Abstract
The molecule of the title hydrate, C16H11ClFN3·H2O, is slightly twisted, as indicated by the dihedral angle of 9.55 (10)° formed between the quinoline ring system and the benzene ring. The conformation about the C=N double bond is E, and the amine-H atom is oriented towards the quinoline residue. In the crystal structure, the water molecule accepts an N—H⋯O and makes two O—H⋯Nquinoline hydrogen bonds, generating a two-dimensional array in the ab plane, which is further stabilized by C—H⋯O interactions. The most significant contacts between layers are of the type C—H⋯F.
Related literature
For background information on the pharmacological activity of quinoline derivatives, see: Elslager et al. (1969 ▶); Font et al. (1997 ▶); Kaminsky & Meltzer (1968 ▶); Musiol et al. (2006 ▶); Nakamura et al. (1999 ▶); Palmer et al. (1993 ▶); Ridley (2002 ▶); Sloboda et al. (1991 ▶); Tanenbaum & Tuffanelli (1980 ▶); Warshakoon et al. (2006 ▶). For recent studies into quinoline-based anti-malarials, see: Andrade et al. (2007 ▶); Cunico et al. (2006 ▶); da Silva et al. (2003 ▶); de Souza et al. (2005 ▶). For crystallographic studies on molecules related to the title compound, see: Kaiser et al. (2009 ▶); de Souza et al. (2009 ▶); de Ferreira et al. (2009 ▶). For the synthesis, see: Pellerano et al. (1976 ▶).
Experimental
Crystal data
C16H11ClFN3·H2O
M r = 317.74
Monoclinic,
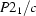
a = 3.7795 (2) Å
b = 15.4188 (11) Å
c = 24.8576 (16) Å
β = 90.286 (4)°
V = 1448.57 (16) Å3
Z = 4
Mo Kα radiation
μ = 0.28 mm−1
T = 120 K
0.90 × 0.04 × 0.04 mm
Data collection
Nonius KappaCCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2007 ▶) T min = 0.614, T max = 0.746
19494 measured reflections
3291 independent reflections
2009 reflections with I > 2σ(I)
R int = 0.098
Refinement
R[F 2 > 2σ(F 2)] = 0.059
wR(F 2) = 0.131
S = 1.04
3291 reflections
205 parameters
3 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.33 e Å−3
Δρmin = −0.37 e Å−3
Data collection: COLLECT (Hooft, 1998 ▶); cell refinement: DENZO (Otwinowski & Minor, 1997 ▶) and COLLECT; data reduction: DENZO and COLLECT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: DIAMOND (Brandenburg, 2006 ▶); software used to prepare material for publication: publCIF (Westrip, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809053367/lh2970sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809053367/lh2970Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1w—H1w⋯N1i | 0.84 (2) | 2.28 (2) | 2.999 (3) | 144 (2) |
| O1w—H2w⋯N1ii | 0.85 (2) | 1.93 (2) | 2.761 (3) | 166 (3) |
| N2—H2n⋯O1wiii | 0.88 | 2.01 | 2.865 (3) | 165 |
| C5—H5⋯O1wiii | 0.95 | 2.45 | 3.379 (3) | 164 |
| C10—H10⋯O1wiii | 0.95 | 2.50 | 3.302 (3) | 142 |
| C1—H1⋯F1iv | 0.95 | 2.56 | 3.399 (3) | 147 |
| C6—H6⋯F1v | 0.95 | 2.56 | 3.477 (3) | 161 |
Symmetry codes: (i) 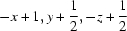 ; (ii)
; (ii) 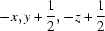 ; (iii)
; (iii) 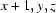 ; (iv)
; (iv) 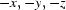 ; (v)
; (v) 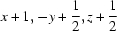 .
.
