Abstract
In the title compound, C9H15NO·C6H2Cl2O4 [sytematic name: 2-azaspiro[4.5]decan-3-one–chloranilic acid (1/1)], the cyclohexane ring of the lactam molecule adopts a slightly distorted normal chair conformation and the five-membered 3-azaspiro ring is in a slightly distorted chair conformation. The dihedral angle between the least-squares planes of the cyclohexane and 3-azaspiro rings is 84.0 (3)°. In the crystal, the chloranilic acid molecule and the gabapentin-lactum molecules are held together by strong intermolecular N—H⋯O and O—H⋯O hydrogen bonds with two bifurcated O acceptor atoms on the chloranilic acid molecule and one on the gabapentin-lactum molecule, each bonding with an inter- and intramolecular hydrogen bond. The molecules are linked into chains parallel to (011) and propagating along the b axis.
Related literature
For the neuroprotective properties of gabapentin-lactam and related compounds, see: Lagreze et al. (2001 ▶); Henle et al. (2006 ▶); Bowery (1993 ▶). For the synthesis and spectroscopic studies of chloranilic acid charge-transfer complexes, see: Al-Attas et al. (2009 ▶). For related structures, see: Gotoh et al. (2008 ▶); Ibers (2001 ▶); Ishida (2004 ▶); Ishida & Kashino (2000 ▶); Jasinski et al. (2009 ▶). For density functional theory (DFT), see: Frisch et al. (2004 ▶); Hehre et al. (1986 ▶); Schmidt & Polik (2007 ▶).
Experimental
Crystal data
C9H15NO·C6H2Cl2O4
M r = 362.20
Triclinic,
a = 6.6127 (9) Å
b = 9.5800 (11) Å
c = 13.0724 (13) Å
α = 102.679 (9)°
β = 91.934 (9)°
γ = 98.481 (10)°
V = 797.23 (16) Å3
Z = 2
Cu Kα radiation
μ = 3.90 mm−1
T = 110 K
0.47 × 0.42 × 0.15 mm
Data collection
Goniometer Xcalibur diffractometer with a Ruby (Gemini Cu) detector
Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2007 ▶) T min = 0.200, T max = 0.557
5138 measured reflections
3123 independent reflections
2731 reflections with I > 2σ(I)
R int = 0.025
Refinement
R[F 2 > 2σ(F 2)] = 0.042
wR(F 2) = 0.119
S = 1.05
3123 reflections
210 parameters
H-atom parameters constrained
Δρmax = 0.45 e Å−3
Δρmin = −0.40 e Å−3
Data collection: CrysAlis PRO (Oxford Diffraction, 2007 ▶); cell refinement: CrysAlis PRO; data reduction: CrysAlis PRO; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809053410/ng2704sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809053410/ng2704Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1A—H1A⋯O2Ai | 0.84 | 1.97 | 2.7493 (19) | 153 |
| O1A—H1A⋯O2A | 0.84 | 2.20 | 2.671 (2) | 115 |
| O3A—H3A⋯O1Bii | 0.84 | 1.70 | 2.4807 (19) | 153 |
| O3A—H3A⋯O4A | 0.84 | 2.26 | 2.7148 (19) | 114 |
| N2B—H2BA⋯O1Bii | 0.88 | 2.07 | 2.913 (2) | 161 |
| N2B—H2BA⋯O4A | 0.88 | 2.53 | 3.091 (2) | 122 |
Symmetry codes: (i) 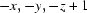 ; (ii)
; (ii) 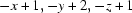 .
.
