Abstract
In the title compound, C10H22NO6P, a staggered conformation is found when the molecule is viewed down the central P—C bond, with the oxo and hydroxy groups gauche to each other. The crystal structure features supramolecular chains of helical topology propagating along the b axis, mediated by O—H⋯O hydrogen bonds.
Related literature
For background to the enantioselective nitroaldol reaction of α-ketophosphonates and nitromethane and for the synthesis, see: Mandal et al. (2007 ▶).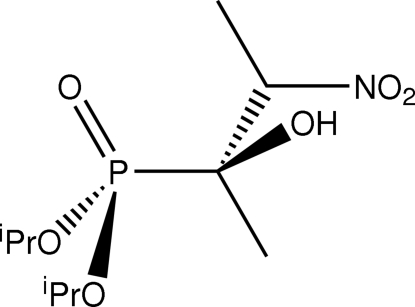
Experimental
Crystal data
C10H22NO6P
M r = 283.26
Orthorhombic,
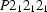
a = 7.8620 (16) Å
b = 11.369 (2) Å
c = 16.920 (3) Å
V = 1512.4 (5) Å3
Z = 4
Mo Kα radiation
μ = 0.20 mm−1
T = 173 K
0.32 × 0.10 × 0.05 mm
Data collection
Rigaku AFC12/SATURN724 diffractometer
Absorption correction: multi-scan (ABSCOR; Higashi, 1995 ▶) T min = 0.884, T max = 1
13441 measured reflections
3072 independent reflections
3020 reflections with I > 2σ(I)
R int = 0.089
Standard reflections: 0
Refinement
R[F 2 > 2σ(F 2)] = 0.040
wR(F 2) = 0.105
S = 1.07
3072 reflections
166 parameters
1 restraint
H-atom parameters constrained
Δρmax = 0.19 e Å−3
Δρmin = −0.25 e Å−3
Absolute structure: Flack (1983 ▶), 1272 Friedel pairs
Flack parameter: 0.05 (11)
Data collection: CrystalClear (Rigaku/MSC, 2005 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: DIAMOND (Brandenburg, 2006 ▶); software used to prepare material for publication: publCIF (Westrip, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809052428/hb5269sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809052428/hb5269Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O4—H4o⋯O1i | 0.84 | 1.90 | 2.7289 (19) | 172 |
Symmetry code: (i)  .
.
Acknowledgments
CGZ thanks the Welch Foundation (grant No. AX-1593) and the NIH-MBRS program (S06 GM08194) for support.
supplementary crystallographic information
Comment
The title compound, (I), was investigated as a part of previous studies on the enantioselective nitroaldol reaction of α-ketophosphonates and nitromethane for the synthesis of optically active α-hydroxy-β-nitrophosphonates (Mandal et al., 2007). The crystal structure analysis of (I), Fig. 1, shows a staggered conformation when the molecule is viewed down the P–C7 axis in which the oxo and hydroxy groups are gauche to each other. The presence of O–H···O hydrogen bonding formed between the hydroxy-O4—H and O═P atoms leads to the formation of supramolecular chains along the b axis, Fig. 2 and Table 1.
Experimental
The title compound was prepared as described in the literature (Mandal et al., 2007).
Refinement
The C-bound H atoms were geometrically placed (C—H = 0.98–1.00 Å) and refined as riding with Uiso(H) = 1.2–1.5Ueq(C). The methyl H-atoms were rotated to fit the electron density. The O–H H atom was located from a difference map and refined with O–H = 0.840±0.001 Å, and with Uiso(H) = 1.5Ueq(O).
Figures
Fig. 1.
Molecular structure of (I), showing displacement ellipsoids at the 35% probability level.
Fig. 2.
Supramolecular chain along the b axis in (I) mediated by O–H···O (orange dashed lines) hydrogen bonding. Colour scheme: P, olive; O, red; N, blue; C, grey; and H, green.
