Abstract
The title compound, C11H11N3O3, crystallizes with two independent molecules of similar geometry in the asymmetric unit. The molecular conformations are stabilized by intramolecular C—H⋯O hydrogen bonds. The crystal packing consists of wave-like layers parallel to the bc plane formed by intermolecular C—H⋯O hydrogen-bonding interactions involving only one independent molecule.
Related literature
For related structures, see: Barlos et al. (1985 ▶); Singh et al. (1988 ▶). For details of the biological activity of benzentriazol-containing compounds, see: Zhang et al. (2002 ▶). For comparative bond lengths, see: Allen et al. (1987 ▶).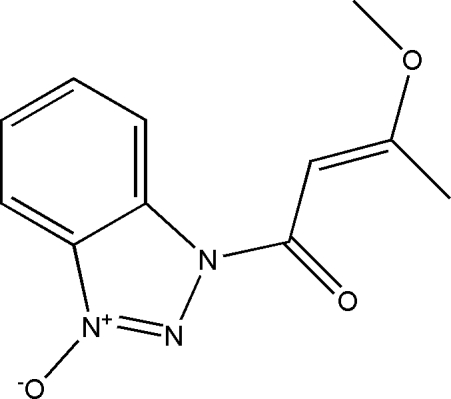
Experimental
Crystal data
C11H11N3O3
M r = 233.23
Monoclinic,
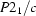
a = 14.011 (3) Å
b = 10.014 (2) Å
c = 15.699 (3) Å
β = 100.13 (3)°
V = 2168.3 (8) Å3
Z = 8
Mo Kα radiation
μ = 0.11 mm−1
T = 173 K
0.40 × 0.40 × 0.30 mm
Data collection
Rigaku Mercury CCD/AFC diffractometer
Absorption correction: multi-scan (CrystalClear; Rigaku, 2007 ▶) T min = 0.959, T max = 0.969
15367 measured reflections
3771 independent reflections
3564 reflections with I > 2σ(I)
R int = 0.050
Refinement
R[F 2 > 2σ(F 2)] = 0.063
wR(F 2) = 0.141
S = 1.17
3771 reflections
308 parameters
H-atom parameters constrained
Δρmax = 0.36 e Å−3
Δρmin = −0.28 e Å−3
Data collection: CrystalClear (Rigaku, 2007 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809050442/rz2397sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809050442/rz2397Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C2—H2B⋯O2 | 0.95 | 2.45 | 2.925 (3) | 111 |
| C13—H13A⋯O5 | 0.95 | 2.49 | 2.961 (3) | 111 |
| C14—H14A⋯O4i | 0.95 | 2.57 | 3.398 (3) | 147 |
| C16—H16A⋯O4ii | 0.95 | 2.45 | 3.379 (3) | 165 |
Symmetry codes: (i) 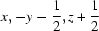 ; (ii)
; (ii)  .
.
supplementary crystallographic information
Comment
Benzotriazole derivatives exhibit good pharmacological activities and a wide spectrum of biological activities (Zhang et al., 2002). In order to search for new benzotriazole compounds with higher bioactivity, we synthesized the title compound and describe its structure here.
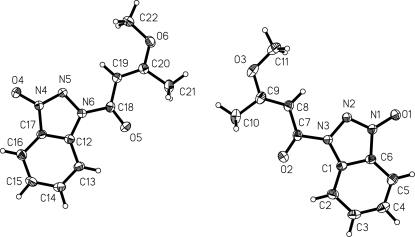
The asymmetric unit of the title compound (Fig. 1) contains two independent molecules of similar geometry. The molecules are almost planar, the maximum deviation being 0.110 (2) Å for atom O2 in one molecule and 0.093 (3) Å for atom C22 in the other molecule. All bond lengths in the molecules are normal (Allen et al., 1987) and in a good agreement with those reported previously for related compound (Barlos et al., 1985; Singh et al., 1988). The molecular conformations are stabilized by intramolecular C—H···O hydrogen bonds (Table 1). In the crystal packing, molecules containing the N4–N6 nitrogen atoms are linked by intermolecular C—H···O hydrogen bonds to form wavy layers parallel to the bc plane intersecting each other.
