Abstract
The title compound, C10H7FO4, is an intermediate in the synthesis of the drug Fidarestat, (2S,4S)-2-aminoformyl-6-fluoro-spiro[chroman-4,4′-imidazolidine]-2′,5′-dione. The dihydropyranone ring adopts an envelope conformation with the asymmetric C atom in the flap position. In the crystal, the molecules are linked into zigzag chains along [100] by O—H⋯O hydrogen bonds and C—H⋯π interactions involving the benzene ring.
Related literature
Fidarestat, which shows strong inhibition to aldose reductases, is used to treat complications of diabetes, see: Mealy (1996 ▶); Mitsuru et al. (2000 ▶). For related structures, see: Kurono et al. (1989 ▶); Yamaguchi et al. (1994 ▶).
Experimental
Crystal data
C10H7FO4
M r = 210.16
Orthorhombic,

a = 5.3472 (11) Å
b = 12.748 (3) Å
c = 12.785 (3) Å
V = 871.5 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.14 mm−1
T = 113 K
0.28 × 0.23 × 0.22 mm
Data collection
Rigaku Saturn CCD area-detector diffractometer
Absorption correction: multi-scan (ABSCOR; Higashi, 1995 ▶) T min = 0.962, T max = 0.970
8640 measured reflections
1212 independent reflections
1049 reflections with I > 2σ(I)
R int = 0.045
Refinement
R[F 2 > 2σ(F 2)] = 0.027
wR(F 2) = 0.073
S = 1.12
1212 reflections
137 parameters
H-atom parameters constrained
Δρmax = 0.17 e Å−3
Δρmin = −0.15 e Å−3
Data collection: CrystalClear (Rigaku/MSC, 2005 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809054555/ci2987sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809054555/ci2987Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O4—H4⋯O2i | 0.84 | 1.81 | 2.6474 (19) | 171 |
| C5—H5B⋯Cg1ii | 0.99 | 2.51 | 3.4521 (19) | 160 |
Symmetry codes: (i)  ; (ii)
; (ii) 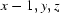 . Cg1 is the centroid of the C1–C3/C7–C9 ring.
. Cg1 is the centroid of the C1–C3/C7–C9 ring.
supplementary crystallographic information
Comment
The optically active (S)-6-fluoro-4-oxochroman-2-carboxylic acid is a key intermediate for synthesizing Fidarestat which shows strong inhibition to aldose reductases to cure incurable complications of diabetes (Mitsuru et al., 2000; Mealy, 1996). Our interests in synthesizing Fidarestat prompted us to develop an efficient methodology for synthesizing (S)-6-fluoro-4-oxochroman-2-carboxylic acid. In our synthetic work, we obtained the title compound, which is similar to those reported in the literature (Kurono et al., 1989; Yamaguchi et al., 1994). Its crystal structure is reported here.
The dihydropyranone ring adopts an envelope conformation with the asymmetric C atom in the flap position (Fig. 1). The molecules are linked into zigzag chains along the [100] by O—H···O hydrogen bonds and C—H···π interactions (Table 1) involving the benzene ring.
Experimental
To a stirred solution of (2S,4R)-2-(1',2'-Dihydroxyethyl)-6-fluoro-chroman-4-one (10.7 g, 0.05 mol) in 300 ml of anhydrous benzene at room temperature was added lead tetraacetate (22.2 g, 0.05 mol). After 30 min, the solution was filtered and the filtrate was evaporated in vacuum to the residue. To the solution of 2% aqueous silver nitrate (651 ml, 0.07 mol) was added 5% aqueous sodium hydroxide (120 ml, 0.16 mol), and then generated the black precipitate immediately. To this stirred solution at room temperature was added, dropwise over 5 min, 4% ammonia water (520 ml, 0.16 mol). The black precipitate disappeared. The residue described above was dissolved in small amounts of THF and then added in this stirred solution at 323–333 K. After 10 min, the solution was filtered, and the precipitate was washed with water. The filtrate was acidified to pH 1 with 6 N aqueous hydrochloric acid, and then extracted with ethyl acetate. The organic extracts were dried (MgSO4) and then concentrated under reduced pressure. The residue was mixed with a mixture of ethanol and water, and left to crystallize 8.7 g (83%) of 6-Fluoro- 4-oxochroman-2-carboxylic acid. Colourless crystals suitable for X-ray analysis were obtained by slow evaporation in ethanol at room temperature.
