Abstract
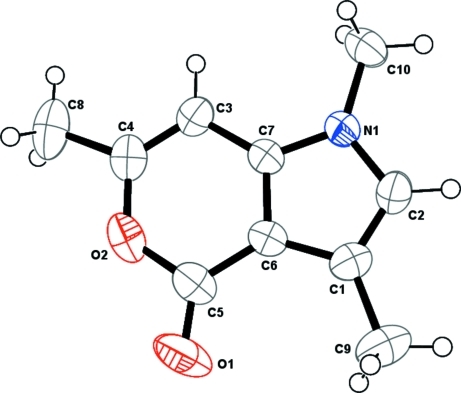
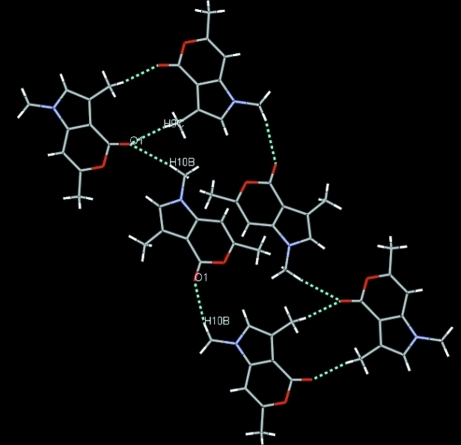
All the non-H atoms of the title compound, C10H11NO2, are almost coplanar [maximum deviation = 0.040 (3) Å]. The crystal structure is stabilized by C—H⋯O hydrogen bonds.
Related literature
For general background to isocoumarins, see: Barry (1964 ▶). For related structures, see: Abid et al. (2006 ▶, 2008 ▶); Hathwar et al. (2007 ▶).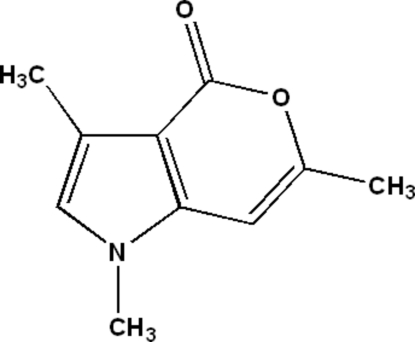
Experimental
Crystal data
C10H11NO2
M r = 177.20
Monoclinic,
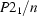
a = 7.5556 (7) Å
b = 8.4819 (8) Å
c = 14.3081 (14) Å
β = 93.870 (6)°
V = 914.86 (15) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 295 K
0.33 × 0.28 × 0.15 mm
Data collection
Oxford Xcalibur Eos (Nova) CCD detector diffractometer
Absorption correction: multi-scan (CrysAlis PRO RED; Oxford Diffraction, 2009 ▶) T min = 0.916, T max = 0.987
7291 measured reflections
1692 independent reflections
1176 reflections with I > 2σ(I)
R int = 0.034
Refinement
R[F 2 > 2σ(F 2)] = 0.051
wR(F 2) = 0.179
S = 1.18
1692 reflections
122 parameters
H-atom parameters constrained
Δρmax = 0.27 e Å−3
Δρmin = −0.18 e Å−3
Data collection: CrysAlis PRO CCD (Oxford Diffraction, 2009 ▶); cell refinement: CrysAlis PRO CCD; data reduction: CrysAlis PRO RED (Oxford Diffraction, 2009 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶) and CAMERON (Watkin et al., 1993 ▶); software used to prepare material for publication: WinGX (Farrugia, 1997 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809055627/bt5154sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809055627/bt5154Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C10—H10B⋯O1i | 0.96 | 2.46 | 3.404 (3) | 170 |
Symmetry code: (i) 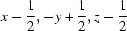 .
.
Acknowledgments
We thank the Department of Science and Technology, India, for use of the CCD facility set up under the IRHPA–DST program at IISc. We thank Professor T. N. Guru Row, IISc, Bangalore, for useful crystallographic discussions. FNK thanks the DST for Fast Track Proposal funding.
supplementary crystallographic information
Comment
Isocoumarins (Barry, 1964) are also useful intermediates in the synthesis of a variety of important compounds including some carbocyclic and heterocyclic compounds. In view of their natural occurrence, biological activities and utility as synthetic intermediates, we have synthesized the title compound, and reported herein its crystal structure.
