | |
| O62A | −0.1032 (3) | 0.32666 (18) | 0.06299 (11) | 0.0427 (5) | |
| N2A | 0.2595 (3) | −0.0043 (2) | 0.08996 (10) | 0.0287 (5) | |
| N4A | −0.2888 (3) | −0.2408 (2) | 0.04636 (10) | 0.0311 (5) | |
| N6A | −0.1906 (3) | 0.2244 (2) | 0.06879 (10) | 0.0301 (5) | |
| C1A | 0.0479 (3) | 0.1211 (2) | 0.07465 (10) | 0.0242 (5) | |
| C2A | 0.0909 (3) | −0.0022 (2) | 0.07761 (11) | 0.0253 (5) | |
| C3A | −0.0144 (3) | −0.1176 (2) | 0.06785 (11) | 0.0265 (5) | |
| H3AA | 0.0223 | −0.1956 | 0.0674 | 0.032* | |
| C4A | −0.1766 (3) | −0.1186 (2) | 0.05852 (11) | 0.0272 (5) | |
| C5A | −0.2330 (3) | −0.0062 (2) | 0.06039 (11) | 0.0261 (5) | |
| H5AA | −0.3445 | −0.0084 | 0.0562 | 0.031* | |
| C6A | −0.1240 (3) | 0.1095 (2) | 0.06843 (11) | 0.0259 (5) | |
| O1B | 0.4787 (2) | 0.68998 (16) | 0.21876 (9) | 0.0303 (4) | |
| O21B | 0.2684 (2) | 0.84672 (18) | 0.19341 (9) | 0.0348 (4) | |
| O22B | 0.3412 (3) | 0.9982 (2) | 0.26585 (11) | 0.0493 (6) | |
| O41B | 0.8703 (3) | 1.24447 (18) | 0.21586 (10) | 0.0441 (5) | |
| O42B | 1.0597 (2) | 1.13663 (18) | 0.20751 (9) | 0.0363 (5) | |
| O61B | 0.9270 (2) | 0.6835 (2) | 0.16693 (9) | 0.0392 (5) | |
| O62B | 0.7440 (2) | 0.58469 (18) | 0.21578 (10) | 0.0405 (5) | |
| N2B | 0.3710 (3) | 0.9196 (2) | 0.22702 (11) | 0.0316 (5) | |
| N4B | 0.9186 (3) | 1.1430 (2) | 0.21198 (10) | 0.0327 (5) | |
| N6B | 0.8091 (3) | 0.6809 (2) | 0.19535 (10) | 0.0285 (5) | |
| C1B | 0.5803 (3) | 0.7898 (2) | 0.21517 (11) | 0.0248 (5) | |
| C2B | 0.5386 (3) | 0.9143 (2) | 0.22026 (11) | 0.0267 (5) | |
| C3B | 0.6441 (3) | 1.0279 (2) | 0.22022 (12) | 0.0298 (6) | |
| H3BA | 0.6099 | 1.1072 | 0.2255 | 0.036* | |
| C4B | 0.8035 (3) | 1.0239 (2) | 0.21222 (11) | 0.0281 (5) | |
| C5B | 0.8561 (3) | 0.9107 (2) | 0.20437 (11) | 0.0263 (5) | |
| H5BA | 0.9650 | 0.9103 | 0.1982 | 0.032* | |
| C6B | 0.7471 (3) | 0.7972 (2) | 0.20563 (11) | 0.0257 (5) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0430 (11) | 0.0197 (9) | 0.0408 (11) | 0.0000 (8) | 0.0037 (9) | 0.0007 (8) |
| N1 | 0.0236 (10) | 0.0124 (10) | 0.0347 (12) | −0.0010 (8) | 0.0009 (8) | 0.0036 (8) |
| N2 | 0.0293 (11) | 0.0130 (10) | 0.0334 (11) | 0.0003 (8) | −0.0010 (9) | 0.0033 (8) |
| C1 | 0.0243 (12) | 0.0182 (12) | 0.0379 (14) | 0.0009 (9) | 0.0048 (10) | 0.0009 (10) |
| C2 | 0.0289 (13) | 0.0207 (12) | 0.0353 (14) | −0.0011 (10) | 0.0052 (11) | 0.0030 (10) |
| C3 | 0.0249 (12) | 0.0173 (12) | 0.0345 (13) | 0.0006 (9) | −0.0003 (10) | 0.0005 (9) |
| C4 | 0.0228 (12) | 0.0179 (11) | 0.0338 (13) | −0.0003 (9) | 0.0027 (10) | 0.0021 (9) |
| C5 | 0.0409 (16) | 0.0195 (13) | 0.0407 (15) | 0.0019 (11) | −0.0042 (12) | 0.0079 (11) |
| C6 | 0.0290 (13) | 0.0189 (12) | 0.0408 (15) | 0.0051 (10) | −0.0005 (11) | 0.0066 (10) |
| C7 | 0.0289 (13) | 0.0214 (12) | 0.0345 (14) | 0.0037 (10) | −0.0034 (10) | 0.0067 (10) |