Acknowledgments
QNMHA thanks the University of Mysore for use of their research facilities. RJB acknowledges the NSF MRI program (grant No. CHE-0619278) for funds to purchase an X-ray diffractometer.
supplementary crystallographic information
Comment
Gabapentin-lactum (systematic name: 3-azaspiro-[4,5]-decan-2-one), is an intermediate for the preparation of gabapentin. Gabapentin-lactam (GBP-L) is also a derivative of the anti-convulsant drug gabapentin. The neuroprotective properties of gabapentin-lactam are described (Lagreze et al. 2001). Gabapentin is currently used as a therapeutic agent against epilepsy as well as neuropathic pain. In contrast to gabapentin, its derivative gabapentin-lactam has a pronounced neuroprotective activity (Henle et al. 2006). Gabapentin is structurally related to the neurotransmitteraminobutyric acid (GABA), which has been widely studied for its significant inhibitory action in the central nervous system (Bowery, 1993). We have recently reported a crystal structure of a second polymorph of gabapentin hydrochloride hemihydrate with a three-center bifurcated hydrogen bond (Jasinski et al. 2009).
Chloranilic acid is a strong dibasic organic acid which exhibits electron-acceptor properties on one hand and acidic properties leading to formation of hydrogen bonds on the other hand. In the case of stronger bases the proton-transfer, hydrogen bonded ion pairs will be formed which is interesting from the point of view of electron transfer reactions in biological systems. Also, protonation of the donor from acidic acceptors are generally a route for the formation of ion pair adducts. The synthesis and spectroscopic studies of charge transfer complexes between chloranilic acid and some heterocyclic amines in ethanol (Al-Attas, Habeeb & Al-Raimi, 2009) have been studied. In view of the importance of gabapentin-lactam, this paper reports the interaction of Gabapentin-lactam as an electron donor with chloranilic acid as an electron acceptor resulting in the formation of a charge transfer complex (I) while the two molecules are held together by intermolecular hydrogen bonding interactions.
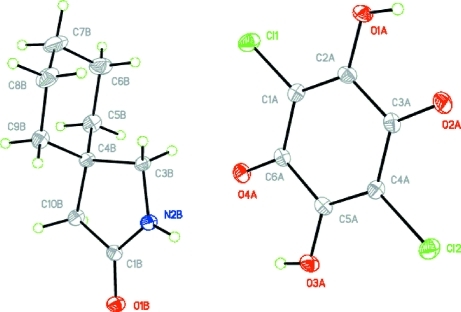
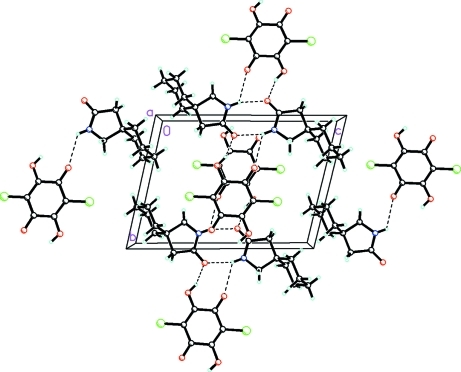
The title compound,C9H15NO.C6H2Cl2O4,(I), is composed of two independent molecules, gabapentin-lactum (C9H15NO) and chloranilic acid (C6H2Cl2O4),in the asymmetric unit (1:1) (Fig.1). In the gabapentin-lactum molecule the cyclohexane ring (C4B—C9B) adopts a slightly distorted normal chair conformation and the 5-membered 3-Azaspiro ring is in a slightly distorted half-chair conformation The N2B and C1B atoms are sp2 hybridized while the C3B, C4B and C10B atoms are sp3. The C10B—C4B—C5B—C6B, N2B—C3B—C4B—C5B and N2B—C3B—C4B—C9B torsion angles are 177.77 (17)°, -136.89 (16)° and 101.19 (17)°, respectively, indicating a significant twist between the 3-azaspiro and cyclohexane rings while sharing a corner C4B atom. The dihedral angle between the least squares planes of these two rings measures 84.0 (3)°. The planar chloranilic acid molecule and gabapentin-lactum molecules are held together by N—H···O and O—H···O intermolecular hydrogen bonds with two bifurcated oxygen acceptor atoms on the chloranilic acid molecule (O2A & O4A) and one on the gabapentin-lactum molecule (O1B), each bifurcating with an inter and intra molecular hydrogen bond, respectively (Fig. 3, Table 1). This produces a set of O—H···O—H···O—H infinite chains along the b axis in (011). The O=C—N—H groups from the 3-azaspiro groups in adjacent gabapentin-lactum molecules form a R2,2(8) graph set motif, while the O=C—C—O—H groups from symmetry related chloranilic acid molecules form a R2,2(10) graph set motif in the unit cell (Fig. 3). The dihedral angles between mean planes of the chloranilic acid molecule and the 3-azaspiro and cyclohexane rings of the gabapentin-lactum molecule are 7.0 (1)° and 77.0 (1)°, respectively. In addition, weak Cg3···Cg3 [= 3.680 (1) Å; slippage = 1.825 Å; -x, 1 - y,1 - z] intermolecular interactions are observed where Cg3 = C1A–C6A, which contribute to crystal packing.
