Abstract
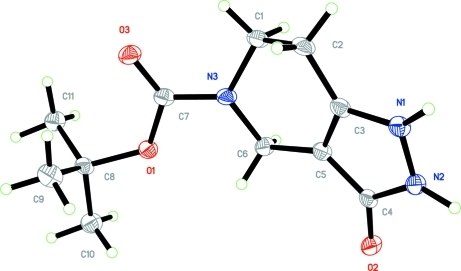
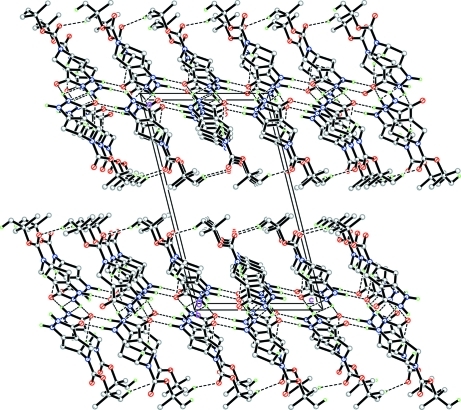
In the title compound, C11H17N3O3, the pyrazole ring is approximately planar, with a maximum deviation of 0.005 (2) Å, and forms a dihedral angle of 5.69 (13)° with the plane through the six atoms of the piperidine ring. In the crystal, pairs of intermolecular N—H⋯O hydrogen bonds form dimers with neighbouring molecules, generating R 2 2(8) ring motifs. These dimers are further linked into two-dimensional arrays parallel to the bc plane by intermolecular N—H⋯O and C—H⋯O hydrogen bonds.
Related literature
For the biological activity of pyrazolone derivatives, see: Al-Haiza et al. (2001 ▶); Brogden, (1986 ▶); Coersmeier et al. (1986 ▶); Gursoy et al. (2000 ▶). For myocardial ischemia, see: Wu et al. (2002 ▶). For brain ischemia, see: Watanabe et al. (1984 ▶); Kawai et al. (1997 ▶). For new compounds with the pyrazolone unit, see: Al-Haiza et al. (2001 ▶). For a related structure, see: Shahani et al. (2009 ▶). For ring conformations, see: Cremer & Pople (1975 ▶). For hydrogen-bond motifs, see: Bernstein et al. (1995 ▶). For bond-length data, see: Allen et al. (1987 ▶). For the stability of the temperature controller used for the data collection, see: Cosier & Glazer (1986 ▶).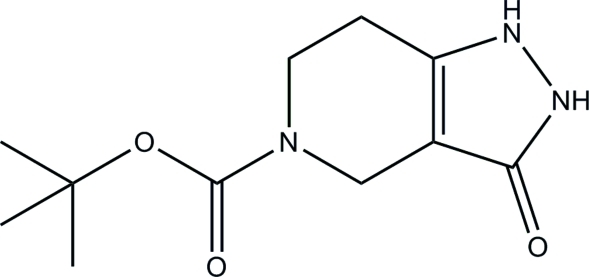
Experimental
Crystal data
C11H17N3O3
M r = 239.28
Monoclinic,
a = 18.6250 (12) Å
b = 6.0893 (5) Å
c = 10.7414 (7) Å
β = 104.100 (4)°
V = 1181.51 (15) Å3
Z = 4
Mo Kα radiation
μ = 0.10 mm−1
T = 100 K
0.97 × 0.35 × 0.14 mm
Data collection
Bruker SMART APEXII CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2009 ▶) T min = 0.910, T max = 0.986
13101 measured reflections
2690 independent reflections
2136 reflections with I > 2σ(I)
R int = 0.035
Refinement
R[F 2 > 2σ(F 2)] = 0.054
wR(F 2) = 0.172
S = 1.24
2690 reflections
222 parameters
All H-atom parameters refined
Δρmax = 0.39 e Å−3
Δρmin = −0.31 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809053021/ng2705sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809053021/ng2705Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N1⋯O2i | 0.99 (3) | 1.77 (3) | 2.748 (3) | 171 (3) |
| N2—H1N2⋯O2ii | 0.94 (3) | 1.75 (4) | 2.665 (3) | 167 (3) |
| C1—H1B⋯O2iii | 0.98 (3) | 2.57 (3) | 3.492 (3) | 157 (3) |
| C11—H11C⋯O3iv | 0.97 (3) | 2.60 (3) | 3.504 (3) | 156 (3) |
Symmetry codes: (i) 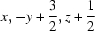 ; (ii)
; (ii) 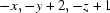 ; (iii)
; (iii) 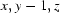 ; (iv)
; (iv) 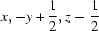 .