| C8 | 0.0268 (13) | 0.0197 (12) | 0.0432 (15) | −0.0011 (10) | −0.0014 (11) | 0.0087 (11) |
| C9 | 0.0315 (14) | 0.0196 (12) | 0.0361 (14) | −0.0020 (10) | −0.0058 (11) | 0.0033 (10) |
| C10 | 0.0273 (13) | 0.0183 (12) | 0.0360 (14) | 0.0027 (10) | −0.0041 (10) | 0.0051 (10) |
| C11 | 0.0279 (13) | 0.0176 (12) | 0.0357 (14) | −0.0021 (10) | −0.0024 (10) | 0.0070 (10) |
| C12 | 0.0310 (13) | 0.0154 (11) | 0.0373 (14) | −0.0012 (10) | −0.0064 (11) | 0.0047 (10) |
| C13 | 0.0250 (12) | 0.0241 (13) | 0.0370 (14) | 0.0040 (10) | −0.0023 (10) | 0.0023 (10) |
| N14 | 0.0307 (12) | 0.0198 (11) | 0.0366 (12) | 0.0066 (9) | 0.0035 (9) | 0.0011 (9) |
| C15 | 0.0225 (12) | 0.0234 (12) | 0.0342 (14) | 0.0018 (10) | −0.0001 (10) | −0.0014 (10) |
| C16 | 0.0321 (14) | 0.0206 (12) | 0.0403 (15) | 0.0038 (10) | 0.0015 (11) | 0.0026 (10) |
| C17 | 0.0354 (15) | 0.0239 (13) | 0.0427 (16) | 0.0090 (11) | 0.0077 (12) | −0.0017 (11) |
| C18 | 0.0299 (14) | 0.0305 (14) | 0.0329 (14) | 0.0057 (11) | 0.0032 (11) | −0.0001 (11) |
| C19 | 0.0502 (19) | 0.0387 (17) | 0.0472 (18) | 0.0180 (14) | 0.0163 (15) | 0.0062 (13) |
| C20 | 0.0224 (12) | 0.0215 (12) | 0.0331 (13) | 0.0004 (9) | −0.0005 (10) | 0.0023 (10) |
| C21 | 0.0235 (12) | 0.0221 (12) | 0.0350 (14) | 0.0036 (10) | 0.0017 (10) | −0.0012 (10) |
| N22 | 0.0268 (11) | 0.0266 (12) | 0.0352 (12) | 0.0034 (9) | 0.0078 (9) | 0.0056 (9) |
| C23 | 0.0272 (13) | 0.0218 (12) | 0.0278 (12) | 0.0018 (10) | −0.0007 (10) | 0.0004 (9) |
| N24 | 0.0291 (11) | 0.0239 (11) | 0.0349 (12) | −0.0015 (9) | 0.0038 (9) | 0.0017 (9) |
| C25 | 0.0356 (15) | 0.0207 (13) | 0.0420 (16) | −0.0040 (11) | 0.0009 (12) | 0.0055 (11) |
| C26 | 0.0339 (14) | 0.0179 (12) | 0.0440 (16) | 0.0042 (10) | 0.0013 (12) | 0.0030 (11) |
| C27 | 0.0295 (13) | 0.0216 (12) | 0.0278 (13) | 0.0046 (10) | −0.0018 (10) | 0.0013 (9) |
| N28 | 0.0268 (11) | 0.0213 (10) | 0.0318 (11) | 0.0029 (8) | 0.0010 (9) | 0.0026 (8) |
| C29 | 0.0270 (13) | 0.0240 (13) | 0.0285 (13) | 0.0054 (10) | −0.0022 (10) | 0.0024 (10) |
| C30 | 0.0319 (14) | 0.0218 (12) | 0.0333 (14) | 0.0062 (10) | 0.0002 (11) | 0.0029 (10) |
| N31 | 0.0322 (12) | 0.0281 (12) | 0.0346 (12) | 0.0100 (9) | 0.0014 (9) | 0.0011 (9) |
| C32 | 0.0258 (13) | 0.0308 (14) | 0.0335 (14) | 0.0049 (11) | 0.0008 (10) | 0.0026 (11) |
| C33 | 0.0266 (13) | 0.0245 (13) | 0.0388 (15) | 0.0014 (10) | −0.0021 (11) | 0.0026 (11) |
| C34 | 0.0271 (13) | 0.0217 (13) | 0.0372 (14) | 0.0052 (10) | −0.0009 (11) | −0.0003 (10) |
| O1A | 0.0314 (10) | 0.0166 (9) | 0.0370 (10) | 0.0004 (7) | 0.0015 (8) | 0.0026 (7) |
| O21A | 0.0308 (10) | 0.0213 (9) | 0.0475 (12) | −0.0027 (8) | −0.0050 (8) | 0.0048 (8) |
| O22A | 0.0385 (11) | 0.0330 (11) | 0.0494 (12) | 0.0142 (9) | 0.0052 (9) | 0.0018 (9) |