Acknowledgments
The use of the EPSRC X-ray crystallographic service at the University of Southampton, England and the valuable assistance of the staff there is gratefully acknowledged. JLW acknowledges support from CAPES (Brazil).
supplementary crystallographic information
Comment
The title compound, crystallized as a hydrate, (I), was prepared as part of continuing studies designed to develop antimalarial compounds based on the quinoline nucleus (Andrade et al., 2007; Cunico et al., 2006; da Silva et al., 2003; de Souza et al., 2005). The systematic examination of quinoline derivatives comes about owing to the fact that the majority of antimalarial agents, including chloroquine (Tanenbaum & Tuffanelli, 1980), mefloquine (Palmer et al., 1993), primaquine (Elslager et al., 1969) and amodiaquine (Ridley, 2002), have a quinoline ring substructure, the mainstay of malaria chemotherapy for much of the past 40 years (Font et al., 1997; Kaminsky & Meltzer, 1968; Musiol et al., 2006; Nakamura et al., 1999; Sloboda et al., 1991; Warshakoon et al., 2006). Allied with these investigations are structural studies aimed at elucidating systematic structural trends in these molecules (Kaiser et al. 2009; de Souza et al. 2009; de Ferreira et al. 2009).
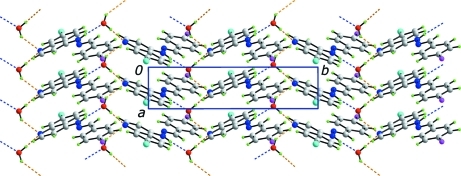
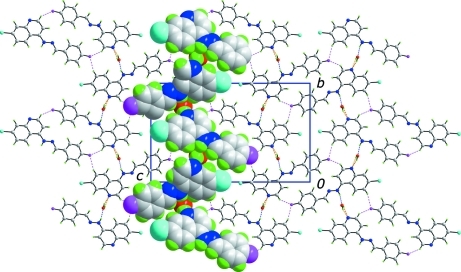
The molecule in (I), Fig. 1, features an effectively planar quinoline residue (maximum deviations of 0.018 (2) Å for atom C4 and -0.025 (2) Å for atom C2) which forms a dihedral angle of 9.55 (10) ° with the C11–C16 benzene ring. Twists in the molecule are evident about the N2–C3 and C10–C11 bonds as seen in the values of the N3–N2–C3–C2 and N3–C10–C11—C12 torsion angles of 6.9 (4) and -6.6 (4) °, respectively. As observed in related systems, the amine-H is orientated over the quinoline residue (Kaiser et al. 2009; de Souza et al. 2009; de Ferreira et al., 2009). The conformation about the N3═C10 double bond is E. The molecule crystallizes as a hydrate and the latter species is pivotal in stabilizing the crystal structure. Thus, the water-H atoms form donor O–H···N hydrogen bonds to quinoline-N atoms derived from two molecules. At the same time, the water-O atom accepts a N–H···O hydrogen bond from the amine-N2 of another molecule. Thus, the water molecule provides links between three molecules, leading to the formation of a 2-D array, Fig. 2 and Table 1. The resultant layer in the ab plane is further stabilized by C–H···O interactions, Table 1, and weak π···π contacts [ring centroid(N1,C1—C4,C9)···ring centroid(C4–C9)i = 3.7070 (14) Å, dihedral angle = 1.45 (11) ° for i: -1 + x, y, z]. Layers stack along the c direction with the most significant contacts between layers being of the type C–H···F whereby the fluoride is bifurcated, Table 1 and Fig. 3.
Experimental
A solution of 7-chloro-4-hydrazinoquinoline (0.20 g, 1.0 mmol) and 4-fluorobenzaldehyde (0.15 g, 1.2 mmol) in EtOH (5 ml) was maintained at room temperature overnight and rotary evaporated. The solid residue, was washed with cold Et2O (3 x 10 ml) and recrystallized from EtOH m.pt. 518–519 K, lit. value 518 K (Pellerano et al., 1976), yield 74%. The sample for the X-ray study was slowly grown from moist EtOH and was found to be the monohydrate. 1H NMR (400 MHz, DMSO-d6) δ: 7.28–7.32 (3H, m), 7.54 (1H, d, J = 8.4 Hz), 7.84–7.88 (3H, m), 8.34–8.40 (3H, m), 11.3 (1H, br.s, NH). MS/ESI: [M+. - H]: 298. IR νmax (cm-1; KBr disc): 3232 (N–H), 1585 (C═N), 817 (C–F).
Refinement
The amine- and C-bound H atoms were geometrically placed (N–H = 0.88 Å and C–H = 0.95 Å) and refined as riding with Uiso(H) = 1.2Ueq(C). The water-bound H atoms were located from a difference map and refined (O–H = 0.84 (1) Å) with Uiso(H) = 1.5Ueq(O).