Experimental
A mixture of 2-(carboxymethyl)-1, 4-dimethyl-1H-pyrrole-3-carboxylic acid (2 mmol) and acetic anhydride (8 mmol) in the presence of pyridine was refluxed for 4 h. Completion of the reaction was monitored by Thin Layer Chromatography. After completing of the reaction, the mixture was poured into crushed ice. The solids were separated and purified by silica gel column chromatography. The product was obtained with 90% yield.
Refinement
All the H atoms were positioned geometrically and refined using a riding model, fixing the bond lengths at 0.96 and 0.93 Å for CH3 aromatic CH, respectively. The displacement parameters of the H atoms were constrained as Uiso(H) = 1.2Ueq (1.5Ueq for methyl) of the carrier atom.
Figures
Fig. 1.
A view of the title complex, showing 50% probability displacement ellipsoids and the atom-numbering scheme.
Fig. 2.
The packing diagram depicting C—H···O intermolecular interactions.
Crystal data
| C10H11NO2 | F(000) = 376 |
| Mr = 177.20 | Dx = 1.287 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 1235 reflections |
| a = 7.5556 (7) Å | θ = 2.9–20.4° |
| b = 8.4819 (8) Å | µ = 0.09 mm−1 |
| c = 14.3081 (14) Å | T = 295 K |
| β = 93.870 (6)° | Block, colorless |
| V = 914.86 (15) Å3 | 0.33 × 0.28 × 0.15 mm |
| Z = 4 |
Data collection
| Oxford Xcalibur Eos (Nova) CCD detector diffractometer | 1692 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 1176 reflections with I > 2σ(I) |
| graphite | Rint = 0.034 |
| ω scans | θmax = 25.5°, θmin = 2.8° |
| Absorption correction: multi-scan (CrysAlis PRO RED; Oxford Diffraction, 2009) | h = −9→9 |
| Tmin = 0.916, Tmax = 0.987 | k = −9→10 |
| 7291 measured reflections | l = −17→17 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.051 | H-atom parameters constrained |
| wR(F2) = 0.179 | w = 1/[σ2(Fo2) + (0.0888P)2 + 0.144P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.18 | (Δ/σ)max < 0.001 |
| 1692 reflections | Δρmax = 0.27 e Å−3 |
| 122 parameters | Δρmin = −0.18 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.004 (1) |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.1293 (2) | 0.4172 (2) | 0.87406 (12) | 0.0412 (5) | |
| O1 | 0.3720 (3) | 0.1280 (3) | 1.12304 (13) | 0.0818 (7) | |
| O2 | 0.3792 (2) | 0.3881 (2) | 1.13262 (10) | 0.0586 (6) | |
| C1 | 0.1778 (3) | 0.1674 (3) | 0.92158 (16) | 0.0466 (6) | |
| C2 | 0.1079 (3) | 0.2591 (3) | 0.85100 (16) | 0.0466 (6) | |
| H2 | 0.0534 | 0.2216 | 0.7951 | 0.056* | |
| C3 | 0.2612 (3) | 0.5620 (3) | 1.01528 (16) | 0.0460 (6) | |
| H3 | 0.2372 | 0.6633 | 0.9929 | 0.055* | |
| C4 | 0.3421 (3) | 0.5382 (3) | 1.09968 (17) | 0.0507 (7) | |
| C5 | 0.3331 (3) | 0.2497 (3) | 1.08310 (16) | 0.0526 (7) | |