| O41A | 0.0412 (11) | 0.0168 (9) | 0.0567 (13) | −0.0008 (8) | −0.0033 (9) | 0.0037 (8) |
| O42A | 0.0267 (11) | 0.0258 (11) | 0.0909 (18) | −0.0039 (8) | 0.0026 (11) | 0.0056 (11) |
| O61A | 0.0363 (12) | 0.0296 (11) | 0.0924 (19) | 0.0103 (9) | 0.0199 (12) | 0.0051 (11) |
| O62A | 0.0376 (11) | 0.0187 (10) | 0.0708 (15) | 0.0010 (8) | −0.0036 (10) | 0.0125 (9) |
| N2A | 0.0293 (11) | 0.0201 (11) | 0.0369 (12) | 0.0033 (9) | 0.0043 (9) | 0.0073 (9) |
| N4A | 0.0311 (12) | 0.0205 (11) | 0.0388 (13) | −0.0022 (9) | 0.0040 (9) | 0.0042 (9) |
| N6A | 0.0331 (12) | 0.0214 (11) | 0.0345 (12) | 0.0043 (9) | 0.0013 (9) | 0.0005 (9) |
| C1A | 0.0299 (13) | 0.0159 (11) | 0.0250 (12) | 0.0012 (10) | 0.0022 (10) | 0.0012 (9) |
| C2A | 0.0272 (13) | 0.0207 (12) | 0.0275 (12) | 0.0036 (10) | 0.0026 (10) | 0.0030 (9) |
| C3A | 0.0318 (13) | 0.0167 (11) | 0.0304 (13) | 0.0036 (10) | 0.0037 (10) | 0.0029 (9) |
| C4A | 0.0324 (14) | 0.0195 (12) | 0.0268 (12) | −0.0014 (10) | 0.0031 (10) | 0.0024 (9) |
| C5A | 0.0256 (12) | 0.0223 (12) | 0.0291 (13) | 0.0016 (10) | 0.0045 (10) | 0.0029 (10) |
| C6A | 0.0310 (13) | 0.0196 (12) | 0.0269 (12) | 0.0043 (10) | 0.0031 (10) | 0.0032 (9) |
| O1B | 0.0242 (9) | 0.0149 (8) | 0.0490 (11) | −0.0015 (7) | 0.0018 (8) | 0.0028 (7) |
| O21B | 0.0261 (9) | 0.0239 (10) | 0.0516 (12) | −0.0020 (7) | −0.0013 (8) | 0.0064 (8) |
| O22B | 0.0426 (12) | 0.0380 (12) | 0.0659 (15) | 0.0149 (10) | 0.0069 (11) | −0.0120 (11) |
| O41B | 0.0470 (13) | 0.0176 (10) | 0.0635 (14) | −0.0038 (9) | 0.0048 (10) | 0.0060 (9) |
| O42B | 0.0337 (11) | 0.0279 (10) | 0.0409 (11) | −0.0099 (8) | 0.0034 (8) | 0.0049 (8) |
| O61B | 0.0339 (11) | 0.0346 (11) | 0.0510 (12) | 0.0102 (9) | 0.0110 (9) | 0.0052 (9) |
| O62B | 0.0268 (10) | 0.0191 (9) | 0.0747 (15) | −0.0013 (8) | 0.0050 (9) | 0.0118 (9) |
| N2B | 0.0314 (12) | 0.0186 (11) | 0.0450 (13) | 0.0038 (9) | 0.0050 (10) | 0.0055 (9) |
| N4B | 0.0387 (14) | 0.0216 (11) | 0.0326 (12) | −0.0061 (10) | 0.0018 (10) | 0.0033 (9) |
| N6B | 0.0217 (10) | 0.0215 (11) | 0.0396 (12) | 0.0005 (8) | −0.0022 (9) | 0.0009 (9) |
| C1B | 0.0266 (13) | 0.0175 (12) | 0.0278 (12) | 0.0003 (10) | −0.0008 (10) | 0.0008 (9) |
| C2B | 0.0261 (13) | 0.0200 (12) | 0.0322 (13) | 0.0020 (10) | 0.0004 (10) | 0.0010 (10) |
| C3B | 0.0367 (14) | 0.0166 (12) | 0.0347 (14) | 0.0035 (10) | −0.0008 (11) | 0.0022 (10) |
| C4B | 0.0323 (14) | 0.0188 (12) | 0.0289 (13) | −0.0047 (10) | 0.0000 (10) | 0.0036 (9) |
| C5B | 0.0244 (12) | 0.0241 (13) | 0.0266 (12) | −0.0033 (10) | 0.0002 (9) | 0.0026 (9) |
| C6B | 0.0267 (13) | 0.0192 (12) | 0.0289 (13) | 0.0010 (10) | −0.0012 (10) | 0.0015 (9) |
Geometric parameters (Å, °)
| O1—C13 | 1.226 (3) | C23—N28 | 1.339 (3) |
| N1—C1 | 1.494 (3) | C23—N24 | 1.352 (3) |
| N1—C4 | 1.498 (3) | N24—C25 | 1.328 (4) |
