Abstract
Serum response factor (SRF) is a widely expressed protein that plays a key role in the regulation of smooth muscle differentiation, proliferation, migration, and apoptosis. It is generally accepted that one mechanism by which SRF regulates these diverse functions is through pathway-specific cofactor interactions. A novel SRF cofactor, chromodomain helicase DNA binding protein 8 (CHD8), was isolated from a yeast two-hybrid screen using SRF as bait. CHD8 is highly expressed in adult smooth muscle tissues. Coimmunoprecipitation assays from A10 smooth muscle cells demonstrated binding of endogenous SRF and CHD8. Data from GST-pulldown assays indicate that the NH2-terminus of CHD8 can interact directly with the MADS domain of SRF. Adenoviral-mediated knockdown of CHD8 in smooth muscle cells resulted in attenuated expression of SRF-dependent, smooth muscle-specific genes. Knockdown of CHD8, SRF, or CTCF, a previously described binding partner of CHD8, in A10 VSMCs also resulted in a marked induction of apoptosis. Mechanistically, apoptosis induced by CHD8 knockdown was accompanied by attenuated expression of the anti-apoptotic proteins, Birc5, and CARD10, whereas SRF knockdown attenuated expression of CARD10 and Mcl-1, but not Birc5, and CTCF knockdown attenuated expression of Birc5. These data suggest that CHD8 plays a dual role in smooth muscle cells modulating SRF activity toward differentiation genes and promoting cell survival through interactions with both SRF and CTCF to regulate expression of Birc5 and CARD10.
Keywords: vascular smooth muscle cells, ternary complex factor
serum response factor (SRF) is a broadly expressed transcription factor that regulates many processes under both physiological and pathological conditions (28). In a variety of cell types SRF has been shown to regulate cell proliferation, migration, differentiation, and death. A number of different signaling pathways regulate these disparate functions of SRF. For example, growth factor or serum signaling through the extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) pathway stimulates the phosphorylation of the ternary complex factor (TCF) family of proteins, such as Elk1, which interact with SRF. Upon this phosphorylation, Elk1/SRF complexes stimulate transcription of immediate-early genes, such as cFos and Egr1, to promote cell proliferation (8, 20, 42). Signaling through the RhoA small GTPase cascade (8, 14) stimulates production of filamentous actin and activates the dissociation of cytoplasmic myocardin-related transcription factor A (MRTFA) (MKL1) from globular actin, leading to nuclear accumulation and activation of MRTFA (31). MRTFA then interacts with SRF to stimulate transcription of genes such as SRF itself, vinculin, junB, and smooth muscle differentiation markers (15, 18, 19, 40, 48, 51). Upregulation of these factors allows SRF to regulate differentiation, migration, and adhesion. SRF is critical for the differentiation of skeletal, cardiac, and smooth muscle lineages (29, 36, 37). SRF regulates smooth muscle differentiation through its interaction with coactivators such as myocardin and the myocardin family members MRTFA and MRTFB (7, 44). SRF has also been implicated in regulating apoptosis through controlling the expression of anti-apoptotic molecules Bcl2 and Mcl-1 (39, 43). SRF has been shown to be required for differentiation-dependent expression of Bcl-2 through a CArG sequence in the Bcl-2 promoter (39), and SRF regulates Mcl-1 transcription through its interaction with the transcription factor Elk-1 (43). Global SRF knockout mice are embryonic lethal due to a failure in gastrulation. The SRF−/− embryos form a misfolded endoderm and ectoderm and fail to form the mesodermal layer, indicating that SRF is critical for formation of the mesoderm from which most muscle is derived (3). To circumvent this embryonic lethality a number of groups have specifically ablated SRF in different muscle tissues. These studies have shown that SRF is critical for the development of each muscle lineage, including smooth muscle (2, 9, 24, 27, 30, 35).
In an attempt to identify other cofactors that regulate SRF activity, we conducted a yeast two-hybrid screen using SRF as bait (13). From this screen we identified the homeodomain protein Barx2 (13) and a cDNA encoding the NH2-terminus of chromodomain helicase DNA-binding protein 8 (CHD8). CHD8 is a member of the CHD family of ATP-dependent chromatin remodeling proteins. CHD proteins, such as CHD8, contain two tandem chromodomains: an ATPase domain and a helicase domain. In addition to these domains, CHD8 contains two homeodomain-like Brk domains. Duplin, an NH2-terminal splice variant of CHD8 orginally discovered as a β-catenin-binding protein (38), contains only the first of the two chromodomains. Chromodomains are regions of 40–50 amino acids that are involved in chromatin remodeling and gene regulation (6). These domains can serve as protein interaction modules, RNA-binding modules, or DNA-binding modules (1, 5, 11). The chromodomains in CHD8 have been shown to specifically interact with histone H3 trimethylated at lysine 4 (49). CHD8 exhibits ATP-dependent chromatin remodeling activity and represses β-catenin target genes (41). CHD8 also binds to the insulator binding protein CTCF (17), and the CHD8/CTCF complex plays an important role in the epigenetic regulation of insulator sites. Duplin/CHD8 knockout mice (simultaneous knockout of duplin and CHD8) are embryonic lethal and fail to form a primative streak or the mesodermal layer during gastrulation, indicating that CHD8/duplin plays a critical role in early development (33). These knockout mice also displayed elevated apoptosis that could be partially rescued by crossing the knockout mice with p53 knockout mice, suggesting that CHD8 plays a role in protecting cells from p53-induced apoptosis (34).
In the current study we demonstrate that CHD8 interacts with SRF and affects expression of SRF-dependent genes characteristic of differentiation. In addition, CHD8, together with SRF and CTCF, plays a significant role in protecting smooth muscle cells from apoptosis.
EXPERIMENTAL PROCEDURES
RNase protection assay.
The RPA III protocol from Ambion was used to perform ribonuclease protection assays. Briefly, a probe was designed to span the alternatively spliced exons that give rise to duplin and CHD8 such that the duplin mRNA will protect a 318nt fragment of the probe, whereas CHD8 mRNA will protect a 150nt fragment. Mouse brain mRNA was used as a template for RT-PCR, and the resultant fragment was cloned, sequenced, and used as the probe for RNase protection analysis. RNA (25 μg) from each mouse tissue was subject to RNase protection analysis.
GST-pull down assays.
A series of SRF deletion constructs fused to GST, which were described previously (13), were utilized. In addition, a series of CHD8 truncations were constructed in the pET vector to perform the reverse experiment; CHD8 truncations employed in experiments are indicated in the figures. Bacterial proteins were isolated, and GST pulldown assays were conducted as previously described (13). Binding interactions were visualized using standard Western blot techniques with primary antibodies against GST or T7 for pET vector fusion proteins.
Coimmunoprecipitation assays.