Refinement
H atoms were positioned geometrically (O-H = 0.84 and C-H = 0.95–1.00 Å) and refined using a riding model, with Uiso(H) = 1.2Ueq(C) and 1.5Ueq(O). In the absence of significant anomalous scattering, Friedel pairs were merged prior to the final refinement. Nine reflections that were affected by the beamstop were discarded.
Figures
Fig. 1.
The molecular structure of the title compound, with displacement ellipsoids drawn at the 50% probability level.
Crystal data
| C10H7FO4 | F(000) = 432 |
| Mr = 210.16 | Dx = 1.602 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 3184 reflections |
| a = 5.3472 (11) Å | θ = 1.6–27.8° |
| b = 12.748 (3) Å | µ = 0.14 mm−1 |
| c = 12.785 (3) Å | T = 113 K |
| V = 871.5 (3) Å3 | Block, colourless |
| Z = 4 | 0.28 × 0.23 × 0.22 mm |
Data collection
| Rigaku Saturn CCD area-detector diffractometer | 1212 independent reflections |
| Radiation source: rotating anode | 1049 reflections with I > 2σ(I) |
| confocal | Rint = 0.045 |
| Detector resolution: 7.31 pixels mm-1 | θmax = 27.9°, θmin = 2.3° |
| ω and φ scans | h = −7→6 |
| Absorption correction: multi-scan (ABSCOR; Higashi, 1995) | k = −16→16 |
| Tmin = 0.962, Tmax = 0.970 | l = −16→16 |
| 8640 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.027 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.073 | H-atom parameters constrained |
| S = 1.12 | w = 1/[σ2(Fo2) + (0.0448P)2] where P = (Fo2 + 2Fc2)/3 |
| 1212 reflections | (Δ/σ)max = 0.001 |
| 137 parameters | Δρmax = 0.17 e Å−3 |
| 0 restraints | Δρmin = −0.15 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| F1 | 0.8260 (2) | 0.32440 (8) | 0.43315 (9) | 0.0336 (3) | |
| O1 | 0.2480 (2) | 0.03466 (8) | 0.23160 (9) | 0.0189 (3) | |
| O2 | 0.1188 (2) | 0.34646 (9) | 0.17373 (10) | 0.0230 (3) | |
| O3 | 0.5538 (2) | 0.03536 (10) | 0.06321 (10) | 0.0230 (3) | |
| O4 | 0.2609 (3) | 0.11076 (10) | −0.03716 (9) | 0.0250 (3) | |
| H4 | 0.3799 | 0.1179 | −0.0795 | 0.037* | |
| C1 | 0.6806 (3) | 0.25426 (12) | 0.38164 (13) | 0.0200 (4) | |
| C2 | 0.5057 (3) | 0.29048 (13) | 0.31336 (12) | 0.0184 (4) | |
| H2 | 0.4862 | 0.3636 | 0.3015 | 0.022* | |
| C3 | 0.3547 (3) | 0.21751 (12) | 0.26079 (12) | 0.0152 (3) | |
| C4 | 0.1561 (3) | 0.25266 (12) | 0.19024 (12) | 0.0154 (3) | |
| C5 | −0.0026 (3) | 0.16808 (13) | 0.14292 (12) | 0.0175 (3) | |
| H5A | −0.0588 | 0.1905 | 0.0726 | 0.021* | |
| H5B | −0.1529 | 0.1573 | 0.1868 | 0.021* | |
| C6 | 0.1404 (3) | 0.06506 (12) | 0.13377 (12) | 0.0167 (3) | |
| H6 | 0.0182 | 0.0095 | 0.1130 | 0.020* | |