| C6 | 0.2454 (3) | 0.2744 (3) | 0.99253 (14) | 0.0420 (6) | |
| C7 | 0.2128 (3) | 0.4269 (2) | 0.96087 (14) | 0.0389 (6) | |
| C8 | 0.4000 (4) | 0.6605 (4) | 1.1693 (2) | 0.0749 (9) | |
| H8A | 0.3742 | 0.7631 | 1.1435 | 0.112* | |
| H8B | 0.5253 | 0.6511 | 1.1842 | 0.112* | |
| H8C | 0.3380 | 0.6465 | 1.2251 | 0.112* | |
| C9 | 0.1882 (4) | −0.0092 (3) | 0.9237 (2) | 0.0702 (9) | |
| H9A | 0.1145 | −0.0518 | 0.8725 | 0.105* | |
| H9B | 0.1480 | −0.0472 | 0.9818 | 0.105* | |
| H9C | 0.3087 | −0.0416 | 0.9181 | 0.105* | |
| C10 | 0.0743 (4) | 0.5500 (3) | 0.81496 (17) | 0.0551 (7) | |
| H10A | 0.0008 | 0.6192 | 0.8487 | 0.083* | |
| H10B | 0.0085 | 0.5123 | 0.7597 | 0.083* | |
| H10C | 0.1771 | 0.6063 | 0.7974 | 0.083* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0466 (11) | 0.0379 (11) | 0.0382 (10) | 0.0025 (8) | −0.0049 (8) | 0.0018 (8) |
| O1 | 0.1038 (17) | 0.0781 (16) | 0.0623 (12) | 0.0317 (12) | −0.0034 (11) | 0.0266 (11) |
| O2 | 0.0562 (11) | 0.0777 (14) | 0.0403 (9) | 0.0065 (9) | −0.0082 (7) | −0.0015 (9) |
| C1 | 0.0506 (13) | 0.0375 (13) | 0.0521 (14) | −0.0005 (11) | 0.0074 (11) | −0.0007 (11) |
| C2 | 0.0488 (13) | 0.0463 (14) | 0.0442 (12) | −0.0021 (11) | −0.0008 (10) | −0.0089 (12) |
| C3 | 0.0474 (13) | 0.0415 (14) | 0.0489 (13) | −0.0035 (10) | 0.0016 (10) | −0.0050 (10) |
| C4 | 0.0443 (13) | 0.0604 (17) | 0.0471 (13) | −0.0021 (12) | 0.0015 (10) | −0.0097 (11) |
| C5 | 0.0541 (15) | 0.0589 (17) | 0.0448 (13) | 0.0102 (13) | 0.0022 (11) | 0.0066 (13) |
| C6 | 0.0432 (12) | 0.0416 (13) | 0.0409 (12) | 0.0030 (10) | 0.0017 (9) | 0.0050 (10) |
| C7 | 0.0392 (12) | 0.0400 (13) | 0.0372 (11) | 0.0023 (10) | 0.0005 (9) | 0.0033 (9) |
| C8 | 0.0654 (18) | 0.096 (2) | 0.0630 (17) | −0.0162 (17) | −0.0004 (13) | −0.0329 (16) |
| C9 | 0.085 (2) | 0.0395 (16) | 0.087 (2) | 0.0000 (15) | 0.0159 (16) | −0.0025 (15) |
| C10 | 0.0634 (16) | 0.0556 (17) | 0.0450 (13) | 0.0061 (13) | −0.0055 (11) | 0.0116 (11) |
Geometric parameters (Å, °)
| N1—C7 | 1.357 (3) | C4—C8 | 1.483 (4) |
| N1—C2 | 1.388 (3) | C5—C6 | 1.431 (3) |
| N1—C10 | 1.452 (3) | C6—C7 | 1.387 (3) |
| O1—C5 | 1.206 (3) | C8—H8A | 0.9600 |
| O2—C4 | 1.380 (3) | C8—H8B | 0.9600 |
| O2—C5 | 1.403 (3) | C8—H8C | 0.9600 |
| C1—C2 | 1.353 (3) | C9—H9A | 0.9600 |
| C1—C6 | 1.430 (3) | C9—H9B | 0.9600 |
| C1—C9 | 1.500 (4) | C9—H9C | 0.9600 |
| C2—H2 | 0.9300 | C10—H10A | 0.9600 |
| C3—C4 | 1.332 (3) | C10—H10B | 0.9600 |
| C3—C7 | 1.419 (3) | C10—H10C | 0.9600 |
| C3—H3 | 0.9300 | ||
| C7—N1—C2 | 108.41 (19) | N1—C7—C6 | 107.66 (19) |
| C7—N1—C10 | 125.64 (19) | N1—C7—C3 | 129.6 (2) |
| C2—N1—C10 | 125.94 (19) | C6—C7—C3 | 122.7 (2) |
| C4—O2—C5 | 124.21 (19) | C4—C8—H8A | 109.5 |