Figures
Fig. 1.
The molecular structure of both components comprising the asymmetric unit of (I) showing the atom-labelling scheme and displacement ellipsoids at the 50% probability level.
Fig. 2.
A view of the 2-D supramolecular array in (I) showing the O–H···N and N–H···O hydrogen bonds as orange and blue dashed lines, respectively. Colour code: Cl, cyan; F, pink; O, red; N, blue; C, grey; and H, green.
Fig. 3.
A view in projection along the a axis of the unit-cell contents in (I) showing the stacking of layers along the c direction. The O–H···N and N–H···O hydrogen bonds are shown as orange and blue dashed lines, respectively, and the C–H···F contacts are represented by pink dashed lines. One of the 2-D arrays, as shown in Fig. 2, has been highlighted in space-filling mode. Colour code: Cl, cyan; F, pink; O, red; N, blue; C, grey; and H, green.
Crystal data
| C16H11ClFN3·H2O | F(000) = 656 |
| Mr = 317.74 | Dx = 1.457 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 13530 reflections |
| a = 3.7795 (2) Å | θ = 2.9–27.5° |
| b = 15.4188 (11) Å | µ = 0.28 mm−1 |
| c = 24.8576 (16) Å | T = 120 K |
| β = 90.286 (4)° | Needle, colourless |
| V = 1448.57 (16) Å3 | 0.90 × 0.04 × 0.04 mm |
| Z = 4 |
Data collection
| Enraf–Nonius KappaCCD area-detector diffractometer | 3291 independent reflections |
| Radiation source: Enraf Nonius FR591 rotating anode | 2009 reflections with I > 2σ(I) |
| 10 cm confocal mirrors | Rint = 0.098 |
| Detector resolution: 9.091 pixels mm-1 | θmax = 27.5°, θmin = 3.1° |
| φ and ω scans | h = −4→4 |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2007) | k = −20→19 |
| Tmin = 0.614, Tmax = 0.746 | l = −32→32 |
| 19494 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.059 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.131 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0463P)2 + 0.5902P] where P = (Fo2 + 2Fc2)/3 |
| 3291 reflections | (Δ/σ)max < 0.001 |
| 205 parameters | Δρmax = 0.33 e Å−3 |
| 3 restraints | Δρmin = −0.37 e Å−3 |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.85154 (18) | −0.01273 (5) | 0.43677 (3) | 0.0319 (2) | |
| F1 | 0.1997 (4) | 0.22958 (11) | −0.11624 (6) | 0.0367 (5) | |
| N1 | 0.3942 (5) | −0.13713 (14) | 0.26095 (9) | 0.0214 (5) | |
| N2 | 0.6585 (5) | 0.08516 (14) | 0.17189 (8) | 0.0221 (5) | |
| H2N | 0.7847 | 0.1268 | 0.1867 | 0.027* | |
| N3 | 0.5476 (5) | 0.09163 (15) | 0.11914 (8) | 0.0212 (5) | |
| C1 | 0.3270 (7) | −0.12969 (18) | 0.20855 (11) | 0.0225 (6) | |