.
Acknowledgments
HKF and TSH thank Universiti Sains Malaysia (USM) for the Research University Golden Goose Grant (1001/PFIZIK/811012). VV is grateful to DST-India for funding through the Young Scientist Scheme (Fast Track Proposal).
supplementary crystallographic information
Comment
Pyrazolone derivatives have a broad spectrum of biological activities being used as analgesic, antipyretic and anti-inflammatory therapeutical drugs (Brogden, 1986; Gursoy et al., 2000). A class of new compounds with pyrazolone moiety was synthesized and reported for their antibacterial, antifungal activities by Al-Haiza et al. (2001). A new pyrazolone derivative, edaravone (3-methyl-1-phenyl-2-pyrazoline-5-one) is being used as a drug in clinical practice for brain ischemia (Watanabe et al., 1984; Kawai et al., 1997) and the same has been found to be effective against myocardial ischemia (Wu et al., 2002).
In the crystal structure (Fig. 1), the pyrazole ring (C3/N1/N2/C4/C5) is approximately planar, with a maximum deviation of 0.005 (2) Å at atom N2. The piperidine ring (C1/C2/C3/C5/C6/N3) adopts a half-boat conformation (Cremer & Pople, 1975) with puckering of Q = 0.465 (2) Å, Θ = 52.9 (2)° & φ = 39.8 (4)°. The maximum deviation in this piperidine ring is 0.286 Å at atom N3. The dihedral angle formed between the mean planes of pyrazole and piperidine rings is 5.69 (13)°. The bond lengths (Allen et al., 1987) and angles are within normal ranges and comparable to a closely related structure (Shahani et al., 2009).
In the crystal packing (Fig. 2), pairs of intermolecular N2—H1N2···O2 hydrogen bonds form dimers with neighbouring molecules, generating R22(8) ring motifs (Bernstein et al., 1995). These dimers are further linked into two-dimensional arrays parallel to the bc plane by intermolecular N1—H1N1···O2, C1—H1B···O2 and C11—H11C···O3 hydrogen bonds.
Experimental
LiHMDS (1.0 M solution in toluene, 11 mmol) was added quickly to a solution of tert-butyl 4-oxopiperidine-1-carboxylate (10 mmol 15 ml of toluene) using syringe at 273 K with stirring for 10 minutes. Ethyl chloro formate (11 mmol) was then added quickly. The reaction mixture was slowly (10 minutes) brought to room temperature and stirred for 10 minutes. Acetic acid (2 ml), ethanol (15 ml), and hydrazine hydrate (30 mmol) were added and refluxed for 15 minutes. The reaction mixture was concentrated to dryness under reduced pressure and re-dissolved in ethyl acetate. The organic layer was washed with saturated brine solution, dried over Na2SO4, evaporated under reduced pressure and purified by crystallizing using ethanol (white solid). The recrystallization was done using 1:1 mixture of ethanol and acetone. Yield: 78%. M.p. 498.5–500.5 K. MS calculated for C11H17N3O3: 239.126. Found: 239.80 (M+).
Refinement
All hydrogen atoms were located in a difference map and were refined freely [range of N—H = 0.94 and 0.99 Å; and C—H = 0.95–1.02 Å].
Figures
Fig. 1.
The molecular structure of the title compound, showing 50% probability displacement ellipsoids and the atom numbering scheme.
Fig. 2.
The crystal packing of the title compound, viewed along b axis, showing two-dimensional arrays parallel to the bc plane.