Experimental
3-Methoxycrotonic acid (20 mmol) was dissolved in dichloromethane and cooled to 273 K, then 1-hydroxybenzotriazole (30 mmol) was added in one portion. After 10 h stirring at room temperature, the solution was washed successively with 1 N HCl and saturated NaCl, and the organic layer was separated, dried with Na2SO4 and evaporated to obtain the primary product. The pure compound was isolated by column chromatography (3.4 g, yield 73%). Single crystals suitable for X-ray measurements were obtained by slow evaporation of an ethyl acetate solution at room temperature. 1H NMR (400 MHz, CDCl3) δ: 8.49 (d, J = 8 Hz, 1H), 7.99 (d, J = 8 Hz, 1H), 7.75 (t, 1H), 7.53 (t, 1H), 6.30 (s, 1H), 3.85 (s, 3H), 2.50 (s, 3H).
Refinement
H atoms were positioned geometrically and refined using a riding model, with C—H = 0.93–0.96 Å and with Uiso(H) = 1.2 Ueq(C) or 1.5 Ueq(C) for methyl H atoms.
Figures
Fig. 1.
The molecular structure of the title compound, with atom-labelling scheme and 40% probability displacement ellipsoids.
Crystal data
| C11H11N3O3 | F(000) = 976 |
| Mr = 233.23 | Dx = 1.429 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 6607 reflections |
| a = 14.011 (3) Å | θ = 1.3–27.5° |
| b = 10.014 (2) Å | µ = 0.11 mm−1 |
| c = 15.699 (3) Å | T = 173 K |
| β = 100.13 (3)° | Block, colourless |
| V = 2168.3 (8) Å3 | 0.40 × 0.40 × 0.30 mm |
| Z = 8 |
Data collection
| Rigaku Mercury CCD/AFC diffractometer | 3771 independent reflections |
| Radiation source: Sealed Tube | 3564 reflections with I > 2σ(I) |
| Graphite Monochromator | Rint = 0.050 |
| φ and ω scans | θmax = 25.0°, θmin = 1.5° |
| Absorption correction: multi-scan (CrystalClear; Rigaku, 2007) | h = −11→16 |
| Tmin = 0.959, Tmax = 0.969 | k = −11→11 |
| 15367 measured reflections | l = −18→18 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.063 | H-atom parameters constrained |
| wR(F2) = 0.141 | w = 1/[σ2(Fo2) + (0.047P)2 + 1.4328P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.17 | (Δ/σ)max < 0.001 |
| 3771 reflections | Δρmax = 0.36 e Å−3 |
| 308 parameters | Δρmin = −0.28 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0028 (7) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.05575 (14) | 1.36306 (16) | −0.08605 (11) | 0.0400 (5) | |
| O2 | 0.14314 (14) | 0.85371 (17) | 0.05299 (11) | 0.0411 (5) | |
| O3 | 0.18500 (13) | 0.72121 (18) | −0.19749 (12) | 0.0436 (5) | |
| O4 | 0.45305 (13) | −0.36283 (16) | −0.08904 (11) | 0.0387 (4) | |
| O5 | 0.33528 (13) | 0.14842 (17) | −0.00327 (10) | 0.0383 (4) | |
