Abstract
The ability to induce pluripotent stem cells from committed, somatic, human cells provides tremendous potential for regenerative medicine. However, there is a defined neoplastic potential inherent to such reprogramming that must be understood and may provide a model for understanding key events in tumorigenesis. Using genome wide assays we identify cancer-related epigenetic abnormalities that arise early during reprogramming and persist in induced pluripotent stem cell (iPS) clones. These include hundreds of abnormal gene silencing events, patterns of aberrant responses to epigenetic modifying drugs resembling those for cancer cells, and presence in iPS and partially reprogrammed cells of cancer-specific, gene promoter, DNA methylation alterations. Our findings suggest that by studying the process of induced reprogramming we may gain significant insight into the origins of epigenetic gene silencing associated with human tumorigenesis and add to means of assessing iPS for safety.
Keywords: reprogramming, induced pluripotent stem cells (iPS), embryonic stem cells (ESC), DNA methylation, chromatin, cancer
INTRODUCTION
Induction of pluripotent stem cells (iPS) from committed, somatic cells encompasses exciting biology with tremendous potential for regenerative medicine (1, 2). However, such cells are currently generated using multiple inducing factors with oncogenic potential (3–6) and mice generated with iPS have increased tumorigenicity and mortality (7). Moreoever, fully reprogrammed human iPS phenotype arise only as rare clonal populations (0.1% or less) among partially reprogrammed cells displaying an immortalized phenotype (8). Thus, many new metrics are needed to characterize iPS (4, 9).
Induction of iPS from committed cells necessitates that a myriad of epigenetic parameters must be reversed and properly re-established. Genome wide studies of chromatin and DNA methylation patterns (10, 11) indicate that this is globally done remarkably well including required reversal of promoter DNA methylation and re-expression of pluripotency related genes such as OCT4 (POU5F1), both of which mark successful reprogramming (10, 12). However, multiple loci can fail in this reversal of DNA methylation (10, 12) and use of drug induced DNA demethylation can improve efficiency of obtaining iPS (10, 13, 14). Another implication of such abnormal loci is that the relatively short duration of the reprogramming process (2–3 weeks) (1, 2, 4, 10) might implicate epigenetic mechanisms in the neoplastic potential of iPS. A recent study compared iPS to cancer (15) but the most cancer specific, proximal promoter epigenetic changes accompanying abnormal gene expression were not outlined. We now address this issue using genome-wide analysis approaches to match gene silencing associated with DNA hypermethylation of normally unmethylated CpG island-containing promoters (16). We identify cancer-related epigenetic abnormalities associated with the timing, and degree, of inducing cellular reprogramming.
Methods and Materials
Reprogramming protocols
MP2 and MP4 cells were reprogrammed from IMR90 fibroblasts by lentiviral vectors and retroviral vectors were used for all other protocols (17).
Expression Data
Global gene expression was analyzed using Agilent whole human genome, 4×44K, microarrays as previously described (18). Heat map color schemes are based on hierarchical clustering by Ward’s algorithm and standard Euclidean distance on log transformed expression measures adjusted for cutoffs for silent (red) and active (green) genes as previously determined (19).
Genome Wide DNA Methylation Analysis
We used the Infinium (Illumina, Inc., San Diego, CA) platform(20) to analyze bisulfite treated DNA (EZ DNA Methylation Kit, Zymo Research, Orange, CA). In this hybridization procedure, β-values are generated as the signal of methylation specific probe over the sum of the signals of the methylation and unmethylated specific probes and assigning a score of 1.0 for full methylation of a specific CpG site, 0 for absence of methylation, and 0 <= β <= 1 for all signals between (21). Probes with poor overall signals (p > 0.05) were removed from analysis. For all genes, only probes positioned from −1000 to +200 bp around transcription start (TSS) are analyzed. In vitro, DNA methylated, genomic DNA (IVD) and DNA from DKO cells genetically deleted for DNA methyltransferases 1 and 3b (22), serve as fully methylated and unmethylated controls respectively. Heat maps are based on hierarchical clustering of β-values using Euclidean distance and Ward’s algorithm and all probes were mapped to the genome (NCBI36.3) using the bowtie algorithm and ultrafast and memory-efficient alignment of short DNA sequences (Genome Biology, 10, R25) with genome annotation via the matching release of the EnsEMBL database. X-linked genes were removed from analyses.
Drug Responses
Drug responsive genes were selected from Agilent expression arrays (see supplemental Fig. 3), based on a 1.41-fold expression (.5 on a log2 scale) difference between mock versus 5-aza-2′-deoxycytidine (DAC) or trichostatin (TSA) treated samples and classified as responsive to DAC alone but not TSA, to TSA alone but not DAC, and to both DAC alone and TSA alone.
RESULTS
Characteristics of reprogrammed human cell lines
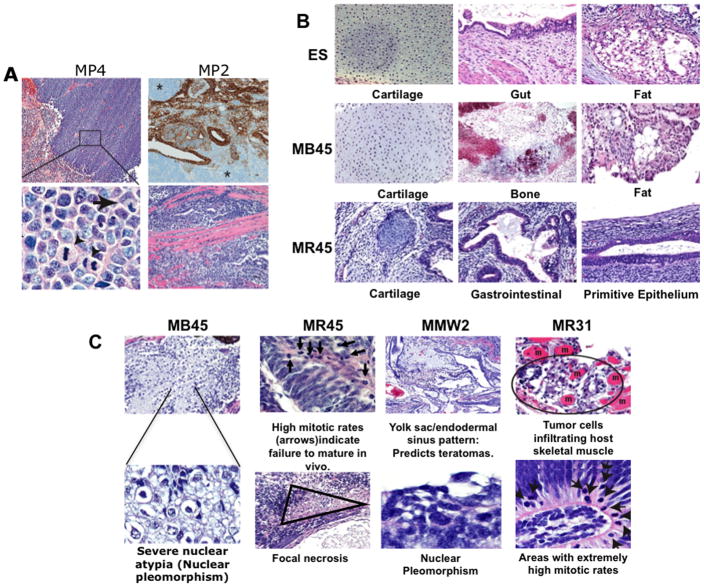
We first examined tumor xenografts from reprogrammed clones (Supplementary Table 1a), for pluripotent potential and neoplastic features. The highest cancer potential was for clone MP4, a fibroblast line induced by lentiviral introduction of OCT4, SOX2, NANOG, LIN28 and SV40 T-antigen(OSNLT)(1, 17), and known to be only partially reprogrammed (17). MP4 cells express undifferentiated markers such as OCT4, but not TRA-1–60, are refractory to differentiation induction in vitro and in vivo (i.e., nullipotent), and possess an abnormal karyotype. Mouse xenografts demonstrate high nuclear to cytoplasmic ratio, an extremely high mitotic rate, areas of necrosis, and the histology of a primitive, aggressive, mesenchymal tumor (Fig. 1a). In comparison, clone MP2, prepared identically to MP4 and a typical iPS expressing TRA-1–60 and all classical iPS markers (17), forms what appears to be a benign, multi-lineage teratoma (Fig. 1a, upper right panel). However, this clone has an abnormal karyotype and upon closer examination, the teratoma contains small foci suggesting malignancy, including regions where cells infiltrate host skeletal muscle bundles (Fig. 1a, lower right panel).