Following a geometry optimization, density functional theory (DFT) calculation at the B3LYP 6–31-G(d) level (Hehre et al., 1986; Schmidt & Polik, 2007) with the Gaussian03 program package (Frisch at al., 2004) the dihederal angle between the least squares planes of the 3-azaspiro and cyclohexane rings in the gabapentin-lactum molecule become 79.2 (9)° compared to 84.0 (3)° in the crystal. The dihedral angles between mean planes of the chloranilic acid molecule and the 3-azaspiro and cyclohexane rings of the gabapentin-lactum molecule become 2.0 (0)° and 77.2 (9)°, respectively, versus 7.0 (1)° and 77.0 (1)° observed in the crystal. Starting geometries were taken from X-ray refinement data. This suggests that strong N—H···O and O—H···O intermolecular hydrogen bonds and weak intermolecular Cg···Cg intermolecular interactions, collectively, influence crystal packing.
Experimental
The title compound was synthesized by adding a solution of chloranilic acid (0.42 g, 2 mmol) in 10 ml me thanol to a solution of gabapentin-lactam (0.21 g, 2 mmol) in 10 ml me thanol. A red color developed and the solution was allowed to evaporate slowly at room temperature. The red colored complex formed was filtered off, washed with diethyl ether and dried under vacuum. X-ray quality crystals were grown from methanol:water (80:20 v/v) solvent mixture (m.p.: 439–442 K). Analysis for C15H17Cl2NO5: Found (Calculated): C:49.68 (49.74); H: 4.70 (4.73); N:3.85 (3.87).
Refinement
The hydroxyl and aza hydrogen atoms (H1A, H3A & H2B) were obtained from a difference fourier map. The rest of the H atoms were placed in their calculated positions and then refined using the riding model with O—H = 0.84 Å, N—H = 0.88 Å, C—H = 0.99 Å, and with Uiso(H) = 1.18–1.22Ueq(C,O,N).
Figures
Fig. 1.
Molecular structure of C9H15NO.C6H2Cl2O4 showing the atom labeling scheme and 50% probability displacement ellipsoids. H atoms are presented as small circles of arbitrary radius.
Fig. 2.
Packing diagram of the title compound, (I), viewed down the a axis. Dashed lines indicate strong N—H···O and O—H···O intermoloecular hydrogen bonds linking the C9H15NO and C6H2Cl2O4 molecules into an infinite O—H···O—H···O—H chain network along the b axis in (011).