| O6 | 0.32908 (13) | 0.26568 (17) | −0.26560 (11) | 0.0380 (4) | |
| N1 | 0.07616 (14) | 1.25933 (19) | −0.03951 (12) | 0.0297 (5) | |
| N2 | 0.09586 (14) | 1.14406 (18) | −0.07177 (12) | 0.0292 (4) | |
| N3 | 0.11216 (14) | 1.05828 (19) | −0.00260 (11) | 0.0272 (4) | |
| N4 | 0.43065 (14) | −0.25590 (19) | −0.05301 (12) | 0.0282 (4) | |
| N5 | 0.40993 (14) | −0.14465 (19) | −0.09592 (12) | 0.0286 (4) | |
| N6 | 0.38814 (13) | −0.05422 (19) | −0.03598 (11) | 0.0264 (4) | |
| C1 | 0.10201 (16) | 1.1226 (2) | 0.07363 (14) | 0.0278 (5) | |
| C2 | 0.10832 (17) | 1.0824 (3) | 0.15936 (15) | 0.0341 (6) | |
| H2B | 0.1237 | 0.9932 | 0.1773 | 0.041* | |
| C3 | 0.09079 (18) | 1.1801 (3) | 0.21672 (15) | 0.0375 (6) | |
| H3A | 0.0940 | 1.1566 | 0.2758 | 0.045* | |
| C4 | 0.06852 (18) | 1.3119 (3) | 0.19175 (16) | 0.0377 (6) | |
| H4A | 0.0574 | 1.3748 | 0.2343 | 0.045* | |
| C5 | 0.06218 (17) | 1.3534 (2) | 0.10725 (16) | 0.0337 (6) | |
| H5B | 0.0473 | 1.4428 | 0.0895 | 0.040* | |
| C6 | 0.07931 (16) | 1.2541 (2) | 0.05007 (14) | 0.0280 (5) | |
| C7 | 0.13675 (16) | 0.9212 (2) | −0.01186 (15) | 0.0286 (5) | |
| C8 | 0.15008 (16) | 0.8828 (2) | −0.09752 (15) | 0.0302 (5) | |
| H8A | 0.1417 | 0.9484 | −0.1419 | 0.036* | |
| C9 | 0.17412 (17) | 0.7568 (2) | −0.11732 (16) | 0.0336 (6) | |
| C10 | 0.1935 (2) | 0.6417 (3) | −0.0578 (2) | 0.0485 (7) | |
| H10A | 0.2091 | 0.5631 | −0.0900 | 0.073* | |
| H10B | 0.1358 | 0.6232 | −0.0323 | 0.073* | |
| H10C | 0.2482 | 0.6625 | −0.0118 | 0.073* | |
| C11 | 0.1690 (2) | 0.8191 (3) | −0.26569 (18) | 0.0473 (7) | |
| H11A | 0.1797 | 0.7781 | −0.3199 | 0.071* | |
| H11B | 0.2143 | 0.8937 | −0.2509 | 0.071* | |
| H11C | 0.1023 | 0.8522 | −0.2726 | 0.071* | |
| C12 | 0.39549 (16) | −0.1127 (2) | 0.04485 (14) | 0.0271 (5) | |
| C13 | 0.38077 (17) | −0.0679 (3) | 0.12604 (15) | 0.0326 (6) | |
| H13A | 0.3613 | 0.0211 | 0.1350 | 0.039* | |
| C14 | 0.39604 (18) | −0.1599 (3) | 0.19193 (16) | 0.0368 (6) | |
| H14A | 0.3867 | −0.1329 | 0.2479 | 0.044* | |
| C15 | 0.42483 (18) | −0.2921 (3) | 0.18018 (16) | 0.0367 (6) | |
| H15A | 0.4350 | −0.3513 | 0.2282 | 0.044* | |
| C16 | 0.43866 (17) | −0.3378 (2) | 0.10031 (15) | 0.0333 (6) | |
| H16A | 0.4572 | −0.4271 | 0.0911 | 0.040* | |
| C17 | 0.42345 (16) | −0.2437 (2) | 0.03457 (14) | 0.0273 (5) | |
| C18 | 0.35556 (16) | 0.0780 (2) | −0.06071 (15) | 0.0293 (5) | |
| C19 | 0.35254 (16) | 0.1095 (2) | −0.15081 (15) | 0.0289 (5) | |
| H19A | 0.3658 | 0.0412 | −0.1891 | 0.035* | |