Figure 1. Features of teratomas from reprogrammed cells.
a. Immunohistochemical stains (upper right) for epithelial cytokeratins or H&E stained sections from MP4 (left, upper and lower panels) and MP2 tumors (right, upper and lower panels). MP4 is positive only for the mesenchymal marker, vimentin (data not shown) and has areas of necrosis (pink area to the left – upper left panel), a malignant feature. Higher magnification (lower left panel, original magnification 600×) shows multiple mitotic figures (arrowheads) and abnormal nuclear shapes. MP2 ( upper right panel) exhibits alternating areas of epithelial and mesenchymal differentiation, as evidenced by cytokeratin staining (brown) but has small areas with cells infiltrating host skeletal muscle bundles (pink staining -lower right panel). b. Differentiated tissues in tumors from H9 ESC and iPS clones, MB45 and MR45, as labeled. c. Morphologic correlates of malignancy in iPS clones (from left to right, MB45, MR45, MMW2, and MR31) including severe nuclear atypia, host muscle infiltration, abnormally high mitotic rates, and necrosis..
We then performed blinded comparison (BS and DMB) of teratomas (Fig. 1 and Supplementary table 1b) from the conventional embryonic stem cell ( ESC) lines, H1, H9, and SC233 versus from multiple additional pluripotent iPS with normal karyotypes and metrics of fully reprogrammed human iPS (23), including expression of the embryonic markers, AP, OCT4, and TRA-1–60. These include, MR45 and MR46, generated from fibroblasts by retrovirally introducing OCT4, SOX2, KLF4 and c-MYC (OSKM)(2), MB41, MB45, MMW1, and MMW2 generated with the same retroviral vectors but from mesenchymal stem cells (MSCs), and two iPS lines (MR31 and MR32) retrovirally derived from IMR90 fibroblasts using 3 factors (OSK, without c-MYC). All formed mouse xenografts with differentiated cell types of multiple embryonic germ layers (Fig. 1b). However, the successfully reprogrammed iPS cell xenografts show a range of degree of maturation (from .067 to .231 lineage structures/ areas examined) of defined structures such as cartilage, bone, intestine, etc. (Fig. 1c), that is lower than the range of values (0.338–0.97) for three ESC derived teratomas (Fig. 1b, 1c –Supplemental Table 1b). Most importantly, all iPS teratomas examined have foci with malignant-like characteristics which include focal necrosis, nuclear pleomorphism, aberrantly high mitotic rates, and infiltration into the mouse musculature (Examples shown in Fig. 1c; Supplemental table 1b). Such foci were generally not found in the three ESC derived teratomas (Supplemental table 1b), save for one small area of focal necrosis in the teratoma derived from H1 ESC.
Overall Gene Expression profiles of reprogrammed clones
We compared, using full transcriptome Agilent arrays, global gene expression between representative reprogrammed clones versus ESC’s and found, as have others (10), highly similar, but not identical, patterns. Even the partially reprogrammed MP4 clone deviated only slightly from the pluripotent clones and ESC and clustered separately from four human cancer cell lines (Supplementary Fig. 1). However, focus upon genes highly expressed in ESC revealed distinct differences. Compared to parent cells, pluripotent clones MR46 and MR45 significantly up-regulated to levels in ESC, 12 of 16 and 13 of 16 such genes, respectively (Supplementary Table 3), MP2 increased only 11 of 16 above levels in parent fibroblasts, and only 8 to levels in ESC. The malignant, partially reprogrammed, MP4 properly increased only 5 of the 16 genes with respect to starting fibroblasts and only 6 reached levels in ESC. Also, partially reprogrammed clone, MP4, expressed both OCT4 and c-MYC, due to incomplete repression of introduced transgenes, significantly higher than in ESC (Supplementary Table 3).
Abnormal gene silencing, and gene responses to epigenetic modifying agents, associated with cellular reprogramming
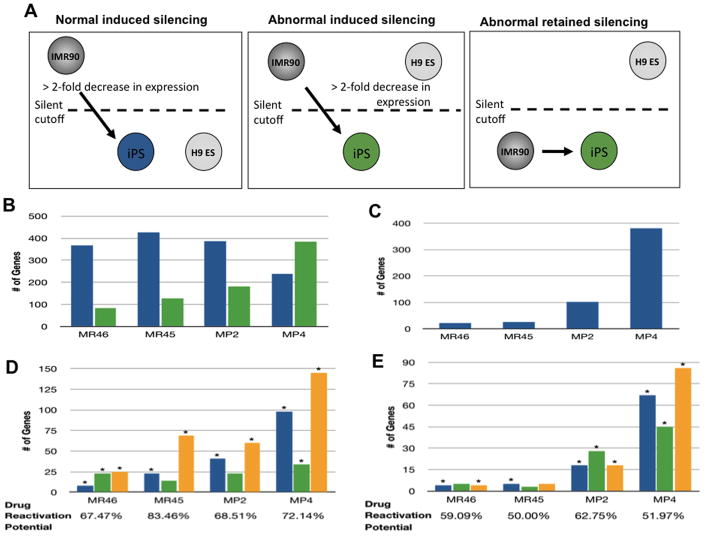
We next linked, in reprogrammed clones, gene expression to epigenetic aspects of neoplasia, with respect to abnormal gene silencing events during reprogramming. In pluripotent clones, MR46, MR45, and MP2, normal silencing events (Fig. 2a, left panel) dominate over abnormal silencing events for both genes that should be silent in fibroblasts but activated in ESC, or which should be active in both cell types (Fig 2a middle and right panels) while the opposite is true for the tumorigenic, MP4 cells (Fig 2b, c – Supplementary Table 4). However, while MP4 has the highest number of abnormal silencing events (~800), even the best performing iPS clone, MR46, has ~ 100 such genes (Fig. 2b, c, combined; Supplementary Table 4). These numbers may be characteristic of iPS since analysis of the best reprogrammed mouse iPS clone (MCV8.1, subclone 8.1(24) from a recent study of Mikkelsen et. al. (10) revealed 418 abnormally silenced genes (Supplementary Fig. 2).