Coimmunoprecipitation assays from mammalian cells were performed using the Active Motif Nuclear coIP Kit, according to the protocol suggested by the manufacturer with a few minor adjustments. Before the cells were harvested, the cells were incubated with 1 mM dithiobis[succinimidyl]propionate (DSP) for 30 min to cross-link intracellular proteins. Cross-linking was stopped by addition of 10 mM Tris, pH 7.5, in PBS for 15 min. Lysates were precleared for 1 h with 50 μl of EZ view Protein A Sepharose (PAS) (Sigma) beads in 900 μl of immunoprecipitate (IP) wash buffer. PAS beads were washed twice for 5 min in IP wash buffer with 1 mg/ml BSA and once for 5 min in wash buffer without BSA before addition to the antibody-lysate mixture. SRF was immunoprecipitated from 500 μg of nuclear extract with ∼5 μg of protein-specific antibody or IgG control. All conditions during the coimmunoprecipitation were low stringency. Standard Western blot techniques were used to visualize the presence of CHD8 (A310-225A, Bethyl Labs, Abnova, AO1) or SRF (G-20, Santa Cruz). A10 vascular smooth muscle cells (VSMCs) were utilized for collection of nuclear lysate. All experiments were repeated at least three times using independent nuclear lysates.
Quantitative chromatin immunoprecipitation assays.
Quantitative chromatin immunoprecipitation assays were performed essentially as described previously (51) except that cells were treated with the chemical cross-linker EGS (1.5 mM in PBS, Pierce) for 20 min at RT before cross-linking with formaldehyde. Cross-linked chromatin was immunoprecipitated with 2 μg of anti-CHD8 (Bethyl, A310-225A) or rabbit IgG control and bound to ssDNA/protein A agarose beads (Millipore). The precipitated DNA was then purified and amplified by real-time PCR for quantification of the target sequences using SYBR green PCR master mix (Roche) with respective gene-specific primers. Rat Birc5 promoter sense: 5′-GCCTGCAAACCCTGAAGAAG; antisense: 5′-GTCCTGGCCTAGGGTACCTCTCT-3′; rat cMyc insulator sense: 5′-GAACCTGGAAGCCCAGCAA-3′; antisense: 5′-TACTGGCCACAGATCACAGCTTT-3′; rat smooth muscle myosin heavy chain (smMHC) promoter sense: 5′-TGCACGGGACCATATTTAGTCA-3′, antisense: 5′-CTCGAAACAAGAATTCCCAGCTT-3′.
Cell culture.
All primary mouse cells and 10T1/2 mouse fibroblasts were cultured in DMEM supplemented with 10% FBS, 2 mM l-glutamine, and 50 units/50 μg penicillin-streptomycin. A10 VSMCs were cultured in DMEM supplemented with 20% FBS, 2 mM l-glutamine, and 50 units/50 μg penicillin-streptomycin.
Adenovirus construction and cell transduction.
Previously published short-hairpin RNA (shRNA) sequences directed to CHD8 (nucleotides 6474-6492 of NM_201637) (17), SRF (47), or CCCTC-binding factor (CTCF) (17) were cloned into the AdenoX vectors (BD Bioscience). The virus was packaged and harvested in HEK cells according to the manufacturer's protocol. Primary mouse colon or bladder SMC or A10 SMC were plated at a density of 5 × 104 cells per well in 12-well plates and transduced with either CHD8, SRF, or CTCF shRNA or control shRNA virus as previously described (12, 47, 52). A minimum of triplicate wells of cells were infected with each virus, and all experiments were repeated at least three times using independent preparations of smooth muscle cells.
TGFβ-induced myofibroblast differentiation.
10T1/2 cells were plated at a density of 3 × 105 cells per well in a standard six-well format. After an overnight incubation at 37°C with 5% CO2, cells were transduced with adenoviral-mediated shRNA targeting CHD8 as well as an shRNA control. After cells were transduced with virus for 4 h, the virus was aspirated and media was replaced with media supplemented with 0.5% FBS. Twenty-four hours later, cells were either given fresh 0.5% FBS media or treated with TGFβ1 at a final concentration of 2 ng/ml under low serum conditions for 24 h and then collected for analysis of gene expression by quantitative RT-PCR (qRT-PCR).
qRT-PCR.
For the RT step, 0.5 μg of RNA was reverse transcribed to cDNA using random hexamer primers (Invitrogen Superscript First-Stand Kit). A 1:10 dilution of the resulting cDNA was used in a quantitative PCR (qRT-PCR) reaction with SYBR green (Roche). Sequence-specific primers for CHD8, SRF, SRF's targets, and controls included the following: CHD8 sense 5′-CCAGCTCCAGCTCCAGCAC-3′; anti-sense 5′-CCTGCAGTAGCAGCAACTCAG-3′; SRF sense 5′-GTTCATCGACAACAAGCTGC-3′; anti-sense 5′-CTGTCAGCGTGGACAGCTCATAG-3′; cyclin D1 sense 5′-GCCAGAGGCGGATGAGAACAAGC-3′; anti-sense 5′-GGTCACACTTGATGACTCTGG-3′; EGR1 sense 5′-GAGCACCTGACCACAGAGTC-3′; anti-sense 5′-CCACAAAGTGTTGCCACTGTTG-3′; SM α-actin sense 5′-CCAGAGTGGAGAAA GCCCAGC-3′; anti-sense 5′-GGCTGTGCTGTC TTCCTCTTCAC-3′; SM22α sense 5′-CGAAGC CAGTGAAGGTGCCTGAGAAC-3′; anti-sense 5′-CCCAAAGCCATTAG AGTCCTCTGCACTGC-3′; telokin sense 5′-GACACCGCCTGAGTCCA ACCTCCG-3′; anti-sense 5′-GGCTTTTCCTCAGCAA CAGCCTCC-3′; cMYC sense 5′-CCACCAGCAGC GACTCTGAA-3′; anti-sense 5′-CTGTGCGGAGGT TTGCTGTG-3′; 36B4 sense 5′-GGACCCGAGA AGACCTCCTT-3′; anti-sense 5′-TGCTGCCGTTGTC AAACACC-3′; and Birc5/survivin sense 5′-CTGGCCCTTCCTGGAGGA-3′; anti-sense 5′-CTCGGTAGGGCAGTGGATGA-3′; CARD10 sense 5′-ATGTCGGATATCACAGGGAGTGT-3′, antisense 5′-CCTTCCGGCTTTCCCAAA-3′; caspase 1 sense 5′-TCTCACAGCTCT GGAGATGACAA-3′; antisense 5′-GACCATGAGA CATGAATACAAGGAA-3′: CTCF sense 5′-CGCGAAGAATGACCACAAATC, antisense CAGATCTCCGGTCCCTAGCTTCAA-3′. Primers for cFOS and vinculin were obtained from Qiagen (QT00147308 and QT00158319, respectively). Each of these primer sets spans introns in their respective genes.
Induction of apoptosis.
A10 vascular SMCs were plated at 5 × 104 cells per well in 12-well plates transduced in quadruplicate with either CHD8 shRNA, SRF shRNA, CTCF shRNA, or control shRNA virus as previously described (12, 47, 52). In some experiments 72 h after transduction, cells were treated with 0.2 mM H2O2 for 6 h. Medium was collected, and wells were washed twice with PBS, which was added to the collected media. The pooled media and washes were spun at 2,000 rpm for 2 min at room temperature to accumulate any nonadherent cells. The PBS and media were then aspirated, and the pellet was washed once with PBS. In the meantime, 100 μl of RIPA lysis buffer were added to each well, and the cells were incubated on ice for 10 min. Cells were then scraped into the corresponding tube with the cell pellet and incubated on ice an additional 5 min. Tubes were then spun at 14,000 rpm for 2 min at 4°C, and resulting supernatants were transferred to a fresh tube. BCA assays were performed to ensure equal loading during protein separation by SDS-PAGE. Standard Western blot techniques were employed to visualize changes in protein expression. TUNEL assays were performed using the TdT-FragEL DNA fragmentation detection kit (Calbiochem) according to the manufacturer's directions.