| H1 | 0.2176 | −0.1776 | 0.1911 | 0.027* | |
| C2 | 0.4043 (6) | −0.05737 (18) | 0.17733 (10) | 0.0210 (6) | |
| H2 | 0.3436 | −0.0566 | 0.1402 | 0.025* | |
| C3 | 0.5701 (6) | 0.01357 (17) | 0.20052 (10) | 0.0178 (6) | |
| C4 | 0.6457 (6) | 0.01003 (16) | 0.25750 (10) | 0.0176 (6) | |
| C5 | 0.8024 (6) | 0.07868 (18) | 0.28696 (11) | 0.0210 (6) | |
| H5 | 0.8680 | 0.1303 | 0.2687 | 0.025* | |
| C6 | 0.8613 (6) | 0.07213 (18) | 0.34118 (10) | 0.0220 (6) | |
| H6 | 0.9655 | 0.1189 | 0.3605 | 0.026* | |
| C7 | 0.7663 (7) | −0.00432 (18) | 0.36790 (11) | 0.0218 (6) | |
| C8 | 0.6151 (6) | −0.07217 (18) | 0.34132 (10) | 0.0214 (6) | |
| H8 | 0.5531 | −0.1233 | 0.3604 | 0.026* | |
| C9 | 0.5501 (6) | −0.06654 (16) | 0.28533 (10) | 0.0179 (6) | |
| C10 | 0.6205 (7) | 0.16293 (18) | 0.09495 (11) | 0.0219 (6) | |
| H10 | 0.7507 | 0.2064 | 0.1136 | 0.026* | |
| C11 | 0.5085 (7) | 0.17884 (18) | 0.03962 (11) | 0.0216 (6) | |
| C12 | 0.3483 (7) | 0.11448 (18) | 0.00825 (11) | 0.0231 (6) | |
| H12 | 0.3087 | 0.0586 | 0.0232 | 0.028* | |
| C13 | 0.2471 (7) | 0.13101 (18) | −0.04411 (11) | 0.0237 (6) | |
| H13 | 0.1409 | 0.0871 | −0.0656 | 0.028* | |
| C14 | 0.3041 (7) | 0.21294 (19) | −0.06453 (11) | 0.0256 (7) | |
| C15 | 0.4572 (7) | 0.27824 (19) | −0.03525 (11) | 0.0269 (7) | |
| H15 | 0.4911 | 0.3342 | −0.0504 | 0.032* | |
| C16 | 0.5614 (7) | 0.26028 (18) | 0.01715 (11) | 0.0232 (6) | |
| H16 | 0.6708 | 0.3044 | 0.0380 | 0.028* | |
| O1W | 0.0566 (5) | 0.23633 (12) | 0.20098 (8) | 0.0287 (5) | |
| H1W | 0.262 (3) | 0.2503 (17) | 0.2111 (12) | 0.043* | |
| H2W | −0.086 (5) | 0.2775 (13) | 0.2069 (12) | 0.043* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0381 (4) | 0.0386 (5) | 0.0191 (4) | −0.0007 (3) | −0.0065 (3) | −0.0001 (3) |
| F1 | 0.0475 (10) | 0.0422 (11) | 0.0205 (9) | 0.0086 (8) | −0.0082 (8) | 0.0037 (8) |
| N1 | 0.0218 (12) | 0.0225 (13) | 0.0200 (13) | 0.0000 (10) | −0.0006 (9) | −0.0006 (10) |
| N2 | 0.0269 (12) | 0.0218 (13) | 0.0177 (12) | −0.0046 (10) | −0.0046 (9) | 0.0001 (10) |
| N3 | 0.0219 (12) | 0.0265 (14) | 0.0153 (12) | 0.0018 (10) | −0.0022 (9) | 0.0005 (10) |
| C1 | 0.0185 (14) | 0.0204 (16) | 0.0286 (17) | 0.0005 (11) | −0.0024 (12) | −0.0026 (13) |
| C2 | 0.0204 (14) | 0.0270 (16) | 0.0156 (14) | 0.0007 (12) | −0.0052 (11) | −0.0005 (12) |
| C3 | 0.0150 (13) | 0.0189 (15) | 0.0195 (14) | 0.0014 (11) | −0.0008 (10) | −0.0011 (12) |
| C4 | 0.0151 (13) | 0.0168 (14) | 0.0209 (14) | 0.0033 (11) | −0.0002 (10) | −0.0018 (12) |