Figure 2. Abnormal epigenetic gene silencing during reprogramming.
a. Induced silencing genes (left and middle panels) have basal expression in reprogrammed clones > 2 fold reduced when compared to the IMR90 parental line, and below an arbitrary silent cutoff point determined for the microarray in previous studies(19). This silencing is normal (left panel) if expression levels in the reprogrammed clone are within .5 log (1.4 fold) of that observed in H9 ES cells and abnormal if expression in the iPS clone differs by greater than .5 logs (1.4 fold) from ESC cells. Abnormal retained silencing (far right panel) = genes silent in the fibroblasts and iPS clone which should have been activated to within .5 log of the expression in ESC. b. Normal (blue bars) and abnormal (green bars) induced silencing genes in MR46, MR45, MP2 and MP4 clones. c. Abnormally retained silenced genes in MR46, MR45, MP2, and MP4 cells. d. Response of abnormally silenced genes to DAC and TSA (from Fig. 2b) in MR46, MR45, MP2, and MP4 clones. Blue bars = reactivated by DAC alone, green bars = reactivated by TSA alone, orange bars = activated by either DAC or TSA alone. (*) = significant enrichment, p-value < 0.05, Fisher exact test, as compared to expected values for the entire genome. e. Drug responsiveness of genes with abnormal retained silencing (from Fig. 2c) in MR46, MR45, MP2 and MP4. Color coding and statistical testing as in figure 2d.
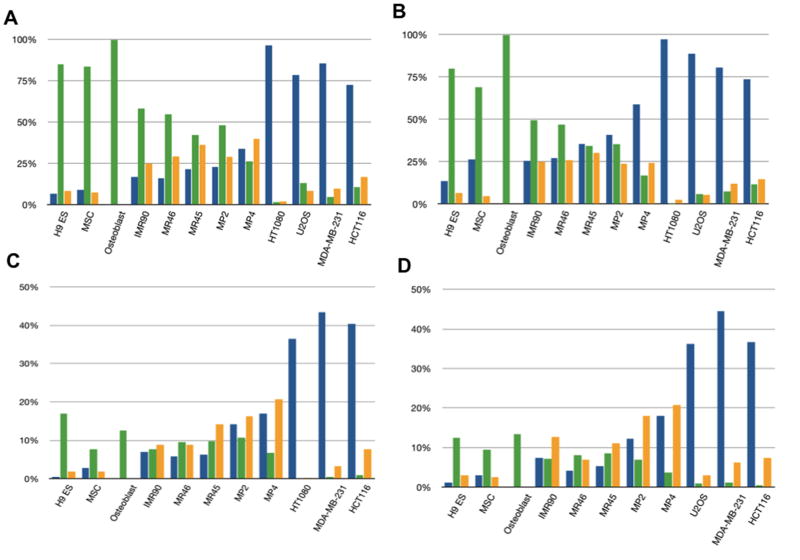
In cancer, DNA hypermethylation of CpG island promoters (16) is a prime candidate for mediating abnormal gene silencing and several abnormally silenced genes in reprogrammed clones are known to have such changes including CDKN2B (Suppl Ref. 1), LXN (Suppl Ref. 2, 3), TIMP3 (Suppl Refs. 2, 4), and PYCARD (Suppl Ref. 5). We thus investigated global gene expression responses to both the DNA demethylation agent, DAC and the histone deactylase inhibitor, TSA. DAC effectively re-expresses genes in cancer with densely hypermethylated, promoter CpG islands while TSA, alone, does not (18)(see supplemental Fig. 3 for array expression responses). The results strikingly separate cancer cells, iPS cells, and especially the partially reprogrammed MP4 clone, from parent fibroblasts, adult MSC, and ESC. Addition of either DAC or TSA (Fig. 2d) reactivates between 67% (MR46) and 84% (MR45) of the abnormal silenced genes in Fig. 2a. A dramatic finding is that, in partially reprogrammed clone MP4, and to a lesser degree other iPS (Fig 2d), more induced silent genes are responsive only to DAC alone, or both DAC and TSA (left to right, Fig. 2d), but not to TSA alone and this is also true, to a slightly less extent (Fig. 2e), for abnormal retained silencing genes (those in Fig. 2a far right panel). This is true for both CpG island and non-CpG island containing genes (Fig. 3a,b respectively). The pattern for MP4 cells begins to resemble that for the cancer cell lines, which have an extraordinary dominance of DAC alone versus TSA alone, responsive genes. Moreoever, genes in normal ESC, adult MSC, and committed bone progenitor cells (osteoblasts), have a starkly dominant response to TSA alone (Fig. 3a, b). These patterns also hold when~ 400 CpG island containing genes responsive to DAC alone in the U2OS osteosarcoma cells (Fig. 3c) and HT1080 fibrosarcoma cells (Fig. 3d) serve as their own controls - ie, for these genes, TSA responses dominate in normal cells, DAC responses dominate in cancer cells, and the reprogrammed clones show a mixed response to DAC and TSA consisting of a predominantly DAC response most apparent in the MP4 clone. Finally normal committed IMR90 fibroblasts have more of a mixture of DAC and TSA responsive genes, but with responses still skewed towards TSA (Fig 3a, b) and the increased frequency of DAC responsive genes even separates all of the fully reprogrammed clones from ESC and adult MSC (Fig. 3a, b).
Figure 3. DAC and TSA responsive genes in reprogrammed clones.
a. Expression response of all silent CpG island containing genes to DAC (blue bars), TSA (green bars), or both DAC and TSA alone (orange bars). b. Response of all silent, non-CpG island containing genes as in 3a. c. Overall drug responsiveness of 430 silent genes for U2OS, i.e. which are not expressed in this cell line, responsive to DAC alone, but not TSA alone, in the other cell lines shown. Color coding as in 3a and 3b. d. Same analysis as in Fig. 3c, for the 435 silent genes responsive to DAC alone in HT1080 fibrosarcoma cells.
Taken together, the data indicate that re-programmed somatic cells can acquire both abnormal gene silencing events and aberrant responses to epigenetic modifying drugs that deviate from normal and are reminiscent of changes observed in cancer cells.
Cancer-related promoter DNA methylation changes arise, early, during reprogramming
To further examine abnormal gene silencing and cancer specific promoter, DNA methylation on a global scale, we used the Infinium platform(25) which queries ~27,000 CpG sites, most, but not all, in annotated, promoter, CpG islands. When focused on analyzing CpG’s located from −1,000 to +200bp from gene transcription start, we find, as have others in genome – wide DNA methylation studies (10, 26, 27), that the vast majority (~ 90%), of ~ 7500 loci in ~ 5700 different genes with well annotated, promoter, CpG islands, are unmethylated in all normal cells (ESC, MSC, fibroblasts -Supplemental Fig 4). In contrast, for ~3,750 probes in ~ 800 autosomal genes not containing dense CpG island promoters, CpG poor promoters are far more methylated, with a varied tissue pattern, in normal cells (Supplemental Fig. 5), consistent with studies of others(12, 26) and verifying a long held biological premise. Finally, consistent with our previous studies (18), gene promoter CpG island hypermethylation in cancer is starkly apparent (Supplemental Fig 4). The number of unmethylated CpG island promoters falls to ~ 78% in 4 adult cancer cell lines and ~ 87% in two germ line teratocarcinoma lines representing 900 hypermethylated genes in the former and over 200 in the latter. Non-CpG island promoters have not been as carefully examined in cancer but the cancer cell lines still cluster separately from normal cells largely because of multiple genes that have gained methylation but also due to many that have lost normal methylation (Supplemental Fig. 5). Importantly, for cancer cell lines such as HCT 116 colorectal cells, the Infinium analyses correctly identified (data not shown) ~90% of genes validated in our laboratory to have cancer specific DNA hypermethylation(18).