| N1—C6 | 1.505 (3) | C25—C26 | 1.380 (4) |
| N1—H1 | 0.85 (3) | C25—H25A | 0.9500 |
| N2—C3 | 1.490 (3) | C26—C27 | 1.390 (4) |
| N2—C5 | 1.491 (3) | C26—H26A | 0.9500 |
| N2—C2 | 1.500 (3) | C27—N28 | 1.341 (3) |
| N2—H2 | 0.89 (4) | C27—C29 | 1.482 (4) |
| C1—C2 | 1.511 (4) | C29—C30 | 1.391 (4) |
| C1—H1A | 0.9900 | C29—C34 | 1.395 (4) |
| C1—H1B | 0.9900 | C30—N31 | 1.338 (4) |
| C2—H2A | 0.9900 | C30—H30A | 0.9500 |
| C2—H2B | 0.9900 | N31—C32 | 1.340 (4) |
| C3—C4 | 1.506 (3) | C32—C33 | 1.379 (4) |
| C3—H3A | 0.9900 | C32—H32A | 0.9500 |
| C3—H3B | 0.9900 | C33—C34 | 1.378 (4) |
| C4—H4A | 0.9900 | C33—H33A | 0.9500 |
| C4—H4B | 0.9900 | C34—H34A | 0.9500 |
| C5—H5A | 0.9800 | O1A—C1A | 1.245 (3) |
| C5—H5B | 0.9800 | O21A—N2A | 1.226 (3) |
| C5—H5C | 0.9800 | O22A—N2A | 1.228 (3) |
| C6—C7 | 1.503 (4) | O41A—N4A | 1.231 (3) |
| C6—H6A | 0.9900 | O42A—N4A | 1.221 (3) |
| C6—H6B | 0.9900 | O61A—N6A | 1.221 (3) |
| C7—C12 | 1.399 (4) | O62A—N6A | 1.225 (3) |
| C7—C8 | 1.403 (4) | N2A—C2A | 1.463 (3) |
| C8—C9 | 1.383 (4) | N4A—C4A | 1.452 (3) |
| C8—H8A | 0.9500 | N6A—C6A | 1.457 (3) |
| C9—C10 | 1.393 (4) | C1A—C2A | 1.451 (3) |
| C9—H9A | 0.9500 | C1A—C6A | 1.451 (4) |
| C10—C11 | 1.397 (3) | C2A—C3A | 1.367 (3) |
| C10—C13 | 1.506 (4) | C3A—C4A | 1.393 (4) |
| C11—C12 | 1.377 (4) | C3A—H3AA | 0.9500 |
| C11—H11A | 0.9500 | C4A—C5A | 1.382 (4) |
| C12—H12A | 0.9500 | C5A—C6A | 1.384 (3) |
| C13—N14 | 1.356 (3) | C5A—H5AA | 0.9500 |
| N14—C15 | 1.419 (3) | O1B—C1B | 1.252 (3) |
| N14—H14 | 0.85 (3) | O21B—N2B | 1.227 (3) |
| C15—C21 | 1.385 (4) | O22B—N2B | 1.228 (3) |
| C15—C16 | 1.394 (4) | O41B—N4B | 1.235 (3) |
| C16—C17 | 1.389 (4) | O42B—N4B | 1.232 (3) |
| C16—H16A | 0.9500 | O61B—N6B | 1.226 (3) |
| C17—C18 | 1.388 (4) | O62B—N6B | 1.229 (3) |
| C17—H17A | 0.9500 | N2B—C2B | 1.462 (3) |
| C18—C20 | 1.396 (4) | N4B—C4B | 1.451 (3) |
| C18—C19 | 1.508 (4) | N6B—C6B | 1.450 (3) |
| C19—H19A | 0.9800 | C1B—C6B | 1.443 (4) |
| C19—H19B | 0.9800 | C1B—C2B | 1.443 (3) |
| C19—H19C | 0.9800 | C2B—C3B | 1.366 (4) |
| C20—C21 | 1.394 (4) | C3B—C4B | 1.394 (4) |
| C20—N22 | 1.419 (3) | C3B—H3BA | 0.9500 |
| C21—H21A | 0.9500 | C4B—C5B | 1.373 (4) |
| N22—C23 | 1.369 (3) | C5B—C6B | 1.384 (3) |
| N22—H22 | 0.87 (4) | C5B—H5BA | 0.9500 |
| C1—N1—C4 | 108.6 (2) | C18—C20—N22 | 118.4 (2) |
| C1—N1—C6 | 111.1 (2) | C15—C21—C20 | 121.1 (2) |
| C4—N1—C6 | 111.62 (19) | C15—C21—H21A | 119.5 |
| C1—N1—H1 | 107 (2) | C20—C21—H21A | 119.5 |
| C4—N1—H1 | 109 (2) | C23—N22—C20 | 129.0 (2) |
| C6—N1—H1 | 109 (2) | C23—N22—H22 | 116 (2) |
| C3—N2—C5 | 110.9 (2) | C20—N22—H22 | 113 (2) |
| C3—N2—C2 | 110.4 (2) | N28—C23—N24 | 125.8 (2) |
| C5—N2—C2 | 110.8 (2) | N28—C23—N22 | 119.1 (2) |
| C3—N2—H2 | 105 (2) | N24—C23—N22 | 115.2 (2) |
| C5—N2—H2 | 110 (2) | C25—N24—C23 | 115.0 (2) |