Crystal data
| C11H17N3O3 | F(000) = 512 |
| Mr = 239.28 | Dx = 1.345 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 7287 reflections |
| a = 18.6250 (12) Å | θ = 2.3–33.5° |
| b = 6.0893 (5) Å | µ = 0.10 mm−1 |
| c = 10.7414 (7) Å | T = 100 K |
| β = 104.100 (4)° | Plate, colourless |
| V = 1181.51 (15) Å3 | 0.97 × 0.35 × 0.14 mm |
| Z = 4 |
Data collection
| Bruker SMART APEXII CCD area-detector diffractometer | 2690 independent reflections |
| Radiation source: fine-focus sealed tube | 2136 reflections with I > 2σ(I) |
| graphite | Rint = 0.035 |
| φ and ω scans | θmax = 27.5°, θmin = 1.1° |
| Absorption correction: multi-scan (SADABS; Bruker, 2009) | h = −24→24 |
| Tmin = 0.910, Tmax = 0.986 | k = −7→7 |
| 13101 measured reflections | l = −13→13 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.054 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.172 | All H-atom parameters refined |
| S = 1.24 | w = 1/[σ2(Fo2) + (0.0688P)2 + 1.2823P] where P = (Fo2 + 2Fc2)/3 |
| 2690 reflections | (Δ/σ)max < 0.001 |
| 222 parameters | Δρmax = 0.39 e Å−3 |
| 0 restraints | Δρmin = −0.31 e Å−3 |
Special details
| Experimental. The crystal was placed in the cold stream of an Oxford Cyrosystems Cobra open-flow nitrogen cryostat (Cosier & Glazer, 1986) operating at 100.0 (1) K. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.30793 (9) | 0.5995 (3) | 0.33483 (16) | 0.0205 (4) | |
| O2 | 0.04609 (9) | 0.9437 (3) | 0.36561 (15) | 0.0205 (4) | |
| O3 | 0.34752 (9) | 0.2971 (3) | 0.45801 (16) | 0.0231 (4) | |
| N1 | 0.07659 (11) | 0.5955 (4) | 0.62882 (18) | 0.0191 (4) | |
| N2 | 0.04170 (11) | 0.7682 (4) | 0.55702 (19) | 0.0186 (4) | |
| N3 | 0.23023 (11) | 0.4313 (3) | 0.43120 (19) | 0.0191 (4) | |
| C1 | 0.20586 (14) | 0.2460 (4) | 0.4972 (2) | 0.0215 (5) | |
| C2 | 0.17706 (14) | 0.3233 (4) | 0.6127 (2) | 0.0225 (5) | |
| C3 | 0.12708 (12) | 0.5145 (4) | 0.5701 (2) | 0.0185 (5) | |
| C4 | 0.06920 (12) | 0.7945 (4) | 0.4514 (2) | 0.0167 (5) | |
| C5 | 0.12428 (12) | 0.6319 (4) | 0.4598 (2) | 0.0172 (5) | |
| C6 | 0.17364 (13) | 0.5912 (4) | 0.3715 (2) | 0.0190 (5) | |
| C7 | 0.30029 (12) | 0.4308 (4) | 0.4123 (2) | 0.0180 (5) | |
| C8 | 0.38126 (12) | 0.6603 (4) | 0.3140 (2) | 0.0186 (5) | |
| C9 | 0.43236 (15) | 0.7313 (5) | 0.4403 (3) | 0.0258 (6) | |
| C10 | 0.36107 (14) | 0.8534 (5) | 0.2231 (3) | 0.0249 (6) | |
| C11 | 0.41358 (14) | 0.4723 (5) | 0.2519 (2) | 0.0215 (5) | |
| H1A | 0.2501 (16) | 0.144 (5) | 0.527 (3) | 0.027 (7)* | |
| H1B | 0.1645 (16) | 0.173 (5) | 0.437 (3) | 0.025 (7)* | |