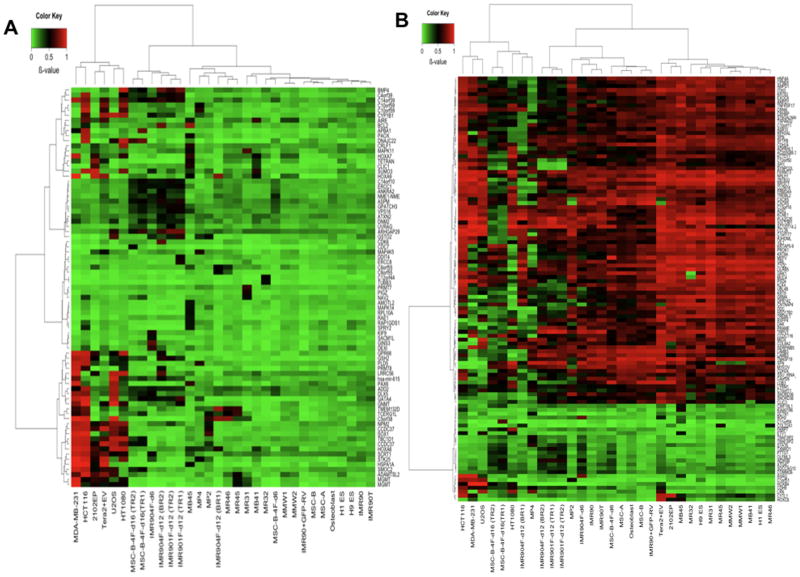
With this above background, we examined not only our reprogrammed cell clones, but also pools of cells (Supplementary Table 1 for listing) from early stages (days 6 to 18) following insertion of four factors into both our fibroblast and MSC parent cells and after introduction of OCT4 alone into fibroblasts. In these pools, cells are in a dynamic, meta-stable, state with a wide range of reprogramming stages (10, 28) wherein most clones do not exhibit ESC morphology and many cells can be cultured indefinitely. Overall, cells in these pools maintain the normally unmethylated state of CpG island promoters, harboring many less abnormalities than seen in cancer cells (Supplemental Fig 4). However, we observe that among normally unmethylated CpG island genes, 50 show abnormal methylation in one or more of the early cell pools by day 6, and 38 have abnormally added methylation in individual reprogrammed clones (Fig. 4a -individual clone genes in Table 1 and pool genes, Supplemental table 5). While the genes generally differ between the pools and clones, ~15% are shared between the two. Of 10 clones examined, including partially reprogrammed MP4 cells, only 2 (MMW1 and MMW2) failed to exhibit a hypermethylated gene while the remainder contained between 3 (MR46) and 17 (MB45). Significantly, over 60% of all these genes (p = 3.055e-07; Fisher’s exact test) in the clones and 50% (p = 7.302e-05; Fisher’s exact test) in the pools are also hypermethylated in one or more cancer cell lines (Fig. 4a; individual genes, Table 1 and Suppl. Table 5). Finally, and very importantly, even pooled cells with introduction of OCT4 alone contained from 12 to 16 hypermethylated genes, and, again, ~ 50% of these were also hypermethylated in at least one cancer cell line (Fig. 4a; Suppl. Table 5).
Figure 4. DNA methylation analysis of all cells studied.
a. Heat map for methylation s-scores from the Infinium platform (color bar at the top left, green = lowest methylation and red= highest methylation) for all aberrantly methylated genes with a CpG island. Upper x-axis = sample clustering; lower x-axis = samples; left y-axis = gene clustering; right y axis = individual genes (those hypermethylated in iPS clones and/or reprogrammed pools are also designated in Table 1 and Supplemental Table 4, respectively). Samples analyzed are: MR46, MR45, MP2 and MP4 clones; parent IMR90fibroblasts; parent adult MS ( MSC-A and MSC-B); adult osteoblasts; ESC lines H1ES and H9ES, cancer cell lines (U2OS, HT1080, MDA-MB-231, HCT 116), reprogrammed clones(MB41, MB45, MMW1, MMW2) derived from MSC using retrovirally introduced OCT4, SOX2, KLF4, and c-MYC (OSKM); two iPS lines (MR31 and MR32) retrovirally derived from IMR90 fibroblasts using 3 factors (OSK, without c-MYC), teratocarcinoma lines, Tera2+EV (empty plasmid vector) and 2102EP; reprogramming pools of IMR90 cells at days 6, 12 and 18( TR= technical repeat; BR = biological repeat ) after retroviral infection using the OSKM cocktail (IMR904F-d6,-d12, and −d18 respectively) and at day 12 after introduction of OCT4 alone (IMR901F-d12); pools of reprogramming cells using adult MSC as the parental line, and after 6 and 18 days for OSKM introduction (MSC-B-4F-d6 and MSC-B-4F-d16 - TR and BR = as above), parent IMR90 cells infected by lentivirus with T-antigen alone (IMR90T), IMR90 infected with GFP (IMR90+GFP-RV), IMR90 cells infected with 4 factors grown for only 6 days (IMR904F-d6). b. Heat map for non-CpG island genes that either gained or lost methylation in clones or reprogramming pools relative to all normal cell types. All samples as for a, and abnormal iPS and/or re-programmed pool genes are in suppl Tables 5 and 6, respectively.
Table 1.
Genes with abnormally methylated CpG island promoters in clones, partially reprogrammed pools, or cancers. Dark squares = beta values above 0.45; light gray squares = values ranging to 0.45.