Western blotting.
For apoptosis studies, 20 μg of protein were loaded for each sample during SDS-PAGE. Antibodies used include anti-SRF (Santa Cruz, G20), anti-CHD8 (Bethyl Labs, A310-225A; Abnova AO1), anti-CTCF (Santa Cruz, H-280), anti-phospho-H2A.X (Upstate), anti-cleaved-caspase-3 (Cell Signaling, 9661), anti-Poly(ADP-ribose) polymerase-1 (PARP, Santa Cruz, H250), anti-GAPDH (Novus), anti-Bcl-2 (BD Biosciences), anti-Bcl-XL (Cell Signaling), anti-Mcl-1 (Abcam), and anti-nonmuscle-MHCIIb (Covance). For GST pull-down assays, antibodies used included anti-T7 (Novagen). GST was visualized via Ponceau staining. Secondary antibodies were used accordingly and included goat-anti-rabbit-horseradish peroxidase and goat-anti-mouse-horseradish peroxidase.
DNA methylation.
DNA methylation assays were performed using a MethylCollector kit (Active Motif) essentially as described by the manufacturer. Briefly, A10 cells were transduced with adenovirus, and 48 h later genomic DNA was prepared (Genlantis). Purified DNA (4 μg) was digested with RsaI to yield an ∼400 bp fragment of the rat Birc5 promoter that includes the CpG island. Digested DNA (400 ng) was bound to his-tagged MBD2b to separate the methylated DNA. His-MBD2b-bound DNA was washed under low stringency conditions. The eluted DNA was analyzed by real-time PCR using Birc5 promoter primers: sense, 5′-TCTGGGAGGCAGTTGAGTAGCT-3′; antisense, 5′-GGCGCCAGGGCTTGCT-3′.
RESULTS
CHD8 is more widely and abundantly expressed than duplin.
From a yeast two-hybrid screen of a mouse intestinal cDNA library (13), we isolated 2 cDNA clones that encoded the amino terminal 529 amino acids of CHD8/duplin. These cDNAs were used to screen a lambda gt11 cDNA library made from mouse intestine. From this screen, three additional cDNAs were isolated. One of these extended further 3′ and included sequence encoding amino acids 282–770 of CHD8. Sequences of these clones together with the sequence from image clone no. 683 (Invitrogen) were used to compile the full-length mouse CHD8 sequence. This sequence was found to correspond to the previously annotated mouse CHD8 sequence NM_201637.2. Duplin, originally discovered in Rattus norvegicus (NM_022933) (38), and CHD8 are alternatively spliced transcripts of the CHD8 gene. Both transcripts are identical in their first 8 exons; however, they diverge in exon 9 (Fig. 1, A and B). CHD8 transcripts utilize an alternative splice donor site in exon 9 such that the stop codon encoded by the 3′ portion of exon 9 is removed when the alternative donor site is spliced to exon 10. The most 3′ cDNA that we isolated from our mouse intestinal cDNA library includes the exon 9–10 splicing pattern characteristic of CHD8 rather than duplin.
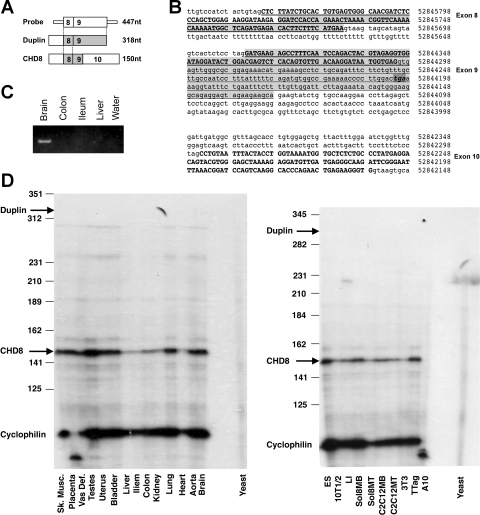
Fig. 1.
Chromodomain helicase DNA binding protein 8 (CHD8) is more ubiquitously expressed than duplin, its NH2-terminal splice variant. A: probe used for RNase protection assays was designed using exons 8 and 9 of duplin as shown in B. B: genomic sequence encompassing CHD8 exons 8–10 as annotated from BLAT on 7-28-08 (only the partial sequence of the introns is shown). Exons of full-length CHD8 are shown in bold, exons included in duplin are underlined, and the stop codon highlighted. The shaded region is the probe used for RNase protection assays. C: probe shown in A was isolated by RT-PCR from RNA isolated from mouse brain. An ethidium bromide-stained gel of the RT-PCR products are shown. D: ribonuclease protection assays were conducted to determine the amount of full-length CHD8 mRNA relative to its splice variant duplin in various mouse tissues (left) and cell lines (right). Arrows point to the expected location of bands protected by CHD8 or dupin. Note that no duplin expression was detected in this assay. Cyclophilin was used as an internal control; yeast RNA was used as a negative control (yeast). Sk. Musc., skeletal muscle; Vas Def, vas deferens; 10T1/2, 10T1/2 mouse fibroblasts; LI, mouse colon smooth muscle cell line; A10, A10 rat aortic smooth muscle; TTag, T-antigen derived colonic tumor cells; 3T3, NIH 3T3 mouse fibroblast; MB, myoblast; MT, myotubes.
To further evaluate the expression of duplin and CHD8, RNase protection assays were performed. A probe was constructed based on the sequence difference between the isoforms in exon 9 (Fig. 1, A and B). Results from this analysis show that CHD8 is the predominant isoform expressed in all mouse tissues, and duplin could only be detected at low levels in brain by RT-PCR (Fig. 1, C and D). The apparent lack of CHD8 in rat aortic A10 cells reflects species differences in the rat and mouse sequences that prevented its detection using RNase protection, as CHD8 can be readily detected by RT-PCR and Western blotting in these cells (Figs. 3–5). From qRT-PCR data, CHD8 mRNA was found to be about 20% more abundent in A10 cells compared with 10T1/2 cells. qRT-PCR analysis of A10 cells using isoform-specific primers for duplin and CHD8 revealed ∼200-fold greater expression of CHD8 compared with duplin in these cells (data not shown).
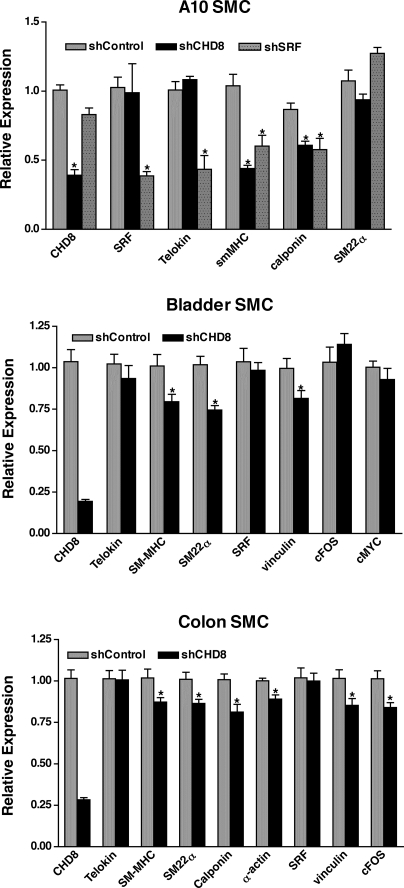
Fig. 3.