| C20 | 0.33127 (17) | 0.2343 (2) | −0.18219 (15) | 0.0307 (5) | |
| C21 | 0.3085 (2) | 0.3529 (2) | −0.13222 (18) | 0.0404 (6) | |
| H21A | 0.2960 | 0.4300 | −0.1710 | 0.061* | |
| H21B | 0.3637 | 0.3723 | −0.0862 | 0.061* | |
| H21C | 0.2510 | 0.3344 | −0.1066 | 0.061* | |
| C22 | 0.3452 (2) | 0.1631 (3) | −0.32498 (16) | 0.0415 (6) | |
| H22A | 0.3413 | 0.2015 | −0.3829 | 0.062* | |
| H22B | 0.2957 | 0.0936 | −0.3265 | 0.062* | |
| H22C | 0.4096 | 0.1240 | −0.3062 | 0.062* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0628 (12) | 0.0248 (9) | 0.0349 (9) | 0.0044 (8) | 0.0158 (9) | 0.0052 (8) |
| O2 | 0.0557 (12) | 0.0323 (10) | 0.0359 (10) | 0.0058 (8) | 0.0098 (8) | 0.0062 (8) |
| O3 | 0.0470 (11) | 0.0364 (10) | 0.0493 (11) | −0.0006 (8) | 0.0138 (9) | −0.0164 (9) |
| O4 | 0.0562 (11) | 0.0268 (9) | 0.0349 (9) | 0.0079 (8) | 0.0128 (8) | −0.0030 (7) |
| O5 | 0.0519 (11) | 0.0336 (9) | 0.0302 (9) | 0.0057 (8) | 0.0090 (8) | −0.0069 (8) |
| O6 | 0.0448 (10) | 0.0327 (10) | 0.0388 (10) | 0.0074 (8) | 0.0132 (8) | 0.0094 (8) |
| N1 | 0.0380 (11) | 0.0234 (10) | 0.0289 (10) | −0.0006 (8) | 0.0090 (9) | −0.0008 (8) |
| N2 | 0.0380 (11) | 0.0235 (10) | 0.0266 (10) | −0.0004 (8) | 0.0075 (8) | 0.0001 (8) |
| N3 | 0.0335 (11) | 0.0257 (10) | 0.0231 (9) | 0.0003 (8) | 0.0066 (8) | 0.0001 (8) |
| N4 | 0.0330 (11) | 0.0247 (10) | 0.0279 (10) | 0.0001 (8) | 0.0082 (8) | 0.0003 (8) |
| N5 | 0.0352 (11) | 0.0260 (10) | 0.0255 (10) | 0.0011 (8) | 0.0078 (8) | −0.0011 (8) |
| N6 | 0.0306 (10) | 0.0257 (10) | 0.0232 (9) | −0.0003 (8) | 0.0054 (8) | −0.0001 (8) |
| C1 | 0.0260 (12) | 0.0314 (13) | 0.0264 (12) | −0.0019 (10) | 0.0057 (9) | −0.0021 (10) |
| C2 | 0.0334 (13) | 0.0417 (14) | 0.0271 (12) | 0.0033 (11) | 0.0050 (10) | 0.0032 (11) |
| C3 | 0.0367 (14) | 0.0526 (16) | 0.0235 (12) | −0.0021 (12) | 0.0063 (10) | −0.0025 (11) |
| C4 | 0.0358 (13) | 0.0477 (16) | 0.0301 (13) | −0.0028 (12) | 0.0073 (11) | −0.0125 (12) |
| C5 | 0.0331 (13) | 0.0307 (13) | 0.0376 (13) | −0.0031 (10) | 0.0068 (11) | −0.0073 (11) |
| C6 | 0.0310 (12) | 0.0288 (12) | 0.0246 (11) | −0.0039 (10) | 0.0056 (10) | −0.0026 (10) |
| C7 | 0.0272 (12) | 0.0248 (12) | 0.0341 (13) | −0.0010 (9) | 0.0066 (10) | 0.0006 (10) |
| C8 | 0.0305 (12) | 0.0269 (12) | 0.0334 (13) | −0.0014 (10) | 0.0064 (10) | −0.0017 (10) |
| C9 | 0.0276 (12) | 0.0302 (13) | 0.0433 (14) | −0.0027 (10) | 0.0073 (11) | −0.0075 (11) |
| C10 | 0.0504 (17) | 0.0285 (14) | 0.0657 (19) | 0.0052 (12) | 0.0080 (15) | −0.0031 (13) |