| chromosome | Location | Gene Name | dTSS | Normals (6 samples) | IMR904F-d6 | MSC-B-4F-d6 | IMR901F-d12 (TR1) | IMR901F-d12 (TR2) | IMR904F-d12 (BR1) | IMR904F-d12 (BR2) | MSC-B-4F-d16 (TR1) | MSC-B-4F-d16 (TR2) | iPS (10 samples) | Cancer (6 cell lines) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 70849059 | ADD2 | −197 | 0.15–0.22 | 0.46 | 0.35 | 0.37 | 0.41 | 0.29 | 0.4 | 0.39 | 0.39 | 0.09–0.32 | 0.24–0.85 |
| 5 | 72897370 | ANKRA2 | −91 | 0.08–0.15 | 0.26 | 0.28 | 0.4 | 0.38 | 0.16 | 0.5 | 0.48 | 0.35 | 0.08–0.22 | 0.1–0.21 |
| 9 | 71476967 | APBA1 | 51 | 0.04–0.1 | 0.04 | 0.06 | 0.08 | 0.08 | 0.05 | 0.09 | 0.73 | 0.09 | 0.03–0.08 | 0.03–0.84 |
| 1 | 94475710 | ARHGAP29 | 43 | 0.11–0.22 | 0.3 | 0.26 | 0.75 | 0.76 | 0.28 | 0.64 | 0.64 | 0.46 | 0.12–0.28 | 0.11–0.58 |
| 1 | 195382611 | ASPM | −299 | 0.13–0.23 | 0.42 | 0.34 | 0.57 | 0.44 | 0.17 | 0.57 | 0.51 | 0.47 | 0.1–0.26 | 0.15–0.29 |
| 12 | 110521815 | ATXN2 | 24 | 0.15–0.24 | 0.23 | 0.19 | 0.33 | 0.43 | 0.14 | 0.48 | 0.32 | 0.25 | 0.12–0.28 | 0.14–0.28 |
| 18 | 59137867 | BCL2 | −249 | 0.08–0.23 | 0.2 | 0.13 | 0.19 | 0.16 | 0.08 | 0.74 | 0.14 | 0.14 | 0.07–0.83 | 0.07–0.43 |
| 14 | 53493207 | BMP4 | 48 | 0.06–0.17 | 0.2 | 0.11 | 0.75 | 0.62 | 0.21 | 0.74 | 0.5 | 0.42 | 0.1–0.24 | 0.05–0.2 |
| 14 | 60022515 | C14orf39 | 28 | 0.1–0.2 | 0.53 | 0.31 | 0.58 | 0.54 | 0.26 | 0.63 | 0.5 | 0.38 | 0.09–0.29 | 0.14–0.86 |
| 4 | 166097517 | C4orf39 | −84 | 0.05–0.11 | 0.25 | 0.15 | 0.59 | 0.54 | 0.09 | 0.47 | 0.39 | 0.35 | 0.04–0.13 | 0.07–0.84 |
| 5 | 2804517 | C5orf38 | −717 | 0.05–0.14 | 0.04 | 0.03 | 0.02 | 0.02 | 0.79 | 0.02 | 0.05 | 0.05 | 0.04–0.8 | 0.03–0.97 |
| 8 | 143805792 | C8orf55 | 144 | 0.03–0.05 | 0.03 | 0.04 | 0.03 | 0.04 | 0.5 | 0.04 | 0.04 | 0.05 | 0.04–0.28 | 0.03–0.07 |
| 8 | 143805240 | C8orf55 | −359 | 0.03–0.05 | 0.11 | 0.04 | 0.1 | 0.1 | 0.5 | 0.11 | 0.08 | 0.08 | 0.09–0.4 | 0.06–0.3 |
| 3 | 127596450 | CCDC37 | −47 | 0.06–0.15 | 0.04 | 0.1 | 0.07 | 0.05 | 0.09 | 0.13 | 0.48 | 0.47 | 0.06–0.39 | 0.53–0.91 |
| 1 | 91739228 | CDC7 | 171 | 0.02–0.07 | 0.03 | 0.02 | 0.06 | 0.06 | 0.02 | 0.09 | 0.09 | 0.49 | 0.01–0.05 | 0.01–0.06 |
| 7 | 92300941 | CDK6 | 183 | 0.05–0.14 | 0.08 | 0.08 | 0.11 | 0.11 | 0.06 | 0.7 | 0.15 | 0.14 | 0.03–0.15 | 0.04–0.15 |
| 2 | 38156747 | CYP1B1 | −49 | 0.06–0.24 | 0.46 | 0.06 | 0.14 | 0.36 | 0.07 | 0.33 | 0.1 | 0.09 | 0.05–0.32 | 0.05–0.89 |
| 10 | 73703385 | DDIT4 | −274 | 0.09–0.13 | 0.08 | 0.06 | 0.53 | 0.07 | 0.09 | 0.07 | 0.07 | 0.08 | 0.06–0.16 | 0.05–0.15 |
| 16 | 10944051 | DEXI | −209 | 0.09–0.23 | 0.61 | 0.05 | 0.1 | 0.09 | 0.04 | 0.26 | 0.09 | 0.07 | 0.02–0.23 | 0.04–0.29 |
| 7 | 96492095 | DLX5 | −40 | 0.14–0.2 | 0.52 | 0.33 | 0.23 | 0.24 | 0.28 | 0.16 | 0.44 | 0.37 | 0.11–0.2 | 0.18–0.83 |
| 12 | 48027131 | DNAJC22 | −153 | 0.04–0.17 | 0.03 | 0.18 | 0.03 | 0.04 | 0.02 | 0.13 | 0.73 | 0.65 | 0.02–0.19 | 0.01–0.91 |
| 19 | 10689824 | DNM2 | −40 | 0.12–0.18 | 0.33 | 0.23 | 0.44 | 0.42 | 0.23 | 0.46 | 0.46 | 0.24 | 0.1–0.27 | 0.11–0.39 |
| 19 | 50618615 | ERCC1 | −36 | 0.1–0.15 | 0.47 | 0.25 | 0.52 | 0.48 | 0.25 | 0.46 | 0.47 | 0.39 | 0.06–0.22 | 0.09–0.21 |
| 5 | 60276598 | ERCC8 | 26 | 0.07–0.11 | 0.05 | 0.07 | 0.56 | 0.06 | 0.04 | 0.06 | 0.08 | 0.08 | 0.05–0.19 | 0.05–0.11 |
| 8 | 11602736 | GATA4 | −231 | 0.04–0.23 | 0.48 | 0.03 | 0.65 | 0.45 | 0.03 | 0.45 | 0.51 | 0.32 | 0.03–0.32 | 0.07–0.94 |
| 16 | 56983684 | GINS3 | −216 | 0.04–0.08 | 0.71 | 0.06 | 0.1 | 0.13 | 0.04 | 0.16 | 0.19 | 0.12 | 0.03–0.12 | 0.03–0.15 |
| 6 | 43036449 | GNMT | −5 | 0.04–0.18 | 0.23 | 0.05 | 0.51 | 0.29 | 0.07 | 0.19 | 0.11 | 0.07 | 0.03–0.17 | 0.11–0.96 |
| 1 | 27099543 | GPATCH3 | −23 | 0.14–0.19 | 0.36 | 0.31 | 0.53 | 0.54 | 0.18 | 0.46 | 0.45 | 0.36 | 0.15–0.28 | 0.11–0.2 |
| 14 | 90790004 | GPR68 | −2 | 0.04–0.1 | 0.11 | 0.11 | 0.12 | 0.05 | 0.04 | 0.53 | 0.07 | 0.05 | 0.05–0.08 | 0.24–0.92 |
| 10 | 106018794 | GSTO2 | 148 | 0.08–0.23 | 0.41 | 0.08 | 0.6 | 0.43 | 0.04 | 0.31 | 0.08 | 0.09 | 0.03–0.47 | 0.02–0.68 |