Knockdown of CHD8 causes reduced expression of SRF-dependent genes. Rat A10 vascular smooth muscle cells (VSMCs) or passage 1 primary mouse bladder or colon SMCs were transduced with an adenovirus containing either CHD8-specific short hairpin RNA (shRNA) or control shRNA. At 72 h after transduction, RNA was isolated, and transcripts were quantitated by real-time RT-PCR. Transcript levels were firstly normalized to 36B4 internal loading control and then normalized to their respective shControl group. Relative expression = 2−ΔΔCt and ΔΔCt = (Ctexperimental − Ct36B4) − (Ctcontrol − Ct36B4). *Samples were statistically different from controls (P < 0.05, n = 12–17 for colon and bladder and n = 6–10 for A10 cells).
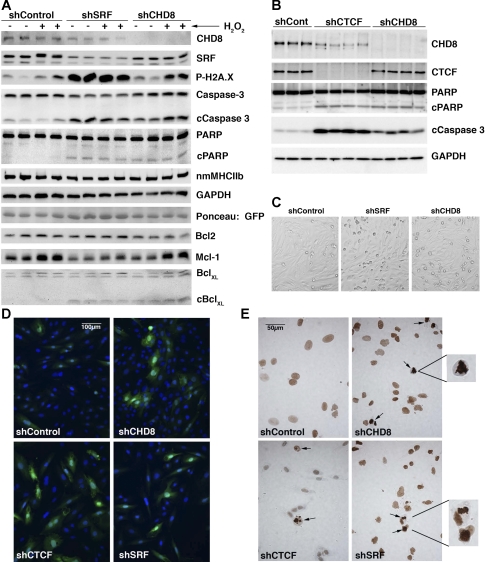
Fig. 4.
CHD8 imparts a prosurvival effect on A10 VSMCs. A: A10 VSMCs were transduced in quadruplicate with either CHD8 shRNA, SRF shRNA, or control shRNA adenovirus. After 72 h, cells were treated with 0.2 mM H2O2 (+) or vehicle (−) for 6 h. Both adherent and detached cells were washed twice with PBS and lysed in 100 μl of RIPA lysis buffer. 20 μg of protein from each extract were separated on SDS-PAGE and analyzed by Western blot analysis with the antibodies indicated at the right of the blot. Western blotting to detect GAPDH and Ponceau S staining to detect adenoviral encoded GFP were used as loading controls. B: A10 cells were transduced with adenovirus encoding shControl, shCHD8, or shCTCF and analyzed as described in A. Western blotting to detect GAPDH was used as a loading control. C: phase contrast images of cells obtained before H2O2 treatment showing increased numbers of rounded detaching apoptotic cells following knockdown of CHD8 or SRF. D: immunofluorescence staining of active cleaved caspase-3 (Asp175) (Cell Signaling no. 9661) in A10 cells transduced with adenovirus encoding the indicated shRNAs (green). Nuclei were counterstained with Hoechst (blue). E: tunnel staining of cells similar to those shown in D. Apoptotic nuclei identified by intense brown staining and condensed nuclei are indicated by the arrow and examples are enlarged in the insets to the right. Note that the background brown staining in most cells is a result of detection of the linear adenoviral genome by the TUNEL procedure.
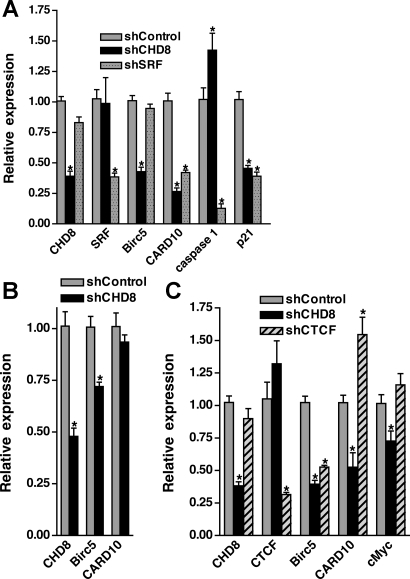
Fig. 5.
Loss of CHD8 causes attenuated expression of Birc5/survivin and Card10. A10 VSMC (A, C) or primary mouse colonic SMCs (B) were transduced with adenovirus encoding shRNA directed against CHD8 (solid bars), SRF (stippled bars), shCTCF (hatched bars), or a nontargeting control (shaded bars). 72 h following transduction, RNA was isolated, and transcripts were analyzed by qRT-PCR as described in Fig. 3. *Transcripts showing statistically significant changes from controls (n = 6–8 for A10 and n = 6 for colon. P < 0.05).
CHD8 binds to the MADS domain of SRF.
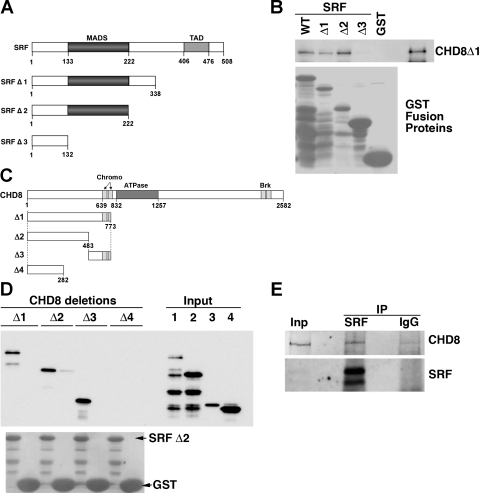
To confirm our yeast two-hybrid result and to map the binding domains of SRF and CHD8, GST pull-down assays were performed (Fig. 2A). Results from this analysis demonstrate that CHD8 binds to the MADS domain of SRF, and SRF binds to two regions in the NH2-terminus of CHD8 between amino acid residues 282–773 (Fig. 2, B and D). SRF binds to a region between 282 and 483 of CHD8, and it also binds to the region encompassing 483–773. To confirm that CHD8 and SRF interact with each other in SMCs, coimmunoprecipitation assays were performed on nuclear extracts isolated from A10 cells. Western blot analysis revealed that endogenous CHD8 and SRF are found together in anti-CHD8 immune complexes in SMCs (Fig. 2E).
Fig. 2.
Serum response factor (SRF) and CHD8 interact both in vitro and in vivo. A: schematic of the structural domains of SRF and the mutant proteins used to identify the binding domain of CHD8. B: GST-SRF-WT, GST-SRF-Δ1, GST-SRF-Δ2, GST-SRF-Δ3, or GST alone were bound to glutathione beads and incubated with CHD8Δ1 pET bacterial lysate to determine the minimal portion of SRF that binds to CHD8. n = 2. C: schematic of the structural domains of CHD8 and the mutant proteins used to identify the binding domain of SRF. D: GST-SRF-Δ2 was incubated with bacterial lysate of various truncations of CHD8. After washing was completed, the glutathione bead-bound proteins were analyzed by Western blot analysis. Presence of GST proteins was confirmed by staining the membrane with Ponceau S (bottom). n = 2. E: proteins were immunoprecipitated with a SRF-specific antibody (Santa Cruz, G20) or an IgG control, from nuclear lysates of A10 cells. Western blot analysis of the immunoprecipitated proteins was conducted using the primary antibodies specific for CHD8 (Bethyl, A310-225A) and SRF (Santa Cruz, G20). The image shown is representative of three separate experiments.