Crystal data
| C10H22NO6P | F(000) = 608 |
| Mr = 283.26 | Dx = 1.244 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 2308 reflections |
| a = 7.8620 (16) Å | θ = 4.0–30.1° |
| b = 11.369 (2) Å | µ = 0.20 mm−1 |
| c = 16.920 (3) Å | T = 173 K |
| V = 1512.4 (5) Å3 | Block, pale-yellow |
| Z = 4 | 0.32 × 0.10 × 0.05 mm |
Data collection
| Rigaku AFC12K/SATURN724 diffractometer | 3072 independent reflections |
| Radiation source: fine-focus sealed tube | 3020 reflections with I > 2σ(I) |
| graphite | Rint = 0.089 |
| ω scans | θmax = 26.5°, θmin = 4.0° |
| Absorption correction: multi-scan (ABSCOR; Higashi, 1995) | h = −9→8 |
| Tmin = 0.884, Tmax = 1 | k = −13→14 |
| 13441 measured reflections | l = −21→20 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.040 | H-atom parameters constrained |
| wR(F2) = 0.105 | w = 1/[σ2(Fo2) + (0.0497P)2 + 0.2822P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.07 | (Δ/σ)max = 0.001 |
| 3072 reflections | Δρmax = 0.19 e Å−3 |
| 166 parameters | Δρmin = −0.25 e Å−3 |
| 1 restraint | Absolute structure: Flack (1983), 1272 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: 0.05 (11) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| P1 | 0.63065 (6) | 0.29509 (4) | 0.74005 (3) | 0.03236 (14) | |
| O1 | 0.51586 (18) | 0.19464 (11) | 0.75316 (9) | 0.0416 (3) | |
| O2 | 0.80539 (18) | 0.26553 (13) | 0.70112 (9) | 0.0415 (3) | |
| O3 | 0.67933 (17) | 0.36408 (13) | 0.81677 (8) | 0.0383 (3) | |
| O4 | 0.41385 (17) | 0.46364 (12) | 0.71762 (9) | 0.0400 (3) | |
| H4O | 0.4448 | 0.5337 | 0.7241 | 0.060* | |
| O5 | 0.3878 (4) | 0.49342 (19) | 0.51776 (13) | 0.0848 (7) | |
| O6 | 0.1751 (3) | 0.4227 (2) | 0.58457 (14) | 0.0861 (7) | |
| N1 | 0.3262 (3) | 0.4256 (2) | 0.56573 (14) | 0.0608 (6) | |
| C1 | 0.9313 (3) | 0.18894 (18) | 0.74033 (13) | 0.0438 (5) | |
| H1 | 0.8736 | 0.1412 | 0.7821 | 0.053* | |
| C2 | 0.9985 (4) | 0.1089 (3) | 0.67718 (17) | 0.0706 (8) | |
| H2A | 0.9051 | 0.0617 | 0.6556 | 0.106* | |
| H2B | 1.0849 | 0.0567 | 0.6998 | 0.106* | |
| H2C | 1.0495 | 0.1560 | 0.6348 | 0.106* | |
| C3 | 1.0653 (3) | 0.2668 (2) | 0.7782 (2) | 0.0656 (8) | |
| H3A | 1.0121 | 0.3162 | 0.8187 | 0.098* | |
| H3B | 1.1173 | 0.3168 | 0.7377 | 0.098* | |
| H3C | 1.1530 | 0.2175 | 0.8026 | 0.098* | |