| C11 | 0.0534 (17) | 0.0480 (17) | 0.0427 (15) | −0.0070 (14) | 0.0149 (13) | −0.0159 (13) |
| C12 | 0.0250 (11) | 0.0314 (13) | 0.0243 (11) | −0.0047 (9) | 0.0029 (9) | 0.0002 (10) |
| C13 | 0.0374 (13) | 0.0348 (13) | 0.0265 (12) | −0.0012 (11) | 0.0085 (10) | −0.0040 (10) |
| C14 | 0.0387 (14) | 0.0459 (15) | 0.0267 (12) | −0.0043 (12) | 0.0084 (11) | −0.0023 (11) |
| C15 | 0.0402 (14) | 0.0419 (15) | 0.0285 (12) | −0.0060 (12) | 0.0073 (11) | 0.0068 (11) |
| C16 | 0.0334 (13) | 0.0321 (13) | 0.0343 (13) | −0.0046 (10) | 0.0060 (11) | 0.0032 (11) |
| C17 | 0.0290 (12) | 0.0296 (12) | 0.0235 (11) | −0.0043 (9) | 0.0049 (9) | −0.0016 (9) |
| C18 | 0.0281 (12) | 0.0259 (12) | 0.0332 (13) | −0.0009 (9) | 0.0035 (10) | −0.0028 (10) |
| C19 | 0.0313 (12) | 0.0269 (12) | 0.0285 (12) | −0.0010 (10) | 0.0047 (10) | −0.0013 (10) |
| C20 | 0.0277 (12) | 0.0312 (13) | 0.0344 (13) | 0.0002 (10) | 0.0085 (10) | 0.0023 (10) |
| C21 | 0.0433 (15) | 0.0274 (13) | 0.0523 (16) | 0.0035 (11) | 0.0131 (13) | 0.0013 (12) |
| C22 | 0.0553 (17) | 0.0390 (15) | 0.0317 (13) | 0.0053 (12) | 0.0118 (12) | 0.0062 (11) |
Geometric parameters (Å, °)
| O1—N1 | 1.273 (2) | C8—C9 | 1.356 (3) |
| O2—C7 | 1.212 (3) | C8—H8A | 0.9500 |
| O3—C9 | 1.342 (3) | C9—C10 | 1.478 (4) |
| O3—C11 | 1.440 (3) | C10—H10A | 0.9800 |
| O4—N4 | 1.276 (2) | C10—H10B | 0.9800 |
| O5—C18 | 1.217 (3) | C10—H10C | 0.9800 |
| O6—C20 | 1.342 (3) | C11—H11A | 0.9800 |
| O6—C22 | 1.431 (3) | C11—H11B | 0.9800 |
| N1—N2 | 1.309 (3) | C11—H11C | 0.9800 |
| N1—C6 | 1.400 (3) | C12—C17 | 1.386 (3) |
| N2—N3 | 1.372 (3) | C12—C13 | 1.400 (3) |
| N3—C1 | 1.388 (3) | C13—C14 | 1.373 (3) |
| N3—C7 | 1.429 (3) | C13—H13A | 0.9500 |
| N4—N5 | 1.308 (3) | C14—C15 | 1.406 (4) |
| N4—C17 | 1.401 (3) | C14—H14A | 0.9500 |
| N5—N6 | 1.378 (3) | C15—C16 | 1.380 (3) |
| N6—C12 | 1.385 (3) | C15—H15A | 0.9500 |
| N6—C18 | 1.432 (3) | C16—C17 | 1.385 (3) |
| C1—C6 | 1.389 (3) | C16—H16A | 0.9500 |
| C1—C2 | 1.393 (3) | C18—C19 | 1.442 (3) |
| C2—C3 | 1.381 (4) | C19—C20 | 1.357 (3) |
| C2—H2B | 0.9500 | C19—H19A | 0.9500 |
| C3—C4 | 1.396 (4) | C20—C21 | 1.488 (3) |
| C3—H3A | 0.9500 | C21—H21A | 0.9800 |
| C4—C5 | 1.378 (4) | C21—H21B | 0.9800 |
| C4—H4A | 0.9500 | C21—H21C | 0.9800 |
| C5—C6 | 1.389 (3) | C22—H22A | 0.9800 |
| C5—H5B | 0.9500 | C22—H22B | 0.9800 |
| C7—C8 | 1.442 (3) | C22—H22C | 0.9800 |
| C9—O3—C11 | 119.2 (2) | H10A—C10—H10C | 109.5 |
| C20—O6—C22 | 119.25 (19) | H10B—C10—H10C | 109.5 |
| O1—N1—N2 | 122.60 (18) | O3—C11—H11A | 109.5 |