CHD8 knockdown attenuates expression of SRF-dependent genes in SMCs.
To investigate the function of CHD8 in smooth muscle, CHD8 was knocked down in A10 VSMCs, using an adenoviral-encoded shRNA, and the subsequent effects on expression of SRF-dependent genes were determined by quantitative RT-PCR. Using this approach, we obtained an ∼60–70% knockdown of CHD8 (Fig. 3). CHD8 knockdown resulted in a significant attenuation in expression of the smooth muscle differentiation markers smooth muscle-MHC and calponin by about 40–50%, without affecting expression of telokin, SM22α, or SRF (Fig. 3, top). The attenuated expression of calponin and smooth muscle-MHC was similar to that seen following knockdown of SRF; however, knockdown of SRF also attenuated expression of telokin ∼50%. Surprisingly, knockdown of either CHD8 or SRF did not affect expression of SM22α, a known SRF-dependent gene. In primary cultures of mouse bladder and colon SMCs, loss of CHD8 also caused a small but significant 20–25% attenuation in expression of differentiation genes, such as smooth muscle-MHC, SM22α, calponin, and SM α-actin (calponin and SM α-actin were only measured in colon) (Fig. 3, middle and bottom). Expression of vinculin also decreased following knockdown of CHD8 in bladder and colon SMCs, whereas expression of SRF was unchanged. cFos was more variable, its expression was unaltered in bladder SMC, whereas it was attenuated in colonic SMCs.
Because of the variable affects of CHD8 knockdown on cFos, we further determined whether CHD8 affected the serum stimulation of immediate early genes in primary colon SMCs. Knockdown of CHD8 was found not to affect the ability of serum to induce expression of cFos or SRF (data not shown). These data suggest that CHD8 is not required for serum induction of SRF-dependent growth/proliferation genes in colonic SMCs.
As SRF is important for both smooth muscle and myofibroblast differentiation, we next determined whether CHD8 is required for myofibroblast differentiation. We utilized transforming growth factor-β (TGFβ)-treated 10T1/2 cells as a model for myofibroblast differentiation. We found that knockdown of CHD8 in this system did not prevent TGFβ-dependent induction of smooth muscle markers such as SM22α and SM α-actin (data not shown). Similarly, knockdown of CHD8 did not significantly blunt the ability of myocardin to stimulate expression of smooth muscle-specific genes in 10T1/2 cells (data not shown).
Loss of CHD8 induces apoptosis in A10 VSMC.
In the experiments described above we noted that in SMCs in which CHD8 levels were reduced there was a significant increase in the number of detached dead cells. Previous studies have linked SRF to apoptosis through its ability to activate the transcription of anti-apoptotic Bcl2 family members (39, 43). To determine whether CHD8 plays a role in regulating apoptosis in SMCs, we knocked down expression of CHD8 or SRF in A10 VSMC and sensitized the cells to apoptosis with H2O2. Analysis of markers of apoptosis such as phospho-H2A.X, cleaved caspase-3, and cleaved PARP, in these cells, revealed that knockdown of either SRF or CHD8 induced apoptosis in the absence of any additional stimuli (Fig. 4, A and B). Apoptosis was further confirmed by increased numbers of small rounded detaching cells following CHD8 or SRF knockdown (Fig. 4C), by immunofluorescence staining for active caspase-3 (Fig. 4D), and by deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) staining (Fig. 4E). Induction of low levels of apoptosis by H2O2 to mimic free radical damage created during vascular injury enhanced the level of apoptosis in the knockdown cells, although H2O2 alone was not sufficient to induce significant apoptosis under the conditions used. Surprisingly, neither SRF nor CHD8 depletion appeared to affect the expression of Bcl-2 (Fig. 4A). While loss of SRF caused a small reduction in Mcl-1 expression and appeared to prevent the H2O2-mediated increase in Mcl-1, loss of CHD8 did not significantly affect its expression (Fig. 4A). In addition, even though both SRF and CHD8 depletion cause apoptosis-dependent cleavage of Bcl-XL, its total expression appears unchanged (Fig. 4A). These data indicate that CHD8 plays an anti-apoptotic role in A10 VSMCs, but this is not due to altered expression of the known SRF-regulated anti-apoptotic genes in the Bcl2 family. This result also lead us to determine whether the anti-apoptotic effects of CHD8 may involve its previously described binding partner CTCF (17). In support of this proposal, we found that CTCF knockdown also induced apoptosis in A10 SMC (Fig. 4B). CTCF knockdown also resulted in a small reproducible decrease in CHD8 protein levels without affecting CHD8 mRNA levels (Figs. 4B and 5C). The decreased CHD8 protein may reflect apoptosis-induced proteolysis of CHD8 as lower molecular mass bands were detectable on these blots with CHD8 antibodies (data not shown).
To identify possible targets of CHD8 that could be inhibiting apoptosis in A10 SMCs, we performed a PCR-based array screen of 84 apoptosis-related genes (SABiosciences catalog no. PARN-012A). Results from this screen, which were confirmed by direct qRT-PCR analysis, revealed decreased expression of two anti-apoptotic proteins CARD10 (75% decrease) and Birc5/survivin (a 50% decrease) in CHD8 knockdown cells. We also detected an increase (40%) in mRNA encoding the proapoptotic protein caspase 1 (Fig. 5A). Knockdown of CHD8 in primary cultures of colonic SMCs also resulted in a decrease in Birc5 expression, although there was no significant decrease in CARD10 (Fig. 5B). In contrast, we observed that knockdown of SRF attenuated expression of CARD10 but not Birc5 in A10 cells (Fig. 5A), whereas knockdown of CTCF decreased Birc5 but not CARD10 (Fig. 5C). These data suggest that CHD8 interacts with SRF and CTCF to regulate Birc5 and CARD10 expression and thereby promote survival of SMCs.
CHD8 directly binds to the Birc5 and smMHC promoters.
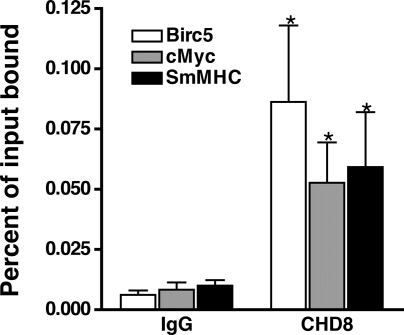
Data described above suggest that CHD8 regulates expression of anti-apoptotic and smooth muscle-specific genes. To confirm that CHD8 can directly regulate these genes, we examined CHD8 binding to the Birc5 and smMHC promoters using quantitative chromatin immunoprecipitation assays (ChIP). These assays revealed similar levels of CHD8 binding to the Birc5 promoter, the smMHC promoter, and the cMyc insulator region, which has previously been shown to bind CHD8 (17) (Fig. 6).