| H2A | 0.1514 (16) | 0.203 (6) | 0.639 (3) | 0.032 (8)* | |
| H2B | 0.2187 (16) | 0.364 (5) | 0.682 (3) | 0.026 (7)* | |
| H6A | 0.1980 (15) | 0.727 (5) | 0.353 (3) | 0.023 (7)* | |
| H6B | 0.1445 (14) | 0.539 (5) | 0.286 (3) | 0.021 (7)* | |
| H9A | 0.4124 (16) | 0.854 (5) | 0.476 (3) | 0.026 (8)* | |
| H9B | 0.4808 (16) | 0.779 (5) | 0.428 (3) | 0.026 (7)* | |
| H9C | 0.4426 (16) | 0.611 (6) | 0.504 (3) | 0.035 (9)* | |
| H10A | 0.4080 (17) | 0.909 (5) | 0.201 (3) | 0.035 (8)* | |
| H10B | 0.3254 (17) | 0.809 (5) | 0.139 (3) | 0.033 (8)* | |
| H10C | 0.3394 (19) | 0.970 (6) | 0.264 (3) | 0.049 (10)* | |
| H11A | 0.4596 (16) | 0.530 (5) | 0.227 (3) | 0.031 (8)* | |
| H11B | 0.4299 (15) | 0.344 (5) | 0.313 (3) | 0.027 (7)* | |
| H11C | 0.3819 (18) | 0.421 (6) | 0.172 (3) | 0.042 (9)* | |
| H1N1 | 0.0676 (18) | 0.566 (6) | 0.714 (3) | 0.043 (9)* | |
| H1N2 | 0.0081 (18) | 0.852 (6) | 0.589 (3) | 0.046 (10)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0230 (8) | 0.0192 (9) | 0.0230 (8) | 0.0028 (7) | 0.0124 (6) | 0.0066 (7) |
| O2 | 0.0288 (8) | 0.0201 (9) | 0.0164 (8) | 0.0054 (7) | 0.0128 (6) | 0.0019 (7) |
| O3 | 0.0267 (8) | 0.0206 (10) | 0.0245 (9) | 0.0053 (7) | 0.0109 (7) | 0.0061 (8) |
| N1 | 0.0267 (10) | 0.0186 (11) | 0.0152 (9) | −0.0001 (8) | 0.0111 (7) | 0.0008 (8) |
| N2 | 0.0246 (9) | 0.0184 (11) | 0.0160 (9) | 0.0018 (8) | 0.0110 (7) | 0.0002 (8) |
| N3 | 0.0250 (10) | 0.0148 (11) | 0.0210 (10) | 0.0028 (8) | 0.0121 (8) | 0.0043 (9) |
| C1 | 0.0285 (12) | 0.0144 (12) | 0.0253 (12) | 0.0000 (10) | 0.0138 (10) | 0.0016 (11) |
| C2 | 0.0299 (12) | 0.0177 (13) | 0.0237 (12) | 0.0001 (10) | 0.0134 (10) | 0.0069 (11) |
| C3 | 0.0235 (11) | 0.0153 (12) | 0.0192 (11) | −0.0036 (9) | 0.0103 (9) | −0.0027 (10) |
| C4 | 0.0212 (10) | 0.0163 (12) | 0.0153 (10) | −0.0027 (9) | 0.0099 (8) | −0.0018 (9) |
| C5 | 0.0224 (10) | 0.0151 (12) | 0.0164 (10) | −0.0015 (9) | 0.0094 (8) | −0.0019 (9) |
| C6 | 0.0259 (11) | 0.0182 (13) | 0.0166 (11) | 0.0041 (10) | 0.0121 (9) | 0.0014 (10) |
| C7 | 0.0244 (11) | 0.0150 (12) | 0.0167 (10) | 0.0001 (10) | 0.0089 (8) | −0.0012 (10) |
| C8 | 0.0222 (11) | 0.0162 (12) | 0.0213 (11) | −0.0014 (9) | 0.0127 (9) | 0.0010 (10) |
| C9 | 0.0327 (13) | 0.0248 (15) | 0.0224 (13) | −0.0028 (11) | 0.0115 (10) | −0.0041 (12) |
| C10 | 0.0298 (12) | 0.0237 (14) | 0.0250 (13) | 0.0024 (11) | 0.0140 (10) | 0.0053 (11) |
| C11 | 0.0265 (12) | 0.0208 (13) | 0.0198 (12) | 0.0023 (10) | 0.0106 (9) | −0.0032 (11) |
Geometric parameters (Å, °)