| O1—N1—C6 | 124.83 (19) | O3—C11—H11B | 109.5 |
| N2—N1—C6 | 112.57 (18) | H11A—C11—H11B | 109.5 |
| N1—N2—N3 | 105.25 (17) | O3—C11—H11C | 109.5 |
| N2—N3—C1 | 111.34 (18) | H11A—C11—H11C | 109.5 |
| N2—N3—C7 | 122.05 (18) | H11B—C11—H11C | 109.5 |
| C1—N3—C7 | 126.62 (19) | N6—C12—C17 | 105.70 (19) |
| O4—N4—N5 | 122.44 (18) | N6—C12—C13 | 134.4 (2) |
| O4—N4—C17 | 125.01 (19) | C17—C12—C13 | 119.9 (2) |
| N5—N4—C17 | 112.55 (18) | C14—C13—C12 | 116.2 (2) |
| N4—N5—N6 | 105.29 (16) | C14—C13—H13A | 121.9 |
| N5—N6—C12 | 110.97 (18) | C12—C13—H13A | 121.9 |
| N5—N6—C18 | 121.38 (18) | C13—C14—C15 | 122.9 (2) |
| C12—N6—C18 | 127.49 (19) | C13—C14—H14A | 118.6 |
| N3—C1—C6 | 105.28 (19) | C15—C14—H14A | 118.6 |
| N3—C1—C2 | 134.4 (2) | C16—C15—C14 | 121.4 (2) |
| C6—C1—C2 | 120.3 (2) | C16—C15—H15A | 119.3 |
| C3—C2—C1 | 115.9 (2) | C14—C15—H15A | 119.3 |
| C3—C2—H2B | 122.0 | C15—C16—C17 | 115.1 (2) |
| C1—C2—H2B | 122.0 | C15—C16—H16A | 122.5 |
| C2—C3—C4 | 122.9 (2) | C17—C16—H16A | 122.5 |
| C2—C3—H3A | 118.5 | C16—C17—C12 | 124.5 (2) |
| C4—C3—H3A | 118.5 | C16—C17—N4 | 130.1 (2) |
| C5—C4—C3 | 121.9 (2) | C12—C17—N4 | 105.49 (19) |
| C5—C4—H4A | 119.0 | O5—C18—N6 | 116.1 (2) |
| C3—C4—H4A | 119.0 | O5—C18—C19 | 129.0 (2) |
| C4—C5—C6 | 114.7 (2) | N6—C18—C19 | 114.9 (2) |
| C4—C5—H5B | 122.7 | C20—C19—C18 | 121.6 (2) |
| C6—C5—H5B | 122.7 | C20—C19—H19A | 119.2 |
| C5—C6—C1 | 124.3 (2) | C18—C19—H19A | 119.2 |
| C5—C6—N1 | 130.1 (2) | O6—C20—C19 | 122.4 (2) |
| C1—C6—N1 | 105.56 (19) | O6—C20—C21 | 111.0 (2) |
| O2—C7—N3 | 115.7 (2) | C19—C20—C21 | 126.7 (2) |
| O2—C7—C8 | 129.2 (2) | C20—C21—H21A | 109.5 |
| N3—C7—C8 | 115.1 (2) | C20—C21—H21B | 109.5 |
| C9—C8—C7 | 122.7 (2) | H21A—C21—H21B | 109.5 |
| C9—C8—H8A | 118.7 | C20—C21—H21C | 109.5 |
| C7—C8—H8A | 118.7 | H21A—C21—H21C | 109.5 |
| O3—C9—C8 | 122.4 (2) | H21B—C21—H21C | 109.5 |
| O3—C9—C10 | 110.3 (2) | O6—C22—H22A | 109.5 |
| C8—C9—C10 | 127.3 (2) | O6—C22—H22B | 109.5 |
| C9—C10—H10A | 109.5 | H22A—C22—H22B | 109.5 |
| C9—C10—H10B | 109.5 | O6—C22—H22C | 109.5 |
| H10A—C10—H10B | 109.5 | H22A—C22—H22C | 109.5 |
| C9—C10—H10C | 109.5 | H22B—C22—H22C | 109.5 |
| O1—N1—N2—N3 | −179.03 (19) | C11—O3—C9—C8 | −1.1 (3) |
| C6—N1—N2—N3 | 0.4 (2) | C11—O3—C9—C10 | 179.8 (2) |
| N1—N2—N3—C1 | 0.1 (2) | C7—C8—C9—O3 | 179.0 (2) |
| N1—N2—N3—C7 | −179.59 (19) | C7—C8—C9—C10 | −2.1 (4) |
| O4—N4—N5—N6 | 178.92 (19) | N5—N6—C12—C17 | 0.4 (2) |
| C17—N4—N5—N6 | 0.1 (2) | C18—N6—C12—C17 | 175.7 (2) |