Crystal data
| C9H15NO·C6H2Cl2O4 | Z = 2 |
| Mr = 362.20 | F(000) = 376 |
| Triclinic, P1 | Dx = 1.509 Mg m−3 |
| Hall symbol: -P 1 | Cu Kα radiation, λ = 1.54184 Å |
| a = 6.6127 (9) Å | Cell parameters from 3438 reflections |
| b = 9.5800 (11) Å | θ = 4.8–74.0° |
| c = 13.0724 (13) Å | µ = 3.90 mm−1 |
| α = 102.679 (9)° | T = 110 K |
| β = 91.934 (9)° | Irregular plate, red-brown |
| γ = 98.481 (10)° | 0.47 × 0.42 × 0.15 mm |
| V = 797.23 (16) Å3 |
Data collection
| Goniometer Xcalibur diffractometer with a Ruby (Gemini Cu) detector | 3123 independent reflections |
| Radiation source: Enhance (Cu) X-ray Source | 2731 reflections with I > 2σ(I) |
| graphite | Rint = 0.025 |
| Detector resolution: 10.5081 pixels mm-1 | θmax = 74.2°, θmin = 4.8° |
| ω scans | h = −8→5 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2007) | k = −10→11 |
| Tmin = 0.200, Tmax = 0.557 | l = −15→16 |
| 5138 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.042 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.119 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0762P)2 + 0.4046P] where P = (Fo2 + 2Fc2)/3 |
| 3123 reflections | (Δ/σ)max = 0.001 |
| 210 parameters | Δρmax = 0.45 e Å−3 |
| 0 restraints | Δρmin = −0.40 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.07819 (8) | 0.35536 (5) | 0.25360 (3) | 0.02058 (15) | |
| Cl2 | 0.30441 (7) | 0.39346 (5) | 0.73387 (3) | 0.01894 (15) | |
| O1A | 0.0035 (2) | 0.11592 (14) | 0.36708 (11) | 0.0185 (3) | |
| H1A | −0.0101 | 0.0550 | 0.4048 | 0.022* | |
| O2A | 0.1030 (2) | 0.13961 (14) | 0.57058 (11) | 0.0198 (3) | |
| O3A | 0.3781 (2) | 0.63101 (14) | 0.62265 (11) | 0.0168 (3) | |
| H3A | 0.3800 | 0.6958 | 0.5885 | 0.020* | |
| O4A | 0.2633 (2) | 0.61192 (14) | 0.41806 (11) | 0.0174 (3) | |
| C1A | 0.1342 (3) | 0.3619 (2) | 0.38385 (15) | 0.0142 (4) | |
| C2A | 0.0914 (3) | 0.2440 (2) | 0.42497 (15) | 0.0149 (4) | |
| C3A | 0.1454 (3) | 0.25105 (19) | 0.53885 (15) | 0.0142 (4) | |
| C4A | 0.2436 (3) | 0.3875 (2) | 0.60388 (14) | 0.0139 (4) | |
| C5A | 0.2870 (3) | 0.50807 (19) | 0.56472 (15) | 0.0134 (4) | |
| C6A | 0.2301 (3) | 0.50147 (19) | 0.45017 (15) | 0.0132 (4) | |
| O1B | 0.5433 (2) | 1.13507 (14) | 0.43096 (11) | 0.0189 (3) | |
| C1B | 0.4590 (3) | 1.03933 (19) | 0.35294 (15) | 0.0147 (4) | |
| N2B | 0.3806 (3) | 0.90640 (16) | 0.35775 (12) | 0.0158 (3) | |
| H2BA | 0.3923 | 0.8731 | 0.4149 | 0.019* | |