| C2—N2—H2 | 109 (2) | N24—C25—C26 | 124.4 (2) |
| N1—C1—C2 | 110.9 (2) | N24—C25—H25A | 117.8 |
| N1—C1—H1A | 109.5 | C26—C25—H25A | 117.8 |
| C2—C1—H1A | 109.5 | C25—C26—C27 | 116.1 (2) |
| N1—C1—H1B | 109.5 | C25—C26—H26A | 121.9 |
| C2—C1—H1B | 109.5 | C27—C26—H26A | 121.9 |
| H1A—C1—H1B | 108.0 | N28—C27—C26 | 121.4 (2) |
| N2—C2—C1 | 112.1 (2) | N28—C27—C29 | 116.0 (2) |
| N2—C2—H2A | 109.2 | C26—C27—C29 | 122.6 (2) |
| C1—C2—H2A | 109.2 | C23—N28—C27 | 117.3 (2) |
| N2—C2—H2B | 109.2 | C30—C29—C34 | 117.4 (2) |
| C1—C2—H2B | 109.2 | C30—C29—C27 | 121.1 (2) |
| H2A—C2—H2B | 107.9 | C34—C29—C27 | 121.5 (2) |
| N2—C3—C4 | 111.5 (2) | N31—C30—C29 | 123.9 (2) |
| N2—C3—H3A | 109.3 | N31—C30—H30A | 118.0 |
| C4—C3—H3A | 109.3 | C29—C30—H30A | 118.0 |
| N2—C3—H3B | 109.3 | C30—N31—C32 | 117.1 (2) |
| C4—C3—H3B | 109.3 | N31—C32—C33 | 123.3 (3) |
| H3A—C3—H3B | 108.0 | N31—C32—H32A | 118.4 |
| N1—C4—C3 | 111.2 (2) | C33—C32—H32A | 118.4 |
| N1—C4—H4A | 109.4 | C34—C33—C32 | 119.0 (2) |
| C3—C4—H4A | 109.4 | C34—C33—H33A | 120.5 |
| N1—C4—H4B | 109.4 | C32—C33—H33A | 120.5 |
| C3—C4—H4B | 109.4 | C33—C34—C29 | 119.2 (2) |
| H4A—C4—H4B | 108.0 | C33—C34—H34A | 120.4 |
| N2—C5—H5A | 109.5 | C29—C34—H34A | 120.4 |
| N2—C5—H5B | 109.5 | O21A—N2A—O22A | 123.7 (2) |
| H5A—C5—H5B | 109.5 | O21A—N2A—C2A | 118.4 (2) |
| N2—C5—H5C | 109.5 | O22A—N2A—C2A | 117.9 (2) |
| H5A—C5—H5C | 109.5 | O42A—N4A—O41A | 123.1 (2) |
| H5B—C5—H5C | 109.5 | O42A—N4A—C4A | 118.6 (2) |
| C7—C6—N1 | 112.6 (2) | O41A—N4A—C4A | 118.2 (2) |
| C7—C6—H6A | 109.1 | O61A—N6A—O62A | 122.0 (2) |
| N1—C6—H6A | 109.1 | O61A—N6A—C6A | 118.4 (2) |
| C7—C6—H6B | 109.1 | O62A—N6A—C6A | 119.6 (2) |
| N1—C6—H6B | 109.1 | O1A—C1A—C2A | 121.3 (2) |
| H6A—C6—H6B | 107.8 | O1A—C1A—C6A | 126.9 (2) |
| C12—C7—C8 | 118.6 (3) | C2A—C1A—C6A | 111.7 (2) |
| C12—C7—C6 | 119.2 (2) | C3A—C2A—C1A | 124.6 (2) |
| C8—C7—C6 | 122.2 (2) | C3A—C2A—N2A | 117.3 (2) |
| C9—C8—C7 | 120.4 (2) | C1A—C2A—N2A | 118.0 (2) |
| C9—C8—H8A | 119.8 | C2A—C3A—C4A | 118.7 (2) |
| C7—C8—H8A | 119.8 | C2A—C3A—H3AA | 120.6 |
| C8—C9—C10 | 120.9 (2) | C4A—C3A—H3AA | 120.6 |
| C8—C9—H9A | 119.5 | C5A—C4A—C3A | 121.5 (2) |
| C10—C9—H9A | 119.5 | C5A—C4A—N4A | 119.4 (2) |
| C9—C10—C11 | 118.4 (3) | C3A—C4A—N4A | 119.1 (2) |
| C9—C10—C13 | 119.6 (2) | C4A—C5A—C6A | 118.8 (2) |
| C11—C10—C13 | 122.1 (2) | C4A—C5A—H5AA | 120.6 |
| C12—C11—C10 | 121.2 (2) | C6A—C5A—H5AA | 120.6 |
| C12—C11—H11A | 119.4 | C5A—C6A—C1A | 124.0 (2) |
| C10—C11—H11A | 119.4 | C5A—C6A—N6A | 116.2 (2) |
| C11—C12—C7 | 120.4 (2) | C1A—C6A—N6A | 119.7 (2) |
| C11—C12—H12A | 119.8 | O21B—N2B—O22B | 123.7 (2) |
| C7—C12—H12A | 119.8 | O21B—N2B—C2B | 118.5 (2) |
| O1—C13—N14 | 123.9 (3) | O22B—N2B—C2B | 117.8 (2) |
| O1—C13—C10 | 121.8 (2) | O42B—N4B—O41B | 123.7 (2) |