| O1—C7 | 1.351 (3) | C3—C5 | 1.374 (3) |
| O1—C8 | 1.484 (3) | C4—C5 | 1.413 (3) |
| O2—C4 | 1.290 (3) | C5—C6 | 1.494 (3) |
| O3—C7 | 1.212 (3) | C6—H6A | 0.98 (3) |
| N1—C3 | 1.347 (3) | C6—H6B | 1.00 (3) |
| N1—N2 | 1.371 (3) | C8—C10 | 1.516 (4) |
| N1—H1N1 | 0.99 (3) | C8—C9 | 1.518 (4) |
| N2—C4 | 1.364 (3) | C8—C11 | 1.521 (3) |
| N2—H1N2 | 0.94 (4) | C9—H9A | 0.96 (3) |
| N3—C7 | 1.369 (3) | C9—H9B | 0.99 (3) |
| N3—C1 | 1.462 (3) | C9—H9C | 0.99 (3) |
| N3—C6 | 1.463 (3) | C10—H10A | 1.02 (3) |
| C1—C2 | 1.540 (3) | C10—H10B | 1.02 (3) |
| C1—H1A | 1.02 (3) | C10—H10C | 0.97 (4) |
| C1—H1B | 0.98 (3) | C11—H11A | 1.02 (3) |
| C2—C3 | 1.491 (4) | C11—H11B | 1.02 (3) |
| C2—H2A | 0.95 (3) | C11—H11C | 0.97 (3) |
| C2—H2B | 0.97 (3) | ||
| C7—O1—C8 | 121.42 (18) | N3—C6—H6A | 109.1 (16) |
| C3—N1—N2 | 107.86 (19) | C5—C6—H6A | 111.8 (17) |
| C3—N1—H1N1 | 132 (2) | N3—C6—H6B | 111.4 (16) |
| N2—N1—H1N1 | 120 (2) | C5—C6—H6B | 110.9 (15) |
| C4—N2—N1 | 109.62 (19) | H6A—C6—H6B | 105 (2) |
| C4—N2—H1N2 | 132 (2) | O3—C7—O1 | 125.9 (2) |
| N1—N2—H1N2 | 118 (2) | O3—C7—N3 | 124.4 (2) |
| C7—N3—C1 | 119.4 (2) | O1—C7—N3 | 109.70 (19) |
| C7—N3—C6 | 123.2 (2) | O1—C8—C10 | 101.39 (18) |
| C1—N3—C6 | 116.77 (19) | O1—C8—C9 | 109.63 (18) |
| N3—C1—C2 | 111.4 (2) | C10—C8—C9 | 111.0 (2) |
| N3—C1—H1A | 107.4 (17) | O1—C8—C11 | 110.8 (2) |
| C2—C1—H1A | 110.2 (16) | C10—C8—C11 | 111.3 (2) |
| N3—C1—H1B | 108.7 (17) | C9—C8—C11 | 112.2 (2) |
| C2—C1—H1B | 107.2 (16) | C8—C9—H9A | 111.1 (17) |
| H1A—C1—H1B | 112 (2) | C8—C9—H9B | 110.9 (16) |
| C3—C2—C1 | 107.8 (2) | H9A—C9—H9B | 106 (2) |
| C3—C2—H2A | 111.7 (19) | C8—C9—H9C | 112.3 (19) |
| C1—C2—H2A | 107.4 (19) | H9A—C9—H9C | 110 (2) |
| C3—C2—H2B | 111.2 (19) | H9B—C9—H9C | 106 (2) |
| C1—C2—H2B | 108.9 (17) | C8—C10—H10A | 108.3 (18) |
| H2A—C2—H2B | 110 (3) | C8—C10—H10B | 111.7 (19) |
| N1—C3—C5 | 109.2 (2) | H10A—C10—H10B | 107 (2) |
| N1—C3—C2 | 126.6 (2) | C8—C10—H10C | 110 (2) |
| C5—C3—C2 | 124.1 (2) | H10A—C10—H10C | 110 (3) |
| O2—C4—N2 | 123.3 (2) | H10B—C10—H10C | 110 (3) |
| O2—C4—C5 | 130.5 (2) | C8—C11—H11A | 107.7 (18) |
| N2—C4—C5 | 106.2 (2) | C8—C11—H11B | 112.6 (17) |
| C3—C5—C4 | 107.13 (19) | H11A—C11—H11B | 107 (2) |
| C3—C5—C6 | 124.1 (2) | C8—C11—H11C | 114 (2) |
| C4—C5—C6 | 128.7 (2) | H11A—C11—H11C | 105 (3) |
| N3—C6—C5 | 108.74 (19) | H11B—C11—H11C | 110 (3) |
| C3—N1—N2—C4 | 1.0 (3) | O2—C4—C5—C6 | 2.0 (4) |
| C7—N3—C1—C2 | 125.7 (2) | N2—C4—C5—C6 | −177.9 (2) |
| C6—N3—C1—C2 | −63.6 (3) | C7—N3—C6—C5 | −148.5 (2) |