| C5 | 0.0204 (14) | 0.0189 (15) | 0.0237 (15) | 0.0012 (11) | 0.0001 (11) | −0.0018 (12) |
| C6 | 0.0242 (15) | 0.0210 (16) | 0.0208 (15) | −0.0003 (12) | −0.0035 (11) | −0.0059 (12) |
| C7 | 0.0204 (14) | 0.0276 (17) | 0.0175 (14) | 0.0030 (12) | −0.0016 (11) | −0.0034 (12) |
| C8 | 0.0198 (14) | 0.0223 (16) | 0.0220 (15) | 0.0050 (12) | −0.0007 (11) | 0.0046 (12) |
| C9 | 0.0154 (13) | 0.0149 (14) | 0.0233 (15) | 0.0003 (11) | −0.0026 (10) | −0.0030 (12) |
| C10 | 0.0222 (15) | 0.0215 (16) | 0.0221 (16) | 0.0019 (12) | 0.0000 (11) | −0.0046 (13) |
| C11 | 0.0185 (14) | 0.0253 (16) | 0.0209 (15) | 0.0030 (12) | 0.0004 (11) | 0.0010 (12) |
| C12 | 0.0261 (15) | 0.0194 (15) | 0.0238 (16) | −0.0004 (12) | 0.0010 (12) | −0.0001 (12) |
| C13 | 0.0240 (15) | 0.0241 (16) | 0.0229 (16) | 0.0009 (12) | −0.0018 (12) | −0.0045 (13) |
| C14 | 0.0273 (15) | 0.0350 (18) | 0.0145 (14) | 0.0051 (13) | −0.0025 (11) | 0.0012 (13) |
| C15 | 0.0265 (15) | 0.0256 (17) | 0.0287 (17) | 0.0025 (13) | −0.0004 (12) | 0.0043 (13) |
| C16 | 0.0244 (15) | 0.0240 (16) | 0.0213 (15) | −0.0016 (12) | −0.0012 (11) | −0.0037 (13) |
| O1W | 0.0255 (11) | 0.0227 (11) | 0.0377 (13) | −0.0003 (9) | −0.0020 (9) | −0.0045 (9) |
Geometric parameters (Å, °)
| Cl1—C7 | 1.745 (3) | C6—H6 | 0.9500 |
| F1—C14 | 1.367 (3) | C7—C8 | 1.362 (4) |
| N1—C1 | 1.331 (3) | C8—C9 | 1.414 (3) |
| N1—C9 | 1.377 (3) | C8—H8 | 0.9500 |
| N2—C3 | 1.356 (3) | C10—C11 | 1.458 (4) |
| N2—N3 | 1.378 (3) | C10—H10 | 0.9500 |
| N2—H2N | 0.8800 | C11—C16 | 1.389 (4) |
| N3—C10 | 1.284 (3) | C11—C12 | 1.398 (4) |
| C1—C2 | 1.390 (4) | C12—C13 | 1.379 (4) |
| C1—H1 | 0.9500 | C12—H12 | 0.9500 |
| C2—C3 | 1.385 (4) | C13—C14 | 1.379 (4) |
| C2—H2 | 0.9500 | C13—H13 | 0.9500 |
| C3—C4 | 1.445 (3) | C14—C15 | 1.369 (4) |
| C4—C5 | 1.415 (4) | C15—C16 | 1.387 (4) |
| C4—C9 | 1.416 (4) | C15—H15 | 0.9500 |
| C5—C6 | 1.369 (3) | C16—H16 | 0.9500 |
| C5—H5 | 0.9500 | O1W—H1W | 0.841 (10) |
| C6—C7 | 1.401 (4) | O1W—H2W | 0.845 (10) |
| C1—N1—C9 | 116.2 (2) | C7—C8—H8 | 120.0 |
| C3—N2—N3 | 118.9 (2) | C9—C8—H8 | 120.0 |
| C3—N2—H2N | 120.5 | N1—C9—C8 | 117.2 (2) |
| N3—N2—H2N | 120.5 | N1—C9—C4 | 123.6 (2) |
| C10—N3—N2 | 116.3 (2) | C8—C9—C4 | 119.2 (2) |
| N1—C1—C2 | 125.2 (3) | N3—C10—C11 | 121.6 (2) |
| N1—C1—H1 | 117.4 | N3—C10—H10 | 119.2 |
| C2—C1—H1 | 117.4 | C11—C10—H10 | 119.2 |
| C3—C2—C1 | 119.8 (2) | C16—C11—C12 | 118.7 (2) |
| C3—C2—H2 | 120.1 | C16—C11—C10 | 119.3 (2) |