| N14—C13—C10 | 114.3 (2) | O42B—N4B—C4B | 117.7 (2) |
| C13—N14—C15 | 129.0 (2) | O41B—N4B—C4B | 118.7 (2) |
| C13—N14—H14 | 117 (2) | O61B—N6B—O62B | 122.5 (2) |
| C15—N14—H14 | 113 (2) | O61B—N6B—C6B | 118.1 (2) |
| C21—C15—C16 | 120.2 (2) | O62B—N6B—C6B | 119.4 (2) |
| C21—C15—N14 | 116.1 (2) | O1B—C1B—C6B | 126.3 (2) |
| C16—C15—N14 | 123.6 (2) | O1B—C1B—C2B | 121.7 (2) |
| C17—C16—C15 | 117.5 (2) | C6B—C1B—C2B | 112.0 (2) |
| C17—C16—H16A | 121.2 | C3B—C2B—C1B | 125.1 (2) |
| C15—C16—H16A | 121.2 | C3B—C2B—N2B | 117.4 (2) |
| C18—C17—C16 | 123.6 (2) | C1B—C2B—N2B | 117.6 (2) |
| C18—C17—H17A | 118.2 | C2B—C3B—C4B | 117.9 (2) |
| C16—C17—H17A | 118.2 | C2B—C3B—H3BA | 121.1 |
| C17—C18—C20 | 117.6 (3) | C4B—C3B—H3BA | 121.1 |
| C17—C18—C19 | 120.0 (3) | C5B—C4B—C3B | 122.3 (2) |
| C20—C18—C19 | 122.2 (3) | C5B—C4B—N4B | 118.5 (2) |
| C18—C19—H19A | 109.5 | C3B—C4B—N4B | 119.3 (2) |
| C18—C19—H19B | 109.5 | C4B—C5B—C6B | 118.6 (2) |
| H19A—C19—H19B | 109.5 | C4B—C5B—H5BA | 120.7 |
| C18—C19—H19C | 109.5 | C6B—C5B—H5BA | 120.7 |
| H19A—C19—H19C | 109.5 | C5B—C6B—C1B | 124.1 (2) |
| H19B—C19—H19C | 109.5 | C5B—C6B—N6B | 115.8 (2) |
| C21—C20—C18 | 119.8 (2) | C1B—C6B—N6B | 120.1 (2) |
| C21—C20—N22 | 121.6 (2) | ||
| C4—N1—C1—C2 | 58.2 (2) | C27—C29—C30—N31 | −179.5 (2) |
| C6—N1—C1—C2 | −178.6 (2) | C29—C30—N31—C32 | −2.2 (4) |
| C3—N2—C2—C1 | 53.6 (3) | C30—N31—C32—C33 | −0.4 (4) |
| C5—N2—C2—C1 | 176.8 (2) | N31—C32—C33—C34 | 2.2 (4) |
| N1—C1—C2—N2 | −56.6 (3) | C32—C33—C34—C29 | −1.4 (4) |
| C5—N2—C3—C4 | −177.2 (2) | C30—C29—C34—C33 | −1.0 (4) |
| C2—N2—C3—C4 | −54.0 (3) | C27—C29—C34—C33 | −178.6 (2) |
| C1—N1—C4—C3 | −59.2 (3) | O1A—C1A—C2A—C3A | −170.2 (2) |
| C6—N1—C4—C3 | 177.9 (2) | C6A—C1A—C2A—C3A | 8.4 (4) |
| N2—C3—C4—N1 | 58.1 (3) | O1A—C1A—C2A—N2A | 7.6 (4) |
| C1—N1—C6—C7 | −179.7 (2) | C6A—C1A—C2A—N2A | −173.8 (2) |
| C4—N1—C6—C7 | −58.3 (3) | O21A—N2A—C2A—C3A | −140.1 (2) |
| N1—C6—C7—C12 | 107.2 (3) | O22A—N2A—C2A—C3A | 37.9 (3) |
| N1—C6—C7—C8 | −71.7 (3) | O21A—N2A—C2A—C1A | 41.9 (3) |
| C12—C7—C8—C9 | −0.3 (4) | O22A—N2A—C2A—C1A | −140.1 (2) |
| C6—C7—C8—C9 | 178.6 (2) | C1A—C2A—C3A—C4A | −4.9 (4) |
| C7—C8—C9—C10 | 1.5 (4) | N2A—C2A—C3A—C4A | 177.2 (2) |
| C8—C9—C10—C11 | −1.3 (4) | C2A—C3A—C4A—C5A | −1.6 (4) |
| C8—C9—C10—C13 | −179.5 (2) | C2A—C3A—C4A—N4A | 178.2 (2) |
| C9—C10—C11—C12 | 0.0 (4) | O42A—N4A—C4A—C5A | 1.1 (4) |
| C13—C10—C11—C12 | 178.1 (2) | O41A—N4A—C4A—C5A | 179.9 (2) |
| C10—C11—C12—C7 | 1.2 (4) | O42A—N4A—C4A—C3A | −178.7 (3) |
| C8—C7—C12—C11 | −1.0 (4) | O41A—N4A—C4A—C3A | 0.1 (4) |
| C6—C7—C12—C11 | −180.0 (2) | C3A—C4A—C5A—C6A | 3.7 (4) |
| C9—C10—C13—O1 | 23.7 (4) | N4A—C4A—C5A—C6A | −176.1 (2) |
| C11—C10—C13—O1 | −154.5 (3) | C4A—C5A—C6A—C1A | 0.5 (4) |
| C9—C10—C13—N14 | −156.9 (2) | C4A—C5A—C6A—N6A | 178.8 (2) |