| C4 | 0.5800 (3) | 0.3614 (2) | 0.89018 (13) | 0.0551 (6) | |
| H4 | 0.4659 | 0.3254 | 0.8803 | 0.066* | |
| C5 | 0.5601 (6) | 0.4848 (3) | 0.91703 (18) | 0.0879 (11) | |
| H5A | 0.4970 | 0.5295 | 0.8771 | 0.132* | |
| H5B | 0.6726 | 0.5202 | 0.9246 | 0.132* | |
| H5C | 0.4976 | 0.4862 | 0.9671 | 0.132* | |
| C6 | 0.6764 (7) | 0.2898 (3) | 0.94900 (18) | 0.1056 (15) | |
| H6A | 0.6874 | 0.2089 | 0.9296 | 0.158* | |
| H6B | 0.6152 | 0.2896 | 0.9995 | 0.158* | |
| H6C | 0.7897 | 0.3238 | 0.9565 | 0.158* | |
| C7 | 0.5409 (2) | 0.40550 (16) | 0.67209 (11) | 0.0336 (4) | |
| C8 | 0.4436 (3) | 0.33911 (18) | 0.60636 (12) | 0.0419 (5) | |
| H8 | 0.3722 | 0.2770 | 0.6318 | 0.050* | |
| C9 | 0.5556 (4) | 0.2804 (2) | 0.54440 (13) | 0.0558 (6) | |
| H9A | 0.4838 | 0.2406 | 0.5053 | 0.084* | |
| H9B | 0.6299 | 0.2227 | 0.5700 | 0.084* | |
| H9C | 0.6251 | 0.3401 | 0.5179 | 0.084* | |
| C10 | 0.6767 (3) | 0.49079 (18) | 0.64308 (14) | 0.0430 (5) | |
| H10A | 0.7312 | 0.5287 | 0.6885 | 0.065* | |
| H10B | 0.6239 | 0.5509 | 0.6095 | 0.065* | |
| H10C | 0.7623 | 0.4477 | 0.6125 | 0.065* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| P1 | 0.0303 (2) | 0.0284 (2) | 0.0384 (2) | 0.00159 (18) | 0.00078 (17) | 0.00062 (19) |
| O1 | 0.0432 (7) | 0.0284 (6) | 0.0532 (8) | −0.0023 (5) | −0.0002 (6) | 0.0044 (7) |
| O2 | 0.0357 (7) | 0.0454 (8) | 0.0435 (7) | 0.0140 (6) | 0.0051 (6) | 0.0031 (6) |
| O3 | 0.0365 (7) | 0.0426 (7) | 0.0359 (7) | −0.0048 (6) | 0.0024 (5) | −0.0017 (6) |
| O4 | 0.0342 (7) | 0.0316 (7) | 0.0541 (8) | 0.0031 (6) | 0.0021 (6) | −0.0080 (6) |
| O5 | 0.1205 (19) | 0.0631 (12) | 0.0708 (13) | 0.0063 (14) | −0.0288 (14) | 0.0171 (10) |
| O6 | 0.0593 (13) | 0.1078 (17) | 0.0912 (16) | 0.0212 (13) | −0.0342 (12) | −0.0093 (14) |
| N1 | 0.0736 (16) | 0.0513 (12) | 0.0576 (12) | 0.0103 (11) | −0.0261 (11) | −0.0073 (11) |
| C1 | 0.0408 (10) | 0.0399 (11) | 0.0507 (11) | 0.0124 (8) | −0.0022 (9) | 0.0007 (10) |
| C2 | 0.0768 (19) | 0.0698 (17) | 0.0650 (16) | 0.0410 (16) | −0.0121 (14) | −0.0150 (14) |
| C3 | 0.0423 (12) | 0.0584 (15) | 0.096 (2) | 0.0113 (11) | −0.0130 (13) | −0.0103 (14) |
| C4 | 0.0576 (14) | 0.0725 (16) | 0.0351 (10) | −0.0157 (12) | 0.0104 (9) | −0.0010 (10) |