Fig. 6.
CHD8 binds to the Birc5 and smooth muscle myosin heavy chain (smMHC) promoters. Quantitative chromatin immunoprecipitation assays (ChIP) analysis of CHD8 binding to the Birc5 promoter, cMyc insulator [a known CHD8 binding site (17)], and smMHC promoter. Equal amounts of chromatin were immunoprecipitated with antibodies to CHD8 (Bethyl, A310-225A) or rabbit IgG control. Data presented are expressed relative to input samples and are means ± SE from 5 to 6 separate IPs.
DISCUSSION
Previous studies have demonstrated that SRF plays important roles in regulating genes required for differentiation, proliferation, migration, and apoptosis through interactions with specific coactivators (8, 14, 20, 29, 36, 37, 39, 43). In the current study we found that SRF associates with the chromatin remodeling enzyme CHD8 in SMCs and that CHD8 is required for expression of several SRF-dependent genes characteristic of differentiated smooth muscle. The importance of CHD8 was both gene and cell-type specific. For example, whereas knockdown of CHD8 attenuated smMHC expression in each of the smooth muscle cell types examined, it did not affect telokin expression. As both of these genes are known to be regulated by SRF and myocardin family members, these data suggest that there are distinct as well as common aspects of the regulation of these two genes, which ultimately leads to their differential expression. Consistent with this, there is only a single CArG box in the telokin promoter to which SRF binds and interacts with myocardin and other factors to regulate transcription (52). In contrast, SRF binds to multiple sites within the smMHC promoter and intronic enhancer to facilitate multimerization of myocardin to drive transcriptional activity. (46) Given that CHD8 binds to the MADS domain of SRF, which is also the main binding domain for many other cofactors such as Elk1 and myocardin, it is perhaps not surprising that CHD8 does not act identically on all SRF target genes. SM22α expression was attenuated in SMC from bladder and colon but not in A10 vascular SMCs following knockdown of CHD8 (Fig. 3). Surprisingly, SRF knockdown also did not attenuate SM22α expression in A10 cells. These data may suggest that SM22α is transcribed by SRF-independent mechanisms in these cells; however, this finding more likely reflects the partial knockdown of SRFs and CHD8 together with the high affinity of the SM22α promoter for SRF (21). Although we observed a small decrease in basal expression of cFos, an SRF-dependent gene required for proliferation, in colon SMCs following CHD8 knockdown, the ability of serum to stimulate cFos expression was not affected by CHD8. This would suggest that CHD8 is perhaps not required for the ets/Elk-dependent activation of SRF target genes.
CHD8 could regulate SRF-dependent genes through a number of different mechanisms. CHD8 has been shown to bind to histone H3 that is di- and tri-methylated at K4, through its chromodomains (49). In addition, this modified histone has been shown to be associated with smooth muscle-specific genes that are actively transcribing and to associate with SRF/myocardin complexes (26). Based on these observations we propose that CHD8 may function to help recruit SRF to the promoters of smooth muscle-specific genes through CHD8's chromodomains interacting with histone H3 K4diMe. CHD8 could also affect expression of differentiation genes directly through its role as an ATP-dependent chromatin remodeling enzyme (41). In support of this proposal, CHD8 has been shown to be present in complexes that also include the switch/sucrose nonfermentable (SWI/SNF) chromatin remodeling enzymes (41), and we have previously shown that the SWI/SNF complex plays a critical role in the induction of smooth muscle differentiation genes by the myocardin family of transcription activators (51, 53).
Loss of CHD8 resulted in a marked increase in apoptosis of A10 VSMCs, indicating that CHD8 normally has an important survival function in these cells (Fig. 4). This is consistent with the massive apoptosis that was observed in global CHD8 knockout embryos (33). It has also been reported that duplin (the short form of CHD8) suppresses p53-mediated apoptosis (34), and simultaneous knockout of p53 and duplin/CHD8 partially rescued the lethal phenotype of duplin/CHD8 null mice (34). Interestingly, these double knockout mice now die due to vascular defects at E10.5. Although we cannot rule out a possible role for p53 in the apoptosis induced by knockdown of CHD8 in A10 cells, we did not observe any elevated expression of the p53 target gene p21 (Fig. 5). In fact, we observed decreased expression of p21 following knockdown of either SRF or CHD8 (Fig. 5A). The decreased expression of p21 is consistent with recent data showing that p21 is activated by myocardin/SRF complexes in SMCs (22). Anti-apoptotic activity has previously been attributed to the short duplin isoform as opposed to the longer CHD8(34); however, we found that CHD8 is the predominant isoform found in adult smooth muscle tissues and cells where duplin is largely undetectable (Fig. 1). This would suggest that in SMCs CHD8 also exhibits anti-apoptotic activity. The unexpected finding that CHD8 is important for expression of the anti-apoptotic proteins Birc5 and CARD10 in A10 VSMCs is consistent with the vascular defects noted in the CHD8/p53 knockout mice and suggests that CHD8 may play an important survival role in VSMCs independent of any protective effects it exerts through regulation of p53. Interestingly, Birc5 transcription and Mcl-1 levels are also repressed by p53, hence in p53 knockout mice Birc5 and Mcl-1 levels would be increased promoting cell survival, thereby contributing to the protective effects of p53 knockout in the p53/CHD8 double knockout mice (10, 16, 32). Previous studies have shown that in a rabbit balloon injury model, Birc5 is upregulated 4 to 7 days postinjury in medial SMCs, and Birc5 expression peaked in the neointima and media at day 14 postinjury (4). Importantly, after stimulating SMCs with serum, PDGF-AB, or HB-EGF, Birc5/survivin expression was increased by 16-fold, 13-fold, and 9-fold, respectively, with peak levels detected at 20 to 24 h poststimulation (4). Moreover, expression of a nonphosphorylatable Birc5/survivin induced apoptosis in proliferating VSMCs. These data suggest that decreased survivin/Birc5 activity in VSMCs is sufficient to induce apoptosis. Although our studies focused on A10 SMCs, our data together with these previously published studies and the observation that Birc5 is also decreased following CHD8 knockdown in colonic SMCs (Fig. 5B), suggest that CHD8 may play an important role in VSMC survival in vivo. In addition to the observed decrease in Birc5 following knockdown of CHD8, we also observed a decrease in the anti-apoptotic protein CARD10. CARD10 is a member of the membrane-associated guanylate kinase family that transmits signals through BCL10 leading to activation of nuclear factor (NF)-κB (45). Decreased expression of CARD10 would thus be predicted to decrease the activity of this survival pathway. The decreased activation of NF-κB survival signaling due to the attenuated CARD10 and increased caspase activation mediated by loss of Birc5 provide mechanisms to account for the apoptosis seen in A10 cells following knockdown of CHD8. The increased expression of caspase 1 observed following CHD8 knockdown may be more associated with an increased inflammatory response as opposed to increased apoptosis as the main function of this caspase is to activate inflammatory cytokines such as IL-1β (25). In support of this, SRF knockdown decreased caspase 1 expression while still inducing apoptosis (Figs. 4 and 5).