| C11—C10—C13—N14 | 25.0 (4) | O1A—C1A—C6A—C5A | 172.4 (2) |
| O1—C13—N14—C15 | 8.6 (4) | C2A—C1A—C6A—C5A | −6.1 (3) |
| C10—C13—N14—C15 | −170.8 (2) | O1A—C1A—C6A—N6A | −5.8 (4) |
| C13—N14—C15—C21 | −160.7 (3) | C2A—C1A—C6A—N6A | 175.8 (2) |
| C13—N14—C15—C16 | 23.3 (4) | O61A—N6A—C6A—C5A | 15.5 (4) |
| C21—C15—C16—C17 | −2.5 (4) | O62A—N6A—C6A—C5A | −164.3 (2) |
| N14—C15—C16—C17 | 173.3 (3) | O61A—N6A—C6A—C1A | −166.2 (3) |
| C15—C16—C17—C18 | −0.2 (4) | O62A—N6A—C6A—C1A | 14.0 (4) |
| C16—C17—C18—C20 | 2.9 (4) | O1B—C1B—C2B—C3B | 176.0 (3) |
| C16—C17—C18—C19 | −173.3 (3) | C6B—C1B—C2B—C3B | −4.2 (4) |
| C17—C18—C20—C21 | −3.0 (4) | O1B—C1B—C2B—N2B | −2.9 (4) |
| C19—C18—C20—C21 | 173.1 (3) | C6B—C1B—C2B—N2B | 176.9 (2) |
| C17—C18—C20—N22 | −178.2 (2) | O21B—N2B—C2B—C3B | 132.4 (3) |
| C19—C18—C20—N22 | −2.1 (4) | O22B—N2B—C2B—C3B | −46.8 (4) |
| C16—C15—C21—C20 | 2.4 (4) | O21B—N2B—C2B—C1B | −48.7 (3) |
| N14—C15—C21—C20 | −173.7 (2) | O22B—N2B—C2B—C1B | 132.2 (3) |
| C18—C20—C21—C15 | 0.4 (4) | C1B—C2B—C3B—C4B | 2.8 (4) |
| N22—C20—C21—C15 | 175.5 (2) | N2B—C2B—C3B—C4B | −178.3 (2) |
| C21—C20—N22—C23 | 17.2 (4) | C2B—C3B—C4B—C5B | 0.2 (4) |
| C18—C20—N22—C23 | −167.7 (3) | C2B—C3B—C4B—N4B | −180.0 (2) |
| C20—N22—C23—N28 | 18.2 (4) | O42B—N4B—C4B—C5B | −3.4 (4) |
| C20—N22—C23—N24 | −162.5 (2) | O41B—N4B—C4B—C5B | 176.1 (2) |
| N28—C23—N24—C25 | −0.6 (4) | O42B—N4B—C4B—C3B | 176.7 (2) |
| N22—C23—N24—C25 | −179.8 (2) | O41B—N4B—C4B—C3B | −3.7 (4) |
| C23—N24—C25—C26 | −1.0 (4) | C3B—C4B—C5B—C6B | −1.3 (4) |
| N24—C25—C26—C27 | 1.0 (4) | N4B—C4B—C5B—C6B | 178.9 (2) |
| C25—C26—C27—N28 | 0.6 (4) | C4B—C5B—C6B—C1B | −0.5 (4) |
| C25—C26—C27—C29 | −178.6 (2) | C4B—C5B—C6B—N6B | 178.3 (2) |
| N24—C23—N28—C27 | 2.1 (4) | O1B—C1B—C6B—C5B | −177.2 (3) |
| N22—C23—N28—C27 | −178.7 (2) | C2B—C1B—C6B—C5B | 3.0 (4) |
| C26—C27—N28—C23 | −2.0 (4) | O1B—C1B—C6B—N6B | 4.1 (4) |
| C29—C27—N28—C23 | 177.2 (2) | C2B—C1B—C6B—N6B | −175.7 (2) |
| N28—C27—C29—C30 | −166.7 (2) | O61B—N6B—C6B—C5B | −25.5 (3) |
| C26—C27—C29—C30 | 12.6 (4) | O62B—N6B—C6B—C5B | 153.6 (2) |
| N28—C27—C29—C34 | 10.8 (4) | O61B—N6B—C6B—C1B | 153.3 (2) |
| C26—C27—C29—C34 | −170.0 (3) | O62B—N6B—C6B—C1B | −27.6 (4) |
| C34—C29—C30—N31 | 3.0 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1B | 0.85 (3) | 1.85 (3) | 2.658 (3) | 157 (3) |
| N1—H1···O62B | 0.85 (3) | 2.35 (3) | 2.890 (3) | 122 (2) |
| N2—H2···O1A | 0.89 (4) | 1.85 (4) | 2.678 (3) | 154 (3) |
| N2—H2···O62A | 0.89 (4) | 2.41 (4) | 3.009 (3) | 125 (3) |
| N14—H14···N31i | 0.85 (3) | 2.23 (3) | 3.069 (3) | 171 (3) |
| C5—H5B···O41Aii | 0.98 | 2.48 | 3.258 (4) | 136 |
| C4—H4B···O42Biii | 0.99 | 2.33 | 3.199 (3) | 146 |
| C3—H3A···O61Biv | 0.99 | 2.57 | 3.199 (3) | 121 |
| C3—H3B···O1B | 0.99 | 2.34 | 3.072 (3) | 130 |