| C5 | 0.128 (3) | 0.085 (2) | 0.0507 (15) | 0.038 (2) | 0.0216 (17) | −0.0070 (15) |
| C6 | 0.189 (5) | 0.081 (2) | 0.0471 (15) | 0.029 (3) | 0.004 (2) | 0.0180 (16) |
| C7 | 0.0344 (9) | 0.0283 (8) | 0.0383 (9) | 0.0022 (7) | −0.0010 (8) | −0.0021 (8) |
| C8 | 0.0498 (12) | 0.0332 (9) | 0.0428 (10) | 0.0031 (9) | −0.0088 (9) | −0.0032 (9) |
| C9 | 0.0772 (17) | 0.0502 (13) | 0.0399 (11) | 0.0049 (13) | 0.0000 (11) | −0.0097 (10) |
| C10 | 0.0451 (11) | 0.0334 (10) | 0.0506 (12) | −0.0024 (9) | 0.0042 (9) | 0.0032 (9) |
Geometric parameters (Å, °)
| P1—O1 | 1.4723 (14) | C4—C5 | 1.483 (4) |
| P1—O2 | 1.5602 (14) | C4—C6 | 1.492 (4) |
| P1—O3 | 1.5643 (15) | C4—H4 | 1.0000 |
| P1—C7 | 1.8428 (19) | C5—H5A | 0.9800 |
| O2—C1 | 1.476 (2) | C5—H5B | 0.9800 |
| O3—C4 | 1.467 (2) | C5—H5C | 0.9800 |
| O4—C7 | 1.424 (2) | C6—H6A | 0.9800 |
| O4—H4O | 0.8400 | C6—H6B | 0.9800 |
| O5—N1 | 1.219 (3) | C6—H6C | 0.9800 |
| O6—N1 | 1.230 (4) | C7—C10 | 1.524 (3) |
| N1—C8 | 1.514 (3) | C7—C8 | 1.546 (3) |
| C1—C2 | 1.500 (3) | C8—C9 | 1.523 (3) |
| C1—C3 | 1.517 (3) | C8—H8 | 1.0000 |
| C1—H1 | 1.0000 | C9—H9A | 0.9800 |
| C2—H2A | 0.9800 | C9—H9B | 0.9800 |
| C2—H2B | 0.9800 | C9—H9C | 0.9800 |
| C2—H2C | 0.9800 | C10—H10A | 0.9800 |
| C3—H3A | 0.9800 | C10—H10B | 0.9800 |
| C3—H3B | 0.9800 | C10—H10C | 0.9800 |
| C3—H3C | 0.9800 | ||
| O1—P1—O2 | 115.86 (9) | C4—C5—H5A | 109.5 |
| O1—P1—O3 | 114.44 (9) | C4—C5—H5B | 109.5 |
| O2—P1—O3 | 104.06 (8) | H5A—C5—H5B | 109.5 |
| O1—P1—C7 | 112.80 (9) | C4—C5—H5C | 109.5 |
| O2—P1—C7 | 102.75 (8) | H5A—C5—H5C | 109.5 |
| O3—P1—C7 | 105.67 (8) | H5B—C5—H5C | 109.5 |
| C1—O2—P1 | 121.86 (13) | C4—C6—H6A | 109.5 |
| C4—O3—P1 | 124.17 (14) | C4—C6—H6B | 109.5 |
| C7—O4—H4O | 108.0 | H6A—C6—H6B | 109.5 |
| O5—N1—O6 | 124.9 (3) | C4—C6—H6C | 109.5 |
| O5—N1—C8 | 118.1 (2) | H6A—C6—H6C | 109.5 |
| O6—N1—C8 | 117.0 (2) | H6B—C6—H6C | 109.5 |
| O2—C1—C2 | 105.91 (18) | O4—C7—C10 | 111.74 (15) |
| O2—C1—C3 | 108.14 (17) | O4—C7—C8 | 105.60 (16) |
| C2—C1—C3 | 114.2 (2) | C10—C7—C8 | 115.18 (18) |
| O2—C1—H1 | 109.5 | O4—C7—P1 | 104.31 (12) |
| C2—C1—H1 | 109.5 | C10—C7—P1 | 111.47 (14) |
| C3—C1—H1 | 109.5 | C8—C7—P1 | 107.80 (13) |
| C1—C2—H2A | 109.5 | N1—C8—C9 | 108.94 (19) |
| C1—C2—H2B | 109.5 | N1—C8—C7 | 108.11 (16) |