| C3B | 0.2729 (3) | 0.8193 (2) | 0.26005 (14) | 0.0163 (4) | |
| H3BA | 0.1226 | 0.8081 | 0.2654 | 0.020* | |
| H3BB | 0.3141 | 0.7220 | 0.2424 | 0.020* | |
| C4B | 0.3395 (3) | 0.90694 (19) | 0.17620 (14) | 0.0155 (4) | |
| C5B | 0.1561 (3) | 0.9111 (2) | 0.10248 (16) | 0.0214 (4) | |
| H5BA | 0.0461 | 0.9484 | 0.1448 | 0.026* | |
| H5BB | 0.1986 | 0.9785 | 0.0568 | 0.026* | |
| C6B | 0.0727 (4) | 0.7610 (2) | 0.03391 (17) | 0.0288 (5) | |
| H6BA | 0.0190 | 0.6956 | 0.0791 | 0.035* | |
| H6BB | −0.0418 | 0.7686 | −0.0143 | 0.035* | |
| C7B | 0.2397 (4) | 0.6977 (3) | −0.02991 (17) | 0.0320 (5) | |
| H7BA | 0.2827 | 0.7577 | −0.0805 | 0.038* | |
| H7BB | 0.1841 | 0.5986 | −0.0705 | 0.038* | |
| C8B | 0.4253 (4) | 0.6919 (2) | 0.04056 (17) | 0.0278 (5) | |
| H8BA | 0.5350 | 0.6579 | −0.0036 | 0.033* | |
| H8BB | 0.3866 | 0.6216 | 0.0847 | 0.033* | |
| C9B | 0.5062 (3) | 0.8409 (2) | 0.11141 (16) | 0.0198 (4) | |
| H9BA | 0.5625 | 0.9073 | 0.0674 | 0.024* | |
| H9BB | 0.6193 | 0.8318 | 0.1598 | 0.024* | |
| C10B | 0.4297 (3) | 1.0593 (2) | 0.24256 (15) | 0.0175 (4) | |
| H10A | 0.5621 | 1.0955 | 0.2170 | 0.021* | |
| H10B | 0.3340 | 1.1288 | 0.2394 | 0.021* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0306 (3) | 0.0172 (2) | 0.0145 (2) | 0.00588 (19) | −0.00172 (18) | 0.00410 (18) |
| Cl2 | 0.0277 (3) | 0.0151 (2) | 0.0153 (2) | 0.00428 (18) | −0.00131 (18) | 0.00606 (17) |
| O1A | 0.0265 (7) | 0.0099 (6) | 0.0176 (7) | −0.0005 (5) | 0.0004 (6) | 0.0026 (5) |
| O2A | 0.0281 (8) | 0.0111 (6) | 0.0211 (7) | 0.0016 (6) | 0.0049 (6) | 0.0057 (5) |
| O3A | 0.0228 (7) | 0.0093 (6) | 0.0179 (7) | −0.0007 (5) | −0.0029 (5) | 0.0048 (5) |
| O4A | 0.0214 (7) | 0.0131 (7) | 0.0194 (7) | 0.0022 (5) | 0.0020 (5) | 0.0076 (5) |
| C1A | 0.0154 (9) | 0.0149 (9) | 0.0141 (9) | 0.0062 (7) | 0.0023 (7) | 0.0042 (7) |
| C2A | 0.0144 (9) | 0.0112 (8) | 0.0183 (9) | 0.0032 (7) | 0.0023 (7) | 0.0008 (7) |
| C3A | 0.0154 (9) | 0.0107 (9) | 0.0184 (9) | 0.0048 (7) | 0.0048 (7) | 0.0047 (7) |
| C4A | 0.0168 (9) | 0.0125 (9) | 0.0138 (9) | 0.0051 (7) | 0.0017 (7) | 0.0042 (7) |
| C5A | 0.0112 (9) | 0.0113 (9) | 0.0184 (9) | 0.0036 (7) | 0.0014 (7) | 0.0037 (7) |
| C6A | 0.0122 (8) | 0.0108 (9) | 0.0179 (9) | 0.0043 (7) | 0.0027 (7) | 0.0044 (7) |
| O1B | 0.0276 (8) | 0.0112 (6) | 0.0166 (7) | −0.0010 (5) | −0.0015 (6) | 0.0036 (5) |