Although we isolated CHD8 as an SRF-associated protein, and either knockdown of CHD8 or SRF induced apoptosis in A10 VSMCs, it is likely that the mechanisms by which SRF and CHD8 protect cells from apoptosis have both common as well as distinct elements/components. For example, knockdown of either protein decreased CARD10 expression, suggesting that SRF/CHD8 complexes may regulate expression of this gene, whereas Birc5 expression decreased only in response to CHD8 knockdown and Mcl-1 expression was decreased only in response to SRF knockdown (Figs. 4 and 5). Previous studies have also demonstrated a role of SRF in regulating cell survival in embryonic stem cells in which the prosurvival activity of SRF was attributed to its ability to regulate differentiation-dependent transcription of the anti-apoptotic factor Bcl-2. In contrast to these studies, we did not observe any change in expression of Bcl-2 following knockdown of SRF in A10 VSMCs, suggesting that the activities of SRF are cell-type specific (Fig. 4). Consistent with our observations in A10 cells, SRF has also been shown to regulate expression of the anti-apoptotic factor Mcl-1 in both HeLa cells and EcR293, transformed human embryonic kidney cells (43). Relevant to this are studies showing that the anti-apoptotic activity of Mcl-1 in VSMCs results from its ability to stabilize fortilin (50). As attenuated Birc5 expression was found following knockdown of CHD8 but not following knockdown of SRF, this would suggest that CHD8 is regulating Birc5 expression through an SRF-independent mechanism. Previously, CHD8 was shown to interact with the chromatin insulator factor CTCF. Through this interaction, CHD8 was able to function in the epigenetic regulation of the reciprocal transcription of H19 and IGF2, as well as to participate in preventing the spread of CpG methylation or condensed chromatin adjacent to the BRCA1 and c-myc promoters, respectively (17). We found that knockdown of CTCF also induced apoptosis in A10 SMCs that was associated with decreased Birc5 expression (Fig. 5C). These data suggest that CHD8/CTCF complexes may be important for regulating Birc5 expression. Interestingly the Birc5 promoter has been reported to contain a CpG island (23) raising the possibility that CHD8/CTCF complexes regulate Birc5 expression through inhibition of promoter methylation. However, although we found that knockdown of CHD8 or CTCF tended to increase the methylation status of the CpG island in the Birc5 promoter, these changes were not significant (data not shown). These data would suggest that this is perhaps not the main mechanism through which these proteins regulate Birc5 expression.
In summary our data demonstrate that SRF, CHD8, and CTCF complexes play important roles in regulating differentiation and survival of SMCs. Our data further demonstrate that SRF activity can be modulated through recruitment of the CHD8 chromatin-remodeling enzyme. Collectively, these data provide new insights into mechanisms regulating survival pathways in smooth muscle cells that have been shown to play important roles in vascular pathophysiology.
GRANTS
This work was supported in part by National Institutes of Health DK61130 and a Biomedical Research Grant from Indiana University School of Medicine.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
REFERENCES
- 1.Akhtar A, Zink D, Becker PB. Chromodomains are protein-RNA interaction modules. Nature 407: 405–409, 2000 [DOI] [PubMed] [Google Scholar]
- 2.Angstenberger M, Wegener JW, Pichler BJ, Judenhofer MS, Feil S, Alberti S, Feil R, Nordheim A. Severe intestinal obstruction on induced smooth muscle-specific ablation of the transcription factor SRF in adult mice. Gastroenterology 133: 1948–1959, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Arsenian S, Weinhold B, Oelgeschlager M, Ruther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J 17: 6289–6299, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanc-Brude OP, Yu J, Simosa H, Conte MS, Sessa WC, Altieri DC. Inhibitor of apoptosis protein survivin regulates vascular injury. Nat Med 8: 987–994, 2002 [DOI] [PubMed] [Google Scholar]
- 5.Bouazoune K, Mitterweger A, Langst G, Imhof A, Akhtar A, Becker PB, Brehm A. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J 21: 2430–2440, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavalli G, Paro R. Chromo-domain proteins: linking chromatin structure to epigenetic regulation. Curr Opin Cell Biol 10: 354–360, 1998 [DOI] [PubMed] [Google Scholar]
- 7.Cen B, Selvaraj A, Prywes R. Myocardin/MKL family of SRF coactivators: key regulators of immediate early and muscle specific gene expression. J Cell Biochem 93: 74–82, 2004 [DOI] [PubMed] [Google Scholar]
- 8.Chai J, Tarnawski AS. Serum response factor: discovery, biochemistry, biological roles and implications for tissue injury healing. J Physiol Pharmacol 53: 147–157, 2002 [PubMed] [Google Scholar]
- 9.Charvet C, Houbron C, Parlakian A, Giordani J, Lahoute C, Bertrand A, Sotiropoulos A, Renou L, Schmitt A, Melki J, Li Z, Daegelen D, Tuil D. New role for serum response factor in postnatal skeletal muscle growth and regeneration via the interleukin 4 and insulin-like growth factor 1 pathways. Mol Cell Biol 26: 6664–6674, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaturvedi V, Sitailo LA, Qin JZ, Bodner B, Denning MF, Curry J, Zhang W, Brash D, Nickoloff BJ. Knockdown of p53 levels in human keratinocytes accelerates Mcl-1 and Bcl-x(L) reduction thereby enhancing UV-light induced apoptosis. Oncogene 24: 5299–5312, 2005 [DOI] [PubMed] [Google Scholar]
- 11.Cowell IG, Austin CA. Self-association of chromo domain peptides. Biochim Biophys Acta 1337: 198–206, 1997 [DOI] [PubMed] [Google Scholar]
- 12.El-Mounayri O, Triplett JW, Yates CW, Herring BP. Regulation of smooth muscle-specific gene expression by homeodomain proteins, Hoxa10 and Hoxb8. J Biol Chem 280: 25854–25863, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Herring BP, Kriegel AM, Hoggatt AM. Identification of Barx2b, a serum response factor-associated homeodomain protein. J Biol Chem 276: 14482–14489, 2001 [DOI] [PubMed] [Google Scholar]
- 14.Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell 81: 1159–1170, 1995 [DOI] [PubMed] [Google Scholar]
- 15.Hinson JS, Medlin MD, Lockman K, Taylor JM, Mack CP. Smooth muscle cell-specific transcription is regulated by nuclear localization of the myocardin-related transcription factors. Am J Physiol Heart Circ Physiol 292: H1170–H1180, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem 277: 3247–3257, 2002 [DOI] [PubMed] [Google Scholar]
- 17.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell 23: 733–742, 2006 [DOI] [PubMed] [Google Scholar]