| H2A—C2—H2B | 109.5 | C9—C8—C7 | 115.0 (2) |
| C1—C2—H2C | 109.5 | N1—C8—H8 | 108.2 |
| H2A—C2—H2C | 109.5 | C9—C8—H8 | 108.2 |
| H2B—C2—H2C | 109.5 | C7—C8—H8 | 108.2 |
| C1—C3—H3A | 109.5 | C8—C9—H9A | 109.5 |
| C1—C3—H3B | 109.5 | C8—C9—H9B | 109.5 |
| H3A—C3—H3B | 109.5 | H9A—C9—H9B | 109.5 |
| C1—C3—H3C | 109.5 | C8—C9—H9C | 109.5 |
| H3A—C3—H3C | 109.5 | H9A—C9—H9C | 109.5 |
| H3B—C3—H3C | 109.5 | H9B—C9—H9C | 109.5 |
| O3—C4—C5 | 107.2 (2) | C7—C10—H10A | 109.5 |
| O3—C4—C6 | 107.8 (2) | C7—C10—H10B | 109.5 |
| C5—C4—C6 | 111.4 (3) | H10A—C10—H10B | 109.5 |
| O3—C4—H4 | 110.1 | C7—C10—H10C | 109.5 |
| C5—C4—H4 | 110.1 | H10A—C10—H10C | 109.5 |
| C6—C4—H4 | 110.1 | H10B—C10—H10C | 109.5 |
| O1—P1—O2—C1 | −63.20 (17) | O3—P1—C7—C10 | 68.93 (15) |
| O3—P1—O2—C1 | 63.34 (17) | O1—P1—C7—C8 | −37.99 (17) |
| C7—P1—O2—C1 | 173.35 (15) | O2—P1—C7—C8 | 87.48 (15) |
| O1—P1—O3—C4 | −21.1 (2) | O3—P1—C7—C8 | −163.73 (13) |
| O2—P1—O3—C4 | −148.58 (17) | O5—N1—C8—C9 | −47.5 (3) |
| C7—P1—O3—C4 | 103.58 (19) | O6—N1—C8—C9 | 133.2 (2) |
| P1—O2—C1—C2 | 136.98 (19) | O5—N1—C8—C7 | 78.1 (3) |
| P1—O2—C1—C3 | −100.2 (2) | O6—N1—C8—C7 | −101.1 (3) |
| P1—O3—C4—C5 | −132.8 (2) | O4—C7—C8—N1 | 51.7 (2) |
| P1—O3—C4—C6 | 107.1 (3) | C10—C7—C8—N1 | −72.1 (2) |
| O1—P1—C7—O4 | 73.94 (14) | P1—C7—C8—N1 | 162.77 (16) |
| O2—P1—C7—O4 | −160.59 (12) | O4—C7—C8—C9 | 173.70 (17) |
| O3—P1—C7—O4 | −51.80 (13) | C10—C7—C8—C9 | 49.9 (2) |
| O1—P1—C7—C10 | −165.33 (14) | P1—C7—C8—C9 | −75.2 (2) |
| O2—P1—C7—C10 | −39.85 (16) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O4—H4o···O1i | 0.84 | 1.90 | 2.7289 (19) | 172 |
Symmetry codes: (i) −x+1, y+1/2, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB5269).
References
- Brandenburg, K. (2006). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Higashi, T. (1995). ABSCOR Rigaku Corporation, Tokyo, Japan.
- Mandal, T., Samanta, S. & Zhao, C.-G. (2007). Org. Lett.9, 943–945. [DOI] [PubMed]
- Rigaku/MSC (2005). CrystalClear Rigaku/MSC Inc., The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2009). publCIF In preparation.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809052428/hb5269sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809052428/hb5269Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