| C1B | 0.0170 (9) | 0.0110 (8) | 0.0166 (9) | 0.0029 (7) | 0.0014 (7) | 0.0037 (7) |
| N2B | 0.0236 (8) | 0.0109 (7) | 0.0130 (8) | 0.0012 (6) | −0.0001 (6) | 0.0044 (6) |
| C3B | 0.0245 (10) | 0.0116 (8) | 0.0123 (9) | 0.0004 (7) | 0.0009 (7) | 0.0032 (7) |
| C4B | 0.0224 (10) | 0.0106 (9) | 0.0138 (9) | 0.0024 (7) | 0.0013 (7) | 0.0038 (7) |
| C5B | 0.0251 (10) | 0.0216 (10) | 0.0192 (10) | 0.0078 (8) | −0.0004 (8) | 0.0058 (8) |
| C6B | 0.0350 (12) | 0.0279 (12) | 0.0208 (10) | 0.0012 (9) | −0.0075 (9) | 0.0034 (9) |
| C7B | 0.0492 (15) | 0.0252 (11) | 0.0174 (10) | 0.0041 (10) | −0.0010 (10) | −0.0022 (8) |
| C8B | 0.0413 (13) | 0.0195 (10) | 0.0229 (10) | 0.0115 (9) | 0.0078 (9) | 0.0002 (8) |
| C9B | 0.0245 (10) | 0.0173 (9) | 0.0193 (9) | 0.0054 (8) | 0.0042 (8) | 0.0060 (8) |
| C10B | 0.0268 (10) | 0.0112 (9) | 0.0155 (9) | 0.0030 (7) | 0.0024 (8) | 0.0049 (7) |
Geometric parameters (Å, °)
| Cl1—C1A | 1.7158 (19) | C3B—H3BB | 0.9900 |
| Cl2—C4A | 1.7196 (18) | C4B—C5B | 1.534 (3) |
| O1A—C2A | 1.330 (2) | C4B—C9B | 1.538 (3) |
| O1A—H1A | 0.8400 | C4B—C10B | 1.546 (2) |
| O2A—C3A | 1.227 (2) | C5B—C6B | 1.530 (3) |
| O3A—C5A | 1.301 (2) | C5B—H5BA | 0.9900 |
| O3A—H3A | 0.8400 | C5B—H5BB | 0.9900 |
| O4A—C6A | 1.215 (2) | C6B—C7B | 1.523 (3) |
| C1A—C2A | 1.352 (3) | C6B—H6BA | 0.9900 |
| C1A—C6A | 1.465 (3) | C6B—H6BB | 0.9900 |
| C2A—C3A | 1.504 (3) | C7B—C8B | 1.525 (3) |
| C3A—C4A | 1.441 (3) | C7B—H7BA | 0.9900 |
| C4A—C5A | 1.361 (3) | C7B—H7BB | 0.9900 |
| C5A—C6A | 1.517 (3) | C8B—C9B | 1.531 (3) |
| O1B—C1B | 1.262 (2) | C8B—H8BA | 0.9900 |
| C1B—N2B | 1.318 (2) | C8B—H8BB | 0.9900 |
| C1B—C10B | 1.507 (2) | C9B—H9BA | 0.9900 |
| N2B—C3B | 1.460 (2) | C9B—H9BB | 0.9900 |
| N2B—H2BA | 0.8800 | C10B—H10A | 0.9900 |
| C3B—C4B | 1.558 (2) | C10B—H10B | 0.9900 |
| C3B—H3BA | 0.9900 | ||
| C2A—O1A—H1A | 109.5 | C10B—C4B—C3B | 103.66 (14) |
| C5A—O3A—H3A | 109.5 | C6B—C5B—C4B | 111.76 (17) |
| C2A—C1A—C6A | 120.62 (17) | C6B—C5B—H5BA | 109.3 |
| C2A—C1A—Cl1 | 121.97 (15) | C4B—C5B—H5BA | 109.3 |
| C6A—C1A—Cl1 | 117.41 (14) | C6B—C5B—H5BB | 109.3 |
| O1A—C2A—C1A | 122.13 (17) | C4B—C5B—H5BB | 109.3 |
| O1A—C2A—C3A | 116.63 (16) | H5BA—C5B—H5BB | 107.9 |
| C1A—C2A—C3A | 121.23 (16) | C7B—C6B—C5B | 110.90 (19) |
| O2A—C3A—C4A | 124.02 (17) | C7B—C6B—H6BA | 109.5 |
| O2A—C3A—C2A | 117.66 (17) | C5B—C6B—H6BA | 109.5 |
| C4A—C3A—C2A | 118.32 (16) | C7B—C6B—H6BB | 109.5 |