| C7 | 0.3894 (3) | 0.10981 (12) | 0.27966 (12) | 0.0155 (4) | |
| C8 | 0.5700 (3) | 0.07625 (13) | 0.35030 (12) | 0.0182 (4) | |
| H8 | 0.5923 | 0.0034 | 0.3630 | 0.022* | |
| C9 | 0.7161 (4) | 0.14813 (12) | 0.40169 (12) | 0.0200 (4) | |
| H9 | 0.8396 | 0.1257 | 0.4502 | 0.024* | |
| C10 | 0.3450 (3) | 0.06823 (12) | 0.05065 (13) | 0.0174 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| F1 | 0.0418 (7) | 0.0231 (5) | 0.0358 (6) | −0.0017 (5) | −0.0248 (5) | −0.0041 (5) |
| O1 | 0.0218 (6) | 0.0139 (5) | 0.0211 (5) | −0.0031 (5) | −0.0003 (5) | 0.0013 (4) |
| O2 | 0.0246 (7) | 0.0158 (6) | 0.0287 (6) | 0.0020 (5) | −0.0091 (5) | 0.0024 (5) |
| O3 | 0.0155 (6) | 0.0265 (6) | 0.0270 (6) | 0.0037 (5) | 0.0012 (5) | −0.0053 (5) |
| O4 | 0.0189 (6) | 0.0340 (6) | 0.0221 (6) | −0.0007 (6) | 0.0050 (6) | 0.0024 (5) |
| C1 | 0.0225 (10) | 0.0191 (8) | 0.0184 (7) | −0.0018 (7) | −0.0048 (7) | −0.0029 (7) |
| C2 | 0.0226 (10) | 0.0146 (7) | 0.0181 (7) | 0.0007 (7) | −0.0026 (7) | −0.0004 (6) |
| C3 | 0.0155 (8) | 0.0147 (7) | 0.0155 (7) | 0.0006 (6) | 0.0004 (6) | 0.0009 (6) |
| C4 | 0.0137 (8) | 0.0165 (8) | 0.0161 (7) | 0.0007 (7) | 0.0018 (6) | −0.0007 (6) |
| C5 | 0.0142 (8) | 0.0182 (8) | 0.0201 (8) | −0.0016 (7) | 0.0011 (6) | −0.0024 (6) |
| C6 | 0.0151 (8) | 0.0149 (7) | 0.0201 (8) | −0.0027 (7) | 0.0004 (6) | −0.0013 (6) |
| C7 | 0.0170 (9) | 0.0140 (7) | 0.0154 (7) | −0.0006 (7) | 0.0045 (7) | −0.0012 (6) |
| C8 | 0.0226 (9) | 0.0137 (7) | 0.0181 (7) | 0.0049 (7) | 0.0033 (7) | 0.0035 (6) |
| C9 | 0.0217 (9) | 0.0229 (8) | 0.0154 (7) | 0.0042 (7) | −0.0006 (7) | 0.0024 (6) |
| C10 | 0.0174 (9) | 0.0133 (7) | 0.0214 (8) | −0.0041 (7) | 0.0010 (7) | −0.0048 (7) |
Geometric parameters (Å, °)
| F1—C1 | 1.3557 (18) | C3—C4 | 1.464 (2) |
| O1—C7 | 1.366 (2) | C4—C5 | 1.499 (2) |
| O1—C6 | 1.4302 (19) | C5—C6 | 1.524 (2) |
| O2—C4 | 1.2305 (19) | C5—H5A | 0.99 |
| O3—C10 | 1.203 (2) | C5—H5B | 0.99 |
| O4—C10 | 1.325 (2) | C6—C10 | 1.526 (2) |
| O4—H4 | 0.84 | C6—H6 | 1.00 |
| C1—C2 | 1.360 (2) | C7—C8 | 1.390 (2) |
| C1—C9 | 1.390 (2) | C8—C9 | 1.372 (2) |
| C2—C3 | 1.403 (2) | C8—H8 | 0.95 |
| C2—H2 | 0.95 | C9—H9 | 0.95 |
| C3—C7 | 1.406 (2) | ||
| C7—O1—C6 | 115.20 (12) | H5A—C5—H5B | 108.0 |
| C10—O4—H4 | 109.5 | O1—C6—C5 | 111.60 (12) |
| F1—C1—C2 | 118.84 (14) | O1—C6—C10 | 109.14 (14) |
| F1—C1—C9 | 118.29 (15) | C5—C6—C10 | 112.98 (13) |
| C2—C1—C9 | 122.87 (16) | O1—C6—H6 | 107.6 |
| C1—C2—C3 | 118.56 (15) | C5—C6—H6 | 107.6 |
| C1—C2—H2 | 120.7 | C10—C6—H6 | 107.6 |
| C3—C2—H2 | 120.7 | O1—C7—C8 | 117.44 (14) |
| C2—C3—C7 | 119.28 (15) | O1—C7—C3 | 122.30 (15) |