| 4 | 54661069 | GSX2 | 89 | 0.04–0.05 | 0.03 | 0.03 | 0.38 | 0.54 | 0.07 | 0.58 | 0.31 | 0.26 | 0.03–0.11 | 0.06–0.88 |
| 7 | 27153921 | HOXA6 | −3 | 0.07–0.14 | 0.11 | 0.04 | 0.12 | 0.17 | 0.57 | 0.19 | 0.14 | 0.16 | 0.08–0.39 | 0.46–0.94 |
| 7 | 27171443 | HOXA9 | 129 | 0.02–0.12 | 0.08 | 0.01 | 0.07 | 0.15 | 0.8 | 0.09 | 0.23 | 0.21 | 0.04–0.5 | 0.02–0.99 |
| 14 | 34661189 | KIAA0391 | −293 | 0.12–0.16 | 0.44 | 0.27 | 0.46 | 0.54 | 0.2 | 0.45 | 0.51 | 0.42 | 0.07–0.18 | 0.1–0.26 |
| 3 | 47298896 | KIF9 | 124 | 0.04–0.11 | 0.57 | 0.06 | 0.12 | 0.1 | 0.05 | 0.1 | 0.11 | 0.08 | 0.03–0.11 | 0.04–0.12 |
| 3 | 47298896 | KLHK18 | −524 | 0.04–0.11 | 0.57 | 0.06 | 0.12 | 0.1 | 0.05 | 0.1 | 0.11 | 0.08 | 0.03–0.11 | 0.04–0.12 |
| 11 | 527614 | IRRC56 | 62 | 0.11–0.21 | 0.08 | 0.16 | 0.14 | 0.12 | 0.05 | 0.7 | 0.06 | 0.07 | 0.05–0.29 | 0.06–0.95 |
| 12 | 52713991 | MIRN615 | −35 | 0.1–0.19 | 0.24 | 0.1 | 0.31 | 0.35 | 0.09 | 0.49 | 0.38 | 0.29 | 0.07–0.18 | 0.1–0.94 |
| 19 | 10689824 | MIRN638 | −232 | 0.12–0.18 | 0.33 | 0.23 | 0.44 | 0.42 | 0.23 | 0.46 | 0.46 | 0.24 | 0.1–0.27 | 0.11–0.39 |
| 5 | 60276598 | NDUFAF2 | −169 | 0.07–0.11 | 0.05 | 0.07 | 0.56 | 0.06 | 0.04 | 0.06 | 0.08 | 0.08 | 0.05–0.19 | 0.05–0.11 |
| 17 | 46585675 | NMEI | −220 | 0.07–0.24 | 0.43 | 0.42 | 0.35 | 0.34 | 0.28 | 0.52 | 0.36 | 0.4 | 0.06–0.22 | 0.07–0.3 |
| 14 | 34661189 | PPP2R3C | 57 | 0.12–0.16 | 0.44 | 0.27 | 0.46 | 0.54 | 0.2 | 0.45 | 0.51 | 0.42 | 0.07–0.18 | 0.1–0.26 |
| 12 | 3470903 | PRMT8 | 192 | 0.05–0.13 | 0.03 | 0.03 | 0.04 | 0.09 | 0.03 | 0.46 | 0.06 | 0.06 | 0.02–0.12 | 0.02–0.92 |
| 3 | 45705779 | SACM1L | −90 | 0.06–0.12 | 0.46 | 0.04 | 0.04 | 0.05 | 0.05 | 0.05 | 0.06 | 0.05 | 0.04–0.12 | 0.04–0.11 |
| 8 | 145530624 | SCRT1 | 103 | 0.21–0.24 | 0.52 | 0.15 | 0.07 | 0.1 | 0.1 | 0.1 | 0.05 | 0.12 | 0.09–0.31 | 0.51–0.91 |
| 6 | 168584280 | SMOC2 | −376 | 0.15–0.18 | 0.12 | 0.12 | 0.13 | 0.1 | 0.07 | 0.16 | 0.52 | 0.41 | 0.07–0.16 | 0.11–0.93 |
| 4 | 37568253 | TBC1D1 | −838 | 0.08–0.15 | 0.05 | 0.18 | 0.07 | 0.03 | 0.06 | 0.08 | 0.47 | 0.34 | 0.08–0.2 | 0.36–0.92 |
| 12 | 128953838 | TMEM132D | −25 | 0.07–0.12 | 0.06 | 0.07 | 0.09 | 0.14 | 0.82 | 0.23 | 0.15 | 0.14 | 0.03–0.63 | 0.02–0.73 |
| 5 | 72897370 | UTP15 | 9 | 0.08–0.15 | 0.26 | 0.28 | 0.4 | 0.38 | 0.16 | 0.5 | 0.48 | 0.35 | 0.08–0.22 | 0.1–0.21 |
| 11 | 75203913 | UVRAG | 28 | 0.08–0.15 | 0.11 | 0.14 | 0.3 | 0.49 | 0.08 | 0.3 | 0.34 | 0.31 | 0.07–0.16 | 0.07–0.13 |
| 15 | 38974052 | VPS18 | 107 | 0.11–0.15 | 0.33 | 0.29 | 0.52 | 0.51 | 0.12 | 0.4 | 0.45 | 0.38 | 0.1–0.38 | 0.11–0.19 |
The fully reprogrammed, iPS clones generally also behave very well with respect to non-CpG island genes. Thus, for all clones, 87 to 95% of such promoters properly either gain or lose DNA methylation properly relative to ESC (Fig. 4b). Interestingly, and as might be expected, most of the genes in these groups do not make the changes required for iPS in the reprogramming pools of fibroblasts or MSC generally retaining the methylation patterns of the starting parent cells (Fig. 4b). Again, despite the global proper behavior, multiple non-CpG island genes, relative to normal cells, both gain and lose promoter methylation. Thirteen genes abnormally gain methylation in the clones (Suppl Table 6), and another 13 in the pools (Suppl Table 7) while 56 and 37 lose methylation respectfully (Suppl Tables 6 and 7). Particularly, MP4, contains 37 abnormal genes, 5 having abnormal gain and 32, abnormal losses, of methylation (Fig 4b; Suppl Tables 6 and 7). A high percentage, 60% for the clones (p = 1.645e-12; Fisher’s exact test), and 43% for the pools, (p = 5.141e-08), of the abnormal gains and losses of DNA methylation also appear in one or more of the cancer cell lines (Suppl Tables 6 and 7). Finally, for MP4, only 35% of non-CpG island genes properly gained or lost DNA methylation relative to ESC.