| C5A—C4A—C3A | 121.67 (17) | C5B—C6B—H6BB | 109.5 |
| C5A—C4A—Cl2 | 120.66 (14) | H6BA—C6B—H6BB | 108.0 |
| C3A—C4A—Cl2 | 117.67 (14) | C6B—C7B—C8B | 111.52 (18) |
| O3A—C5A—C4A | 121.97 (17) | C6B—C7B—H7BA | 109.3 |
| O3A—C5A—C6A | 118.01 (16) | C8B—C7B—H7BA | 109.3 |
| C4A—C5A—C6A | 120.03 (16) | C6B—C7B—H7BB | 109.3 |
| O4A—C6A—C1A | 123.11 (17) | C8B—C7B—H7BB | 109.3 |
| O4A—C6A—C5A | 118.78 (16) | H7BA—C7B—H7BB | 108.0 |
| C1A—C6A—C5A | 118.10 (15) | C7B—C8B—C9B | 111.18 (17) |
| O1B—C1B—N2B | 123.96 (17) | C7B—C8B—H8BA | 109.4 |
| O1B—C1B—C10B | 125.82 (16) | C9B—C8B—H8BA | 109.4 |
| N2B—C1B—C10B | 110.21 (16) | C7B—C8B—H8BB | 109.4 |
| C1B—N2B—C3B | 114.19 (15) | C9B—C8B—H8BB | 109.4 |
| C1B—N2B—H2BA | 122.9 | H8BA—C8B—H8BB | 108.0 |
| C3B—N2B—H2BA | 122.9 | C8B—C9B—C4B | 112.62 (17) |
| N2B—C3B—C4B | 104.11 (15) | C8B—C9B—H9BA | 109.1 |
| N2B—C3B—H3BA | 110.9 | C4B—C9B—H9BA | 109.1 |
| C4B—C3B—H3BA | 110.9 | C8B—C9B—H9BB | 109.1 |
| N2B—C3B—H3BB | 110.9 | C4B—C9B—H9BB | 109.1 |
| C4B—C3B—H3BB | 110.9 | H9BA—C9B—H9BB | 107.8 |
| H3BA—C3B—H3BB | 109.0 | C1B—C10B—C4B | 105.01 (15) |
| C5B—C4B—C9B | 109.57 (15) | C1B—C10B—H10A | 110.7 |
| C5B—C4B—C10B | 111.96 (15) | C4B—C10B—H10A | 110.7 |
| C9B—C4B—C10B | 109.81 (16) | C1B—C10B—H10B | 110.7 |
| C5B—C4B—C3B | 111.08 (16) | C4B—C10B—H10B | 110.7 |
| C9B—C4B—C3B | 110.64 (15) | H10A—C10B—H10B | 108.8 |
| C6A—C1A—C2A—O1A | 179.69 (16) | C4A—C5A—C6A—C1A | −1.8 (3) |
| Cl1—C1A—C2A—O1A | 0.0 (3) | O1B—C1B—N2B—C3B | 174.06 (18) |
| C6A—C1A—C2A—C3A | −1.6 (3) | C10B—C1B—N2B—C3B | −5.0 (2) |
| Cl1—C1A—C2A—C3A | 178.76 (13) | C1B—N2B—C3B—C4B | 14.0 (2) |
| O1A—C2A—C3A—O2A | −0.8 (3) | N2B—C3B—C4B—C5B | −136.89 (16) |
| C1A—C2A—C3A—O2A | −179.58 (18) | N2B—C3B—C4B—C9B | 101.19 (17) |
| O1A—C2A—C3A—C4A | 179.11 (16) | N2B—C3B—C4B—C10B | −16.48 (19) |
| C1A—C2A—C3A—C4A | 0.3 (3) | C9B—C4B—C5B—C6B | 55.7 (2) |
| O2A—C3A—C4A—C5A | −179.95 (18) | C10B—C4B—C5B—C6B | 177.77 (17) |
| C2A—C3A—C4A—C5A | 0.2 (3) | C3B—C4B—C5B—C6B | −66.9 (2) |
| O2A—C3A—C4A—Cl2 | 0.0 (3) | C4B—C5B—C6B—C7B | −57.1 (2) |
| C2A—C3A—C4A—Cl2 | −179.94 (13) | C5B—C6B—C7B—C8B | 55.9 (2) |
| C3A—C4A—C5A—O3A | −179.05 (16) | C6B—C7B—C8B—C9B | −54.4 (3) |
| Cl2—C4A—C5A—O3A | 1.1 (3) | C7B—C8B—C9B—C4B | 54.4 (2) |
| C3A—C4A—C5A—C6A | 0.6 (3) | C5B—C4B—C9B—C8B | −54.5 (2) |
| Cl2—C4A—C5A—C6A | −179.28 (13) | C10B—C4B—C9B—C8B | −177.91 (16) |