| N3—C1—C2—C3 | 45.8 (3) | C1—N3—C6—C5 | 41.2 (3) |
| N2—N1—C3—C5 | −0.9 (3) | C3—C5—C6—N3 | −7.8 (3) |
| N2—N1—C3—C2 | −178.8 (2) | C4—C5—C6—N3 | 170.1 (2) |
| C1—C2—C3—N1 | 162.0 (2) | C8—O1—C7—O3 | −10.1 (4) |
| C1—C2—C3—C5 | −15.7 (3) | C8—O1—C7—N3 | 170.38 (19) |
| N1—N2—C4—O2 | 179.3 (2) | C1—N3—C7—O3 | −8.0 (4) |
| N1—N2—C4—C5 | −0.8 (3) | C6—N3—C7—O3 | −178.1 (2) |
| N1—C3—C5—C4 | 0.4 (3) | C1—N3—C7—O1 | 171.5 (2) |
| C2—C3—C5—C4 | 178.4 (2) | C6—N3—C7—O1 | 1.4 (3) |
| N1—C3—C5—C6 | 178.6 (2) | C7—O1—C8—C10 | −179.8 (2) |
| C2—C3—C5—C6 | −3.4 (4) | C7—O1—C8—C9 | −62.4 (3) |
| O2—C4—C5—C3 | −179.8 (2) | C7—O1—C8—C11 | 61.9 (3) |
| N2—C4—C5—C3 | 0.3 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N1···O2i | 0.99 (3) | 1.77 (3) | 2.748 (3) | 171 (3) |
| N2—H1N2···O2ii | 0.94 (3) | 1.75 (4) | 2.665 (3) | 167 (3) |
| C1—H1B···O2iii | 0.98 (3) | 2.57 (3) | 3.492 (3) | 157 (3) |
| C11—H11C···O3iv | 0.97 (3) | 2.60 (3) | 3.504 (3) | 156 (3) |
Symmetry codes: (i) x, −y+3/2, z+1/2; (ii) −x, −y+2, −z+1; (iii) x, y−1, z; (iv) x, −y+1/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: NG2705).
References
- Al-Haiza, M. A., El-Assiery, S. A. & Sayed, G. H. (2001). Acta Pharm.51, 251–261.
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl.34, 1555–1573.
- Brogden, R. N. (1986). Drugs, 32, 60–70. [DOI] [PubMed]
- Bruker (2009). APEX2, SAINT and SADABS. Bruker AXS Inc., Madison, Wiscosin, USA.
- Coersmeier, C., Wittenberg, H. R., Aehringhaus, U., Dreyling, K. W., Peskar, B. M., Brune, K. & Pesker, B. A. (1986). Agents Actions Suppl.19, 137–153. [PubMed]
- Cosier, J. & Glazer, A. M. (1986). J. Appl. Cryst.19, 105–107.
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc.97, 1354–1358.
- Gursoy, A., Demirayak, S., Capan, G., Erol, K. & Vural, K. (2000). Eur. J. Med. Chem.35, 359–364. [DOI] [PubMed]
- Kawai, H., Nakai, H., Suga, M., Yuki, S., Watanabe, T. & Saito, K. I. (1997). J. Pharmcol. Exp. Ther.281, 921–927. [PubMed]
- Shahani, T., Fun, H.-K., Ragavan, R. V., Vijayakumar, V. & Sarveswari, S. (2009). Acta Cryst. E65, o3249–o3250. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Watanabe, T., Yuki, S., Egawa, M. & Nishi, H. (1984). J. Pharmacol. Exp. Ther.268, 1597–1604. [PubMed]
- Wu, T. W., Zeng, L. H., Wu, J. & Fung, K. P. (2002). Life Sci.71, 2249–2255. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809053021/ng2705sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809053021/ng2705Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report