| N4—N5—N6—C12 | −0.3 (2) | N5—N6—C12—C13 | −179.2 (2) |
| N4—N5—N6—C18 | −175.93 (19) | C18—N6—C12—C13 | −3.9 (4) |
| N2—N3—C1—C6 | −0.5 (2) | N6—C12—C13—C14 | 179.9 (2) |
| C7—N3—C1—C6 | 179.1 (2) | C17—C12—C13—C14 | 0.3 (3) |
| N2—N3—C1—C2 | 178.3 (2) | C12—C13—C14—C15 | 0.0 (4) |
| C7—N3—C1—C2 | −2.0 (4) | C13—C14—C15—C16 | −0.6 (4) |
| N3—C1—C2—C3 | −178.7 (2) | C14—C15—C16—C17 | 0.9 (3) |
| C6—C1—C2—C3 | 0.0 (3) | C15—C16—C17—C12 | −0.6 (3) |
| C1—C2—C3—C4 | −0.4 (4) | C15—C16—C17—N4 | −179.8 (2) |
| C2—C3—C4—C5 | 0.3 (4) | N6—C12—C17—C16 | −179.7 (2) |
| C3—C4—C5—C6 | 0.2 (4) | C13—C12—C17—C16 | 0.0 (4) |
| C4—C5—C6—C1 | −0.5 (4) | N6—C12—C17—N4 | −0.3 (2) |
| C4—C5—C6—N1 | 177.9 (2) | C13—C12—C17—N4 | 179.4 (2) |
| N3—C1—C6—C5 | 179.5 (2) | O4—N4—C17—C16 | 0.7 (4) |
| C2—C1—C6—C5 | 0.4 (4) | N5—N4—C17—C16 | 179.5 (2) |
| N3—C1—C6—N1 | 0.7 (2) | O4—N4—C17—C12 | −178.6 (2) |
| C2—C1—C6—N1 | −178.3 (2) | N5—N4—C17—C12 | 0.2 (3) |
| O1—N1—C6—C5 | 0.0 (4) | N5—N6—C18—O5 | 177.7 (2) |
| N2—N1—C6—C5 | −179.4 (2) | C12—N6—C18—O5 | 2.8 (3) |
| O1—N1—C6—C1 | 178.7 (2) | N5—N6—C18—C19 | −3.0 (3) |
| N2—N1—C6—C1 | −0.7 (3) | C12—N6—C18—C19 | −177.9 (2) |
| N2—N3—C7—O2 | −175.4 (2) | O5—C18—C19—C20 | 5.0 (4) |
| C1—N3—C7—O2 | 5.0 (3) | N6—C18—C19—C20 | −174.2 (2) |
| N2—N3—C7—C8 | 4.4 (3) | C22—O6—C20—C19 | 3.0 (3) |
| C1—N3—C7—C8 | −175.3 (2) | C22—O6—C20—C21 | −177.4 (2) |
| O2—C7—C8—C9 | −0.7 (4) | C18—C19—C20—O6 | 179.3 (2) |
| N3—C7—C8—C9 | 179.6 (2) | C18—C19—C20—C21 | −0.2 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C2—H2B···O2 | 0.95 | 2.45 | 2.925 (3) | 111 |
| C13—H13A···O5 | 0.95 | 2.49 | 2.961 (3) | 111 |
| C14—H14A···O4i | 0.95 | 2.57 | 3.398 (3) | 147 |
| C16—H16A···O4ii | 0.95 | 2.45 | 3.379 (3) | 165 |
Symmetry codes: (i) x, −y−1/2, z+1/2; (ii) −x+1, −y−1, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: RZ2397).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Barlos, K., Papaioannou, D., Voliotis, S., Prewo, R. & Bieri, J. H. (1985). J. Org. Chem. pp. 696–697.
- Rigaku (2007). CrystalClear Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Singh, J., Fox, R., Wong, M., Kissick, T. P., Moniot, J. L., Gougoutas, J. Z., Malley, M. F. & Kocy, O. (1988). J. Org. Chem. pp. 208–210.
- Zhang, Y., Sun, X. W., Hui, X. E. & Zhang, Q. (2002). Chin. J. Chem.20, 168–172.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809050442/rz2397sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809050442/rz2397Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report