| C2A—C1A—C6A—O4A | −176.63 (18) | C3B—C4B—C9B—C8B | 68.3 (2) |
| Cl1—C1A—C6A—O4A | 3.0 (3) | O1B—C1B—C10B—C4B | 174.58 (18) |
| C2A—C1A—C6A—C5A | 2.3 (3) | N2B—C1B—C10B—C4B | −6.4 (2) |
| Cl1—C1A—C6A—C5A | −178.02 (13) | C5B—C4B—C10B—C1B | 133.80 (17) |
| O3A—C5A—C6A—O4A | −3.2 (3) | C9B—C4B—C10B—C1B | −104.25 (17) |
| C4A—C5A—C6A—O4A | 177.16 (17) | C3B—C4B—C10B—C1B | 13.99 (19) |
| O3A—C5A—C6A—C1A | 177.86 (15) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1A—H1A···O2Ai | 0.84 | 1.97 | 2.7493 (19) | 153 |
| O1A—H1A···O2A | 0.84 | 2.20 | 2.671 (2) | 115 |
| O3A—H3A···O1Bii | 0.84 | 1.70 | 2.4807 (19) | 153 |
| O3A—H3A···O4A | 0.84 | 2.26 | 2.7148 (19) | 114 |
| N2B—H2BA···O1Bii | 0.88 | 2.07 | 2.913 (2) | 161 |
| N2B—H2BA···O4A | 0.88 | 2.53 | 3.091 (2) | 122 |
Symmetry codes: (i) −x, −y, −z+1; (ii) −x+1, −y+2, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: NG2704).
References
- Al-Attas, A. S., Habeeb, M. M. & Al-Raimi, D. S. (2009). J. Mol. Struct.928, 158–170.
- Bowery, N. G. (1993). Annu. Rev. Pharmacol. Toxicol.33, 109–147. [DOI] [PubMed]
- Frisch, M. J., et al. (2004). GAUSSIAN03, Revision C01. Gaussian Inc., Wallingford, CT, USA.
- Gotoh, K., Asaji, T. & Ishida, H. (2008). Acta Cryst. C64, o550–o553. [DOI] [PubMed]
- Hehre, W. J., Random, L., Schleyer, P. V. R. & Pople, J. A. (1986). In Ab Initio Molecular Orbital Theory New York: Wiley.
- Henle, F., Leemhuis, J., Fischer, C., Bock, H. H., Lindemeyer, K., Feuerstein, T. J. & Meyer, D. K. (2006). J. Pharmacol. Exp. Ther.319, 181–191. [DOI] [PubMed]
- Ibers, J. A. (2001). Acta Cryst. C57, 641–643. [DOI] [PubMed]
- Ishida, H. (2004). Acta Cryst. E60, o1900–o1901.
- Ishida, H. & Kashino, S. (2000). Acta Cryst. C56, e202–e204. [DOI] [PubMed]
- Jasinski, J. P., Butcher, R. J., Yathirajan, H. S., Mallesha, L., Mohana, K. N. & Narayana, B. (2009). J. Chem. Crystallogr.39, 777–780.
- Lagreze, W. A., Muller-Velten, R. & Feuerstein, T. J. (2001). Graefe’s Arch. Clin. Exp. Ophthalmol.239, 845–849. [DOI] [PubMed]
- Oxford Diffraction (2007). CrysAlis PRO and CrysAlis RED Oxford Diffraction Ltd, Abingdon, England.
- Schmidt, J. R. & Polik, W. F. (2007). WebMO Pro WebMO, LLC: Holland, MI, USA, available from http://www.webmo.net.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809053410/ng2704sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809053410/ng2704Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report