| C12—H12A···O42Biii | 0.95 | 2.63 | 3.423 (3) | 142 |
| C19—H19A···O61Bv | 0.98 | 2.50 | 3.435 (4) | 159 |
| C19—H19A···N6Bv | 0.98 | 2.65 | 3.541 (4) | 152 |
Symmetry codes: (i) −x+1, −y, −z+1; (ii) −x, −y, −z; (iii) x−1, y−1, z; (iv) x−1, y, z; (v) −x+1, −y+1, −z+1.
Table 2 π-Ring hydrogen-bond geometry (Å, °) for (I).
Cg5 is the centroid of the C15–C21 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C33—H33A···Cg5vi | 0.95 | 2.90 | 3.545 (8) | 127 |
Symmetry code: (vi) x+1, y, z.
Table 3 π–π stacking geometry (Å, °) for (I).
| Cg2···Cg7v | 3.740 (4) |
| Cg3···Cg3v | 3.496 (7) |
| Cg6···Cg6vii | 3.396 (0) |
Symmetry codes: (v) -x+1, -y+1, -z+1; (vii) -x+2, -y+2, -z. Notes: Cg2, Cg3, Cg6 and Cg7 are the centroids of the C25/C26/C27/N28/C23/N4, C32/C33/C34/C29//C30/N31, C1A–C6A and C1B–C6B rings, respectively.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT5129).
References
- Allen, F. H., Johnson, O., Shields, G. P., Smith, B. R. & Towler, M. (2004). J. Appl. Cryst.37, 335–338.
- Bindya, S., Wong, W.-T., Ashok, M. A., Yathirajan, H. S. & Rathore, R. S. (2007). Acta Cryst. C63, o546–o548. [DOI] [PubMed]
- Capdeville, R., Buchdunger, E., Zimmermann, J. & Matter, A. (2002). Nat. Rev. Drug Discov.1, 493–502. [DOI] [PubMed]
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc.97, 1354–1358.
- Harrison, W. T. A., Bindya, S., Ashok, M. A., Yathirajan, H. S. & Narayana, B. (2007). Acta Cryst. E63, o3143.
- Harrison, W. T. A., Sreevidya, T. V., Narayana, B., Sarojini, B. K. & Yathirajan, H. S. (2007). Acta Cryst. E63, o3871.
- Jasinski, J. P., Butcher, R. J., Hakim Al-Arique, Q. N. M., Yathirajan, H. S. & Narayana, B. (2009a). Acta Cryst. E65, o1738–o1739. [DOI] [PMC free article] [PubMed]
- Jasinski, J. P., Butcher, R. J., Hakim Al-Arique, Q. N. M., Yathirajan, H. S. & Narayana, B. (2009b). Acta Cryst. E65, o2201–o2202. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst.39, 453–457.
- Moen, M. D., Mckeage, K., Plosker, G. L. & Siddiqui, M. A. A. (2007). Drugs, 67, 299–320. [DOI] [PubMed]
- Oxford Diffraction (2007). CrysAlis PRO and CrysAlis RED Oxford Diffraction Ltd, Abingdon, Oxfordshire, England.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Swamy, M. T., Ashok, M. A., Yathirajan, H. S., Narayana, B. & Bolte, M. (2007). Acta Cryst. E63, o4919.
- Szumma, A., Jurczak, J. & Urbańczyk-Lipkowska, Z. (2000). J. Mol. Struct.526, 165–175.
- Yathirajan, H. S., Ashok, M. A., Narayana Achar, B. & Bolte, M. (2007a). Acta Cryst. E63, o1691–o1692.
- Yathirajan, H. S., Ashok, M. A., Narayana Achar, B. & Bolte, M. (2007b). Acta Cryst. E63, o1693–o1695.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810000577/bt5129sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810000577/bt5129Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report