- 18.Iwasaki K, Hayashi K, Fujioka T, Sobue K. Rho/Rho-associated kinase signal regulates myogenic differentiation via myocardin-related transcription factor-A/Smad-dependent transcription of the Id3 gene. J Biol Chem 283: 21230–21241, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeon ES, Park WS, Lee MJ, Kim YM, Han J, Kim JH. A Rho kinase/myocardin-related transcription factor-A-dependent mechanism underlies the sphingosylphosphorylcholine-induced differentiation of mesenchymal stem cells into contractile smooth muscle cells. Circ Res 103: 635–642, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Johansen FE, Prywes R. Two pathways for serum regulation of the c-fos serum response element require specific sequence elements and a minimal domain of serum response factor. Mol Cell Biol 14: 5920–5928, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S, Ip HS, Lu MM, Clendenin C, Parmacek MS. A serum response factor-dependent transcriptional regulatory program identifies distinct smooth muscle cell sublineages. Mol Cell Biol 17: 2266–2278, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura Y, Morita T, Hayashi K, Miki T, Sobue K. Myocardin functions as an effective inducer of growth arrest and differentiation in human uterine leiomyosarcoma cells. Cancer Res 70: 501–511, 2010 [DOI] [PubMed] [Google Scholar]
- 23.Li F, Altieri DC. Transcriptional analysis of human survivin gene expression. Biochem J 344: 305–311, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Czubryt MP, McAnally J, Bassel-Duby R, Richardson JA, Wiebel FF, Nordheim A, Olson EN. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc Natl Acad Sci USA 102: 1082–1087, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell 10: 417–426, 2002 [DOI] [PubMed] [Google Scholar]
- 26.McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest 116: 36–48, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mericskay M, Blanc J, Tritsch E, Moriez R, Aubert P, Neunlist M, Feil R, Li Z. Inducible mouse model of chronic intestinal pseudo-obstruction by smooth muscle-specific inactivation of the SRF gene. Gastroenterology 133: 1960–1970, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Miano JM. Serum response factor: toggling between disparate programs of gene expression. J Mol Cell Cardiol 35: 577–593, 2003 [DOI] [PubMed] [Google Scholar]
- 29.Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol 292: C70–C81, 2007 [DOI] [PubMed] [Google Scholar]
- 30.Miano JM, Ramanan N, Georger MA, de Mesy Bentley KL, Emerson RL, Balza RO, Jr, Xiao Q, Weiler H, Ginty DD, Misra RP. Restricted inactivation of serum response factor to the cardiovascular system. Proc Natl Acad Sci USA 101: 17132–17137, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113: 329–342, 2003 [DOI] [PubMed] [Google Scholar]
- 32.Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S, Wen SF, Wang L, Kirschmeier P, Bishop WR, Nielsen LL, Pickett CB, Liu S. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene 21: 2613–2622, 2002 [DOI] [PubMed] [Google Scholar]
- 33.Nishiyama M, Nakayama K, Tsunematsu R, Tsukiyama T, Kikuchi A, Nakayama KI. Early embryonic death in mice lacking the beta-catenin-binding protein Duplin. Mol Cell Biol 24: 8386–8394, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishiyama M, Oshikawa K, Tsukada YI, Nakagawa T, Iemura SI, Natsume T, Fan Y, Kikuchi A, Skoultchi AI, Nakayama KI. CHD8 suppresses p53-mediated apoptosis through histone H1 recruitment during early embryogenesis. Nat Cell Biol 11: 172–182, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niu Z, Yu W, Zhang SX, Barron M, Belaguli NS, Schneider MD, Parmacek M, Nordheim A, Schwartz RJ. Conditional mutagenesis of the murine serum response factor gene blocks cardiogenesis and the transcription of downstream gene targets. J Biol Chem 280: 32531–32538, 2005 [DOI] [PubMed] [Google Scholar]
- 36.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev 84: 767–801, 2004 [DOI] [PubMed] [Google Scholar]
- 37.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev 20: 1545–1556, 2006 [DOI] [PubMed] [Google Scholar]
- 38.Sakamoto I, Kishida S, Fukui A, Kishida M, Yamamoto H, Hino S, Michiue T, Takada S, Asashima M, Kikuchi A. A novel beta -catenin-binding protein inhibits beta -catenin-dependent tcf activation and axis formation [In Process Citation]. J Biol Chem 275: 32871–32878, 2000 [DOI] [PubMed] [Google Scholar]
- 39.Schratt G, Philippar U, Hockemeyer D, Schwarz H, Alberti S, Nordheim A. SRF regulates Bcl-2 expression and promotes cell survival during murine embryonic development. EMBO J 23: 1834–1844, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selvaraj A, Prywes R. Expression profiling of serum inducible genes identifies a subset of SRF target genes that are MKL dependent. BMC Mol Biol 5: 13, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson BA, Tremblay V, Lin G, Bochar DA. CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol Cell Biol 28: 3894–3904, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Treisman R. Ternary complex factors: growth factor regulated transcriptional activators. Curr Opin Genet Dev 4: 96–101, 1994 [DOI] [PubMed] [Google Scholar]
- 43.Vickers ER, Kasza A, Kurnaz IA, Seifert A, Zeef LA, O'Donnell A, Hayes A, Sharrocks AD. Ternary complex factor-serum response factor complex-regulated gene activity is required for cellular proliferation and inhibition of apoptotic cell death. Mol Cell Biol 24: 10340–10351, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105: 851–862, 2001 [DOI] [PubMed] [Google Scholar]
- 45.Wang L, Guo Y, Huang WJ, Ke X, Poyet JL, Manji GA, Merriam S, Glucksmann MA, DiStefano PS, Alnemri ES, Bertin J. Card10 is a novel caspase recruitment domain/membrane-associated guanylate kinase family member that interacts with BCL10 and activates NF-kappa B. J Biol Chem 276: 21405–21409, 2001 [DOI] [PubMed] [Google Scholar]
- 46.Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci USA 100: 7129–7134, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin F, Hoggatt AM, Zhou J, Herring BP. 130-kDa smooth muscle myosin light chain kinase is transcribed from a CArG-dependent, internal promoter within the mouse mylk gene. Am J Physiol Cell Physiol 290: C1599–C1609, 2006 [DOI] [PubMed] [Google Scholar]
- 48.Yoshida T, Gan Q, Shang Y, Owens GK. Platelet-derived growth factor-BB represses smooth muscle cell marker genes via changes in binding of MKL factors and histone deacetylases to their promoters. Am J Physiol Cell Physiol 292: C886–C895, 2007 [DOI] [PubMed] [Google Scholar]
- 49.Yuan CC, Zhao X, Florens L, Swanson SK, Washburn MP, Hernandez N. CHD8 associates with human Staf and contributes to efficient U6 RNA polymerase III transcription. Mol Cell Biol 27: 8729–8738, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang D, Li F, Weidner D, Mnjoyan ZH, Fujise K. Physical and functional interaction between myeloid cell leukemia 1 protein (MCL1) and Fortilin. The potential role of MCL1 as a fortilin chaperone. J Biol Chem 277: 37430–37438, 2002 [DOI] [PubMed] [Google Scholar]
- 51.Zhang M, Fang H, Zhou J, Herring BP. A novel role of Brg1 in the regulation of SRF/MRTFA-dependent smooth muscle-specific gene expression. J Biol Chem 282: 25708–25716, 2007 [DOI] [PubMed] [Google Scholar]
- 52.Zhou J, Herring BP. Mechanisms responsible for the promoter-specific effects of myocardin. J Biol Chem 280: 10861–10869, 2005 [DOI] [PubMed] [Google Scholar]
- 53.Zhou J, Zhang M, Fang H, El-Mounayri O, Rodenberg JM, Imbalzano AN, Herring BP. The SWI/SNF chromatin remodeling complex regulates myocardin-induced smooth muscle-specific gene expression. Arterioscler Thromb Vasc Biol 29: 921–928, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]