| C1—C2—H2 | 120.1 | C12—C11—C10 | 122.0 (3) |
| N2—C3—C2 | 122.4 (2) | C13—C12—C11 | 120.8 (3) |
| N2—C3—C4 | 119.8 (2) | C13—C12—H12 | 119.6 |
| C2—C3—C4 | 117.7 (2) | C11—C12—H12 | 119.6 |
| C5—C4—C9 | 118.5 (2) | C12—C13—C14 | 118.3 (3) |
| C5—C4—C3 | 124.0 (2) | C12—C13—H13 | 120.9 |
| C9—C4—C3 | 117.4 (2) | C14—C13—H13 | 120.9 |
| C6—C5—C4 | 121.3 (3) | F1—C14—C15 | 118.8 (3) |
| C6—C5—H5 | 119.3 | F1—C14—C13 | 118.3 (2) |
| C4—C5—H5 | 119.3 | C15—C14—C13 | 123.0 (3) |
| C5—C6—C7 | 119.2 (2) | C14—C15—C16 | 118.0 (3) |
| C5—C6—H6 | 120.4 | C14—C15—H15 | 121.0 |
| C7—C6—H6 | 120.4 | C16—C15—H15 | 121.0 |
| C8—C7—C6 | 121.6 (2) | C15—C16—C11 | 121.2 (3) |
| C8—C7—Cl1 | 119.6 (2) | C15—C16—H16 | 119.4 |
| C6—C7—Cl1 | 118.8 (2) | C11—C16—H16 | 119.4 |
| C7—C8—C9 | 120.0 (2) | H1W—O1W—H2W | 110.0 (16) |
| C3—N2—N3—C10 | 176.0 (2) | C7—C8—C9—N1 | −179.5 (2) |
| C9—N1—C1—C2 | 0.5 (4) | C7—C8—C9—C4 | 0.3 (4) |
| N1—C1—C2—C3 | 1.3 (4) | C5—C4—C9—N1 | 179.6 (2) |
| N3—N2—C3—C2 | 6.9 (4) | C3—C4—C9—N1 | 0.7 (4) |
| N3—N2—C3—C4 | −172.4 (2) | C5—C4—C9—C8 | −0.2 (3) |
| C1—C2—C3—N2 | 178.6 (2) | C3—C4—C9—C8 | −179.1 (2) |
| C1—C2—C3—C4 | −2.0 (4) | N2—N3—C10—C11 | −178.2 (2) |
| N2—C3—C4—C5 | 1.6 (4) | N3—C10—C11—C16 | 173.3 (2) |
| C2—C3—C4—C5 | −177.7 (2) | N3—C10—C11—C12 | −6.6 (4) |
| N2—C3—C4—C9 | −179.5 (2) | C16—C11—C12—C13 | 0.5 (4) |
| C2—C3—C4—C9 | 1.1 (3) | C10—C11—C12—C13 | −179.5 (2) |
| C9—C4—C5—C6 | −0.1 (4) | C11—C12—C13—C14 | −0.8 (4) |
| C3—C4—C5—C6 | 178.7 (2) | C12—C13—C14—F1 | −179.2 (2) |
| C4—C5—C6—C7 | 0.4 (4) | C12—C13—C14—C15 | 0.2 (4) |
| C5—C6—C7—C8 | −0.3 (4) | F1—C14—C15—C16 | 180.0 (2) |
| C5—C6—C7—Cl1 | 178.65 (19) | C13—C14—C15—C16 | 0.5 (4) |
| C6—C7—C8—C9 | 0.0 (4) | C14—C15—C16—C11 | −0.8 (4) |
| Cl1—C7—C8—C9 | −178.98 (18) | C12—C11—C16—C15 | 0.2 (4) |
| C1—N1—C9—C8 | 178.3 (2) | C10—C11—C16—C15 | −179.7 (2) |
| C1—N1—C9—C4 | −1.5 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1w—H1w···N1i | 0.843 (16) | 2.28 (2) | 2.999 (3) | 144 (2) |
| O1w—H2w···N1ii | 0.85 (2) | 1.93 (2) | 2.761 (3) | 166 (3) |
| N2—H2n···O1wiii | 0.88 | 2.01 | 2.865 (3) | 165 |
| C5—H5···O1wiii | 0.95 | 2.45 | 3.379 (3) | 164 |
| C10—H10···O1wiii | 0.95 | 2.50 | 3.302 (3) | 142 |
| C1—H1···F1iv | 0.95 | 2.56 | 3.399 (3) | 147 |
| C6—H6···F1v | 0.95 | 2.56 | 3.477 (3) | 161 |
Symmetry codes: (i) −x+1, y+1/2, −z+1/2; (ii) −x, y+1/2, −z+1/2; (iii) x+1, y, z; (iv) −x, −y, −z; (v) x+1, −y+1/2, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: LH2970).