Importantly, many of the above abnormal CpG island genes are hypermethylated in primary human cancers and have roles in malignancy and/ or embryonic cell fate patterning. GATA4 (Suppl Ref 6), central to proper endodermal differentiation, is hypermethylated in multiple cancer types, O6-MGMT(Suppl Refs 7,8) is also hypermethylated in cancer and loss of function impedes DNA repair, TCERG1L has been recently identified as a low frequency mutated gene in colon cancer (Suppl Ref 9) and is DNA hypermethylated in virtually all such tumors (unpublished data), SPRY2 plays a role in development and differentiation through negatively regulating the extracellular-regulated kinase pathway and is hypermethylated in cancers with prognostic significance(Suppl Ref 10), SOX 1 is DNA hypermethylated in cancer and also plays key roles in development (Suppl Ref 11), BMP4, is an important morphogen for mesenchymal development and is hypermethylated in breast cancer (Suppl Ref 12), and HOXA9 is an important cell pattern control gene frequently hypermethylated in breast cancer (Suppl Ref 13). LXN, hypermethylated in tumorigenic clone MP4, contains a weak CpG island and is hypermethylated in several cancer types (Suppl Refs, 2, 3).
For selected of these important genes, we verified Infinium results by performing the methylation PCR assay, MSP(29), querying 4 to 6 CpG sites positioned near the Infinium probes yielding hypermethylation values. Even though Infinium probes query small numbers of CpG’s in the islands, and development of abnormal methylation may be quite partial over the short time of re-programming, 7 of 10 genes were methylated by MSP, and at one of two promoter sites queried, TCERG1L is fully methylated in iPS clones MP2 and MR45 (Suppl Fig. 6a, b).
Abnormally silenced genes, chromatin, and cellular re-programming
The small fraction of DAC responsive (supplemental Fig. 3), silenced, genes in iPS and reprogramming pools that are DNA methylated in reprogrammed cells is surprising. While, this could represent failure to detect very partial DNA methylation in queried promoters, it may involve links between abnormal gene silencing, DNA methylation in cancer, and chromatin states of embryonic cells. We(30), and others(31, 32), have found high percentages of DNA hypermethylated genes in cancer are marked by polycomb group silencing proteins (PcG) in embryonic cells. Such genes may have an abnormal progression from PcG marking to promoter DNA hypermethylation in cancer (27, 30). In ESC, such promoters are not methylated, but rather maintained in a low, poised expression state by PcG regulation (33–35) and “bivalent” chromatin constituted by transcriptional positive (H3K4me2 and me3), and negative (the PcG mark, H3K27me3) histone modifications (36). Interestingly, abnormally low expression genes in partially reprogrammed cells from mice have bivalent chromatin (28)and aggressive human cancers have increased expression of PcG genes(37) to levels such as those found in our iPS cells (Supplementary Table 2).
We, thus, queried the promoter occupancy of our abnormally silenced genes in iPS clones, reprogramming pools, and cancer cell lines (Table 1, Suppl Tables 4 and 5) in databases of others for embryonic cells (37–40). While there is no significant enrichment at the promoters of these genes for occupancy by the iPS reprogramming factors themselves(37) save, for Nanog at the promoters of the DNA hypermethylated genes in Table 1 and Supplemental Table 5, in the clones and partially reprogrammed pools, there is an enrichment for PcG promoter marking(Fig. 5a). This is confirmed by local chromatin immunoprecipitation (ChIP) of the PcG mark, H3K27me3, for promoter regions of selected of the above genes in ESC, iPS clone MP2, and the partially reprogrammed clone, MP4 (Fig. 5b).
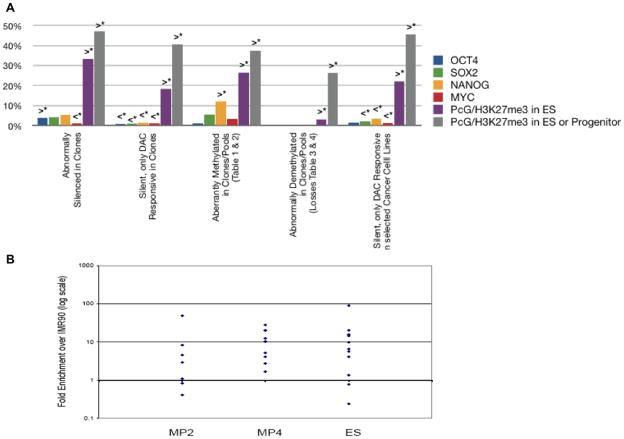
Figure 5. Gene promoter occupancy by transcription factors and polycomb group protein (PcG) repressive histone modification, H3K27me3.
a. Percentage of genes with transcription factor (OCT4, SOX2, NANOG, c-MYC) and H3K27me3 promoter occupancy, in tiling arrays of others(37–40), for all abnormally silenced genes in Fig. 2, abnormally silenced and DAC responsive genes in Fig. 3, abnormally DNA methylated genes in Table 1 and Suppl tables 4–6, and DAC responsive, silenced genes in cancer cell lines. b. Real time chromatin immunoprecipitation (ChIP) of H3K27me3 at promoters of selected, abnormally silenced, genes that are reactivated by DAC, but are not DNA hypermethylated, in MP2 iPS, partially reprogrammed MP4, H9, ESC (ES). Triangles = H3K27me3 enrichment (log scale) relative to a value of 1.0 (horizontal line) for parent IMR90 fibroblasts. Primer sequences and positions provided online.
DISCUSSION
It appears that during the reprogramming process, iPS may be prone to abnormal epigenetic changes characteristic of neoplasia. Our results differ from a recent study (15) in which differential changes both between cancer and normal cells, and iPS and normal ESC, were thought confined to “shores” or regions upstream from promoter CpG islands. In contrast, we find cancer specific promoter CpG island, hyper- methylation is easily visualized on the Infinium arrays in hundreds of genes in the cancer cells (Fig. 4 and Suppl Figs. 4 and 5). These changes can be seen, to a lesser degree, in iPS, and partially reprogrammed cells (Table 1, Suppl. Table 4, and Fig. 4a).
The abnormal gene silencing events in reprogramming and could well involve the inducing factors themselves. We find just one essential re-programming factor, OCT4 (4, 41), produces cancer-specific epigenetic changes and this may help explain why its forced over-expression in the germline of mice produces a striking neoplastic phenotype in skin and intestine (42).
A dramatic finding in our study concerns the dominance of gene responsiveness to TSA in ESC but to DAC in cancer cells and iPS. The chromatin of ESC, especially, exists in a far more open, and histone acetylated, pattern than in more committed embryonic and adult cells (43). Many genes in ESC may be in a poised state for induction in response to commitment signals, and are balanced between very active targeting by histone acetyltransferases and removal of acetylation by HDAC’s. Blocking HDAC’s would then shift the dynamics to allow rapid promoter acetylation to enhance expression of such genes. The more dominant response to DAC in iPS, and especially cancer cells, may reflect an abnormally repressed state of genes which prevents normal cell differentiation and/or full progression of cells during reprogramming to the ESC epigenetic state.