| C2—C1—C6 | 105.5 (2) | C4—C8—H8B | 109.5 |
| C2—C1—C9 | 127.3 (2) | H8A—C8—H8B | 109.5 |
| C6—C1—C9 | 127.2 (2) | C4—C8—H8C | 109.5 |
| C1—C2—N1 | 110.2 (2) | H8A—C8—H8C | 109.5 |
| C1—C2—H2 | 124.9 | H8B—C8—H8C | 109.5 |
| N1—C2—H2 | 124.9 | C1—C9—H9A | 109.5 |
| C4—C3—C7 | 117.4 (2) | C1—C9—H9B | 109.5 |
| C4—C3—H3 | 121.3 | H9A—C9—H9B | 109.5 |
| C7—C3—H3 | 121.3 | C1—C9—H9C | 109.5 |
| C3—C4—O2 | 121.3 (2) | H9A—C9—H9C | 109.5 |
| C3—C4—C8 | 126.8 (3) | H9B—C9—H9C | 109.5 |
| O2—C4—C8 | 111.9 (2) | N1—C10—H10A | 109.5 |
| O1—C5—O2 | 115.6 (2) | N1—C10—H10B | 109.5 |
| O1—C5—C6 | 129.6 (3) | H10A—C10—H10B | 109.5 |
| O2—C5—C6 | 114.8 (2) | N1—C10—H10C | 109.5 |
| C7—C6—C1 | 108.3 (2) | H10A—C10—H10C | 109.5 |
| C7—C6—C5 | 119.6 (2) | H10B—C10—H10C | 109.5 |
| C1—C6—C5 | 132.2 (2) | ||
| C6—C1—C2—N1 | 0.3 (2) | O1—C5—C6—C7 | 179.4 (2) |
| C9—C1—C2—N1 | −177.7 (2) | O2—C5—C6—C7 | 0.0 (3) |
| C7—N1—C2—C1 | −0.5 (2) | O1—C5—C6—C1 | −0.4 (4) |
| C10—N1—C2—C1 | 178.7 (2) | O2—C5—C6—C1 | −179.8 (2) |
| C7—C3—C4—O2 | 0.6 (3) | C2—N1—C7—C6 | 0.4 (2) |
| C7—C3—C4—C8 | −178.2 (2) | C10—N1—C7—C6 | −178.74 (19) |
| C5—O2—C4—C3 | −1.5 (3) | C2—N1—C7—C3 | −178.8 (2) |
| C5—O2—C4—C8 | 177.5 (2) | C10—N1—C7—C3 | 2.1 (4) |
| C4—O2—C5—O1 | −178.4 (2) | C1—C6—C7—N1 | −0.2 (2) |
| C4—O2—C5—C6 | 1.1 (3) | C5—C6—C7—N1 | 179.96 (18) |
| C2—C1—C6—C7 | −0.1 (2) | C1—C6—C7—C3 | 179.06 (19) |
| C9—C1—C6—C7 | 177.9 (2) | C5—C6—C7—C3 | −0.8 (3) |
| C2—C1—C6—C5 | 179.7 (2) | C4—C3—C7—N1 | 179.6 (2) |
| C9—C1—C6—C5 | −2.2 (4) | C4—C3—C7—C6 | 0.5 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C10—H10B···O1i | 0.96 | 2.46 | 3.404 (3) | 170 |
Symmetry codes: (i) x−1/2, −y+1/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT5154).
References
- Abid, O.-U.-R., Qadeer, G., Rama, N. H., Ruzicka, A. & Padelkova, Z. (2008). Acta Cryst. E64, o2018. [DOI] [PMC free article] [PubMed]
- Abid, O., Rama, N. H., Qadeer, G., Khan, G. S. & Lu, X.-M. (2006). Acta Cryst. E62, o2895–o2896.
- Barry, R. D. (1964). Chem. Rev.64, 229–260.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Hathwar, V. R., Manivel, P., Nawaz Khan, F. & Guru Row, T. N. (2007). Acta Cryst. E63, o3707.
- Oxford Diffraction (2009). CrysAlis PRO CCD and CrysAlis PRO RED Oxford Diffraction Ltd, Yarnton, England.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Watkin, D. J., Pearce, L. & Prout, C. K. (1993). CAMERON Chemical Crystallography Laboratory, University of Oxford, England.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809055627/bt5154sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809055627/bt5154Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report