| C2—C3—C4 | 120.64 (14) | C8—C7—C3 | 120.25 (15) |
| C7—C3—C4 | 120.03 (14) | C9—C8—C7 | 120.09 (15) |
| O2—C4—C3 | 121.39 (14) | C9—C8—H8 | 120.0 |
| O2—C4—C5 | 122.54 (15) | C7—C8—H8 | 120.0 |
| C3—C4—C5 | 116.04 (13) | C8—C9—C1 | 118.95 (16) |
| C4—C5—C6 | 111.51 (13) | C8—C9—H9 | 120.5 |
| C4—C5—H5A | 109.3 | C1—C9—H9 | 120.5 |
| C6—C5—H5A | 109.3 | O3—C10—O4 | 124.77 (16) |
| C4—C5—H5B | 109.3 | O3—C10—C6 | 124.27 (15) |
| C6—C5—H5B | 109.3 | O4—C10—C6 | 110.96 (14) |
| F1—C1—C2—C3 | −179.60 (15) | C6—O1—C7—C3 | 24.6 (2) |
| C9—C1—C2—C3 | −0.1 (3) | C2—C3—C7—O1 | 179.64 (14) |
| C1—C2—C3—C7 | −0.4 (2) | C4—C3—C7—O1 | 2.1 (2) |
| C1—C2—C3—C4 | 177.15 (14) | C2—C3—C7—C8 | 0.5 (2) |
| C2—C3—C4—O2 | 1.2 (2) | C4—C3—C7—C8 | −177.00 (14) |
| C7—C3—C4—O2 | 178.73 (16) | O1—C7—C8—C9 | −179.41 (14) |
| C2—C3—C4—C5 | −176.75 (14) | C3—C7—C8—C9 | −0.3 (2) |
| C7—C3—C4—C5 | 0.8 (2) | C7—C8—C9—C1 | −0.2 (2) |
| O2—C4—C5—C6 | 154.54 (16) | F1—C1—C9—C8 | 179.88 (15) |
| C3—C4—C5—C6 | −27.50 (19) | C2—C1—C9—C8 | 0.3 (3) |
| C7—O1—C6—C5 | −52.00 (18) | O1—C6—C10—O3 | 10.0 (2) |
| C7—O1—C6—C10 | 73.59 (16) | C5—C6—C10—O3 | 134.74 (16) |
| C4—C5—C6—O1 | 53.01 (17) | O1—C6—C10—O4 | −170.78 (12) |
| C4—C5—C6—C10 | −70.43 (17) | C5—C6—C10—O4 | −46.00 (17) |
| C6—O1—C7—C8 | −156.25 (14) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O4—H4···O2i | 0.84 | 1.81 | 2.6474 (19) | 171 |
| C5—H5B···Cg1ii | 0.99 | 2.51 | 3.4521 (19) | 160 |
Symmetry codes: (i) x+1/2, −y+1/2, −z; (ii) x−1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: CI2987).
References
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Higashi, T. (1995). ABSCOR Rigaku Corperation, Tokyo, Japan.
- Kurono, M., Kondo, Y., Yamaguchi, T., Miura, K., Usui, T., Terada, N., Asano, K., Mizuno, K., Matsubara, A., Kato, N., Sawai, K., Unno, T., Ozawa, H. & Fukushima, M. (1989). US Patent No. 4861792.
- Mealy, N. (1996). Drugs Future, 21, 261–265.
- Mitsuru, O., Yudiharu, M., Shigeru, S., Oka, M., Matsumoto, Y., Sugiyama, S., Tsuruta, N. & Matsushima, M. (2000). J. Med. Chem.43, 2479–2483. [DOI] [PubMed]
- Rigaku/MSC (2005). CrystalClear Rigaku/MSC, The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Yamaguchi, T., Miura, K., Usui, T., Unno, R., Matsumoto, Y., Fukushima, M., Mizuno, K., Kondo, Y., Baba, Y. & Kurono, M. (1994). Arzneim. Forsch.44, 344–348. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809054555/ci2987sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809054555/ci2987Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