References
- Andrade, A. A., Varotti, F. D., de Freitas, I. Q., de Souza, M. V. N., Vasconcelos, T. R. A., Boechat, N. & Krettli, A. U. (2007). Eur J Pharm 558, 194–198. [DOI] [PubMed]
- Brandenburg, K. (2006). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Cunico, W., Cechinel, C. A., Bonacorso, H. G., Martins, G. M. A. P., Zanetta, N., de Souza, M. V. N., Freitas, I. Q., Soares, R. P. P. & Krettli, A. U. (2006). Bioorg Med Chem Lett 16, 649–653. [DOI] [PubMed]
- Elslager, E. F., Tendick, F. H. & Werbel, L. M. (1969). J Med Chem 12, 600–607. [DOI] [PubMed]
- Ferreira, M. L. de, de Souza, M. V. N., Howie, R. A., Tiekink, E. R. T., Wardell, J. L. & Wardell, S. M. S. V. (2009). Acta Cryst. E65, o3239–o3240. [DOI] [PMC free article] [PubMed]
- Font, M., Monge, A., Ruiz, I. & Heras, B. (1997). Drug Des. Disc.14, 259–272. [PubMed]
- Hooft, R. W. W. (1998). COLLECT Nonius BV, Delft, The Netherlands.
- Kaiser, C. R., Pais, K. C., de Souza, M. V. N., Wardell, J. L., Wardell, S. M. S. V. & Tiekink, E. R. T. (2009). CrystEngComm, 11, 1133–1140.
- Kaminsky, D. & Meltzer, R. I. (1968). J Med Chem 11, 160–163. [DOI] [PubMed]
- Musiol, R., Jampilek, J., Buchta, V., Silva, L., Halina, H., Podeszwa, B., Palka, A., Majerz-Maniecka, K., Oleksyn, B. & Polanski, J. (2006). Bioorg Med Chem 14, 3592–3598. [DOI] [PubMed]
- Nakamura, T., Oka, M., Aizawa, K., Soda, H., Fukuda, M., Terashi, K., Ikeda, K., Mizuta, Y., Noguchi, Y., Kimura, Y., Tsuruo, T. & Kohno, S. (1999). Biochem Biophys Res Commun 255, 618–624. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Palmer, K. J., Holliday, S. M. & Brogden, R. N. (1993). Drugs, 45, 430–475. [DOI] [PubMed]
- Pellerano, C., Savini, L. & Fiorini, I. (1976). Atti Accad Fisiocritic Siena, 8, 43–57.
- Ridley, R. G. (2002). Nature (London), 415, 686–693. [DOI] [PubMed]
- Sheldrick, G. M. (2007). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Silva, A. D. da, de Almeida, M. V., de Souza, M. V. N. & Couri, M. R. C. (2003). Curr Med Chem 10, 21–39. [DOI] [PubMed]
- Sloboda, A. E., Powell, D., Poletto, J. F., Pickett, W. C., Gibbons, J. J., Bell, D. H., Oronsky, A. L. & Kerwar, S. S. (1991). J Rheumatol 18, 855–860. [PubMed]
- Souza, M. V. N. de (2005). Mini-Rev Med Chem 5, 1009–1017.
- Souza, M. V. N. de, Tiekink, E. R. T., Wardell, J. L. & Wardell, S. M. S. V. (2009). Acta Cryst. E65, o3120–o3121. [DOI] [PMC free article] [PubMed]
- Tanenbaum, L. & Tuffanelli, D. L. (1980). Arch Dermatol 116, 587–591. [DOI] [PubMed]
- Warshakoon, N. C., Sheville, J., Bhatt, R. T., Ji, W., Mendez-Andino, J. L., Meyers, K. M., Kim, N., Wos, J. A., Mitchell, C., Paris, J. L., Pinney, B. B. O., Reizes, O. & Hu, X. E. (2006). Bioorg Med Chem Lett 16, 5207–5211. [DOI] [PubMed]
- Westrip, S. P. (2009). publCIF In preparation.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809053367/lh2970sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809053367/lh2970Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report