IPS and cancer cells do differ dramatically in that the Infinium assay validates that a majority (>70%) of basally silent genes which respond to DAC alone but not TSA (i.e., candidate genes in suppl Fig. 3) have promoter DNA methylation in the cancer cells but not the iPS cells. A key explanation may lie in the potential molecular progression from PcG promoter occupancy to DNA promoter hypermethylation that may be ongoing in cancer as suggested by ourselves and others (27). This progression may involve targeting of DNMTs to involved genes (27), and resultant DAC re-expression of even non-DNA methylated genes. DNMT’s bind co-repressors, experimentally can repress gene promoters independently of catalyzing DNA methylation, and can act as scaffolding proteins by utilizing regions distal to their DNA methylation catalytic sites (44–46). To inhibit DNMT’s, DAC incorporates into DNA and covalently bind to DNMTs, resulting in removal of DNMT1 from the soluble nuclear protein pool(47) and its degradation(48). Thus, DAC may re-express genes with or without DNA methylation as indicated by our recent result for over-expressing a polycomb constituent in teratocarcinoma cells (49).
In summary, epigenetic changes may contribute to neoplastic potential of iPS. Mapping these may help provide constructive parameters to monitor the preparation and use of such cells, guide use of epigenetic modifying drugs to enhance efficiency of obtaining iPS (10), and help derive gene marker panels for the above purposes. Finally, cellular reprogramming may provide a model to study how epigenetic abnormalities may be central to the origins of cancer and whether reprogramming might play a role in formation of key subpopulations of cancer cells. For example, cellular reprogramming might underlie the fact that neoplasia can initiate even in mature populations of normal cells(50).
Supplementary Material
Acknowledgments
FUNDING
This work was supported by grants from the National Institutes of Health (SBB) CA116160, UO1 HL099775, and a State of Maryland TEDCO grant.
We thank Dr. Saul Sharkis and members of the Baylin and Cheng labs for reading the manuscript and for helpful discussions and Kathy Bender for manuscript preparation.
Footnotes
Competing Interests
S.B.B. consults for OncoMethylome Sciences. MSP is licensed to OncoMethylome Sciences in agreement with Johns Hopkins University (JHU) and SBB and JHU are entitled to royalty shares received from sales.
References
- 1.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Loh YH, Agarwal S, Park IH, et al. Generation of induced pluripotent stem cells from human blood. Blood. 2009 doi: 10.1182/blood-2009-02-204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–23. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu J, Hu K, Smuga-Otto K, et al. Human Induced Pluripotent Stem Cells Free of Vector and Transgene Sequences. Science. 2009 doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–5. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–7. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 8.Yamanaka S. A fresh look at iPS cells. Cell. 2009;137:13–7. doi: 10.1016/j.cell.2009.03.034. [DOI] [PubMed] [Google Scholar]
- 9.Daley GQ, Lensch MW, Jaenisch R, Meissner A, Plath K, Yamanaka S. Broader implications of defining standards for the pluripotency of iPSCs. Cell Stem Cell. 2009;4:200–1. doi: 10.1016/j.stem.2009.02.009. author reply 2. [DOI] [PubMed] [Google Scholar]
- 10.Mikkelsen TS, Hanna J, Zhang X, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huangfu D, Maehr R, Guo W, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–7. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–74. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Doi A, Park IH, Wen B, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–3. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mali P, Ye Z, Hommond HH, et al. Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem Cells. 2008;26:1998–2005. doi: 10.1634/stemcells.2008-0346. [DOI] [PubMed] [Google Scholar]
- 18.Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–23. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGarvey K, Van Neste L, Cope L, Ohm J, Herman J, Van Criekinge W, Schuebel K, Baylin S. Defining a Chromatin Pattern That Characterizes DNA Hypermethylated Genes in Colon Cancer Cells. Cancer Research. 2008;68:5753–9. doi: 10.1158/0008-5472.CAN-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bibikova M, Fan JB. Golden Gate assay for DNA methylation profiling. Methods Mol Biol. 2009;507:149–63. doi: 10.1007/978-1-59745-522-0_12. [DOI] [PubMed] [Google Scholar]
- 21.Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhee I, Bachman KE, Park BH, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–6. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 23.Chan EM, Ratanasirintrawoot S, Park IH, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009 doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 24.Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177–81. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- 25.Butler H, Ragoussis J. Bead Array-based genotyping. Methods Mol Biol. 2008;439:53–74. doi: 10.1007/978-1-59745-188-8_4. [DOI] [PubMed] [Google Scholar]
- 26.Straussman R, Nejman D, Roberts D, et al. Developmental programming of CpG island methylation profiles in the human genome. Nat Struct Mol Biol. 2009;16:564–71. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- 27.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 28.Sridharan R, Tchieu J, Mason MJ, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–77. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohm JE, McGarvey KM, Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–42. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Widschwendter M, Fiegl H, Egle D, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–8. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 32.Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–6. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 33.Lund Aav LM. Polycomb complexes and silencing mechanisms. Current Opinion in Genetics and Development. 2004;16:1–8. doi: 10.1016/j.ceb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 34.Otte AP, Kwaks TH. Gene repression by Polycomb group protein complexes: a distinct complex for every occasion? Curr Opin Genet Dev. 2003;13:448–54. doi: 10.1016/s0959-437x(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 35.Kuzmichev A, Margueron R, Vaquero A, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci U S A. 2005;102:1859–64. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chi AS, Bernstein BE. Developmental biology. Pluripotent chromatin state. Science. 2009;323:220–1. doi: 10.1126/science.1166261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben-Porath I, Thomson MW, Carey VJ, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006 doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 40.Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa M, Koyanagi M, Tanabe K, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2007 doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 42.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–77. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 43.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–6. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 44.Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. Embo J. 2001;20:2536–44. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–77. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 46.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. NatGenet. 2000;25:338–42. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 47.Cheng JC, Weisenberger DJ, Gonzales FA, et al. Continuous zebularine treatment effectively sustains demethylation in human bladder cancer cells. Mol Cell Biol. 2004;24:1270–8. doi: 10.1128/MCB.24.3.1270-1278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Datta J, Ghoshal K, Denny WA, et al. A new class of quinoline-based DNA hypomethylating agents reactivates tumor suppressor genes by blocking DNA methyltransferase 1 activity and inducing its degradation. Cancer Res. 2009;69:4277–85. doi: 10.1158/0008-5472.CAN-08-3669. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Mohammad HP, Cai Y, McGarvey KM, et al. Polycomb CBX7 promotes initiation of heritable repression of genes frequently silenced with cancer-specific DNA hypermethylation. Cancer Res. 2009;69:6322–30. doi: 10.1158/0008-5472.CAN-09-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gidekel Friedlander SY, Chu GC, Snyder EL, et al. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell. 2009;16:379–89. doi: 10.1016/j.ccr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.