Abstract
Recently, overexpression of the genes TMEM16A and TMEM16B has been shown to produce currents qualitatively similar to native Ca2+-activated Cl− currents (IClCa) in vascular smooth muscle. However, there is no information about this new gene family in vascular smooth muscle, where Cl− channels are a major depolarizing mechanism. Qualitatively similar Cl− currents were evoked by a pipette solution containing 500 nM Ca2+ in smooth muscle cells isolated from BALB/c mouse portal vein, thoracic aorta, and carotid artery. Quantitative PCR using SYBR Green chemistry and primers specific for transmembrane protein (TMEM) 16A or the closely related TMEM16B showed TMEM16A expression as follows: portal vein > thoracic aorta > carotid artery > brain. In addition, several alternatively spliced variant transcripts of TMEM16A were detected. In contrast, TMEM16B expression was very low in smooth muscle. Western blot analysis with different antibodies directed against TMEM16A revealed a number of products with a consistent band at ∼120 kDa, except portal vein, where an 80-kDa band predominated. TMEM16A protein was identified in the smooth muscle layers of 4-μm-thick slices of portal vein, thoracic aorta, and carotid artery. In isolated myocytes, fluorescence specific to a TMEM16A antibody was detected diffusely throughout the cytoplasm, as well as near the membrane. The same antibody used in Western blot analysis of lysates from vascular tissues also recognized an ∼147-kDa mouse TMEM16A-green fluorescent protein (GFP) fusion protein expressed in HEK 293 cells, which correlated to a similar band detected by a GFP antibody. Patch-clamp experiments revealed that IClCa generated by transfection of TMEM16A-GFP in HEK 293 cells displayed remarkable similarities to IClCa recorded in vascular myocytes, including slow kinetics, steep outward rectification, and a response similar to the pharmacological agent niflumic acid. This study shows that TMEM16A expression is robust in murine vascular smooth muscle cells, consolidating the view that this gene is a viable candidate for the native Ca2+-activated Cl− channel in this cell type.
Keywords: calcium-activated chloride channels, anoctamin 1, gene expression, smooth muscle contraction
vascular smooth muscle cells accumulate Cl− (7). Consequently, activation of Cl− channels leads to Cl− efflux, membrane depolarization, and increased Ca2+ influx through voltage-dependent Ca2+ channels, resulting in vasoconstriction (21, 22). Ca2+-activated Cl− channels (CACCs) are the major anionic species in vascular myocytes, but despite the wealth of electrophysiological data, the molecular architecture of CACCs in smooth muscle has not been determined with any confidence (22). Recently, overexpression of transmembrane protein (TMEM) 16A and its closely related paralog TMEM16B, two genes of a family comprising 10 genes in mammals, has been shown to generate Cl− currents with voltage-dependent kinetics and Ca2+ sensitivity similar to those of Ca2+-activated Cl− currents (IClCa) recorded from smooth muscle cells (6, 8, 14, 25, 30, 33, 35). Molecular interference studies [small interfering RNA (siRNA) and transgenic mice] have consolidated the view that TMEM16A is a component of Ca2+-activated Cl− fluxes in airway epithelial cells (16, 19, 24, 28), but there is no information on the expression of TMEM16A in smooth muscles, and IClCa in airway epithelium display little resemblance to IClCa in vascular smooth muscle with respect to voltage-dependent kinetics, regulation by Ca2+/calmodulin-dependent kinase II (CaMKII), and Ca2+ sensitivity (9, 22). TMEM16A immunoreactivity has been identified in gastrointestinal smooth muscles, but it resides almost exclusively in the interstitial cells of Cajal, and not in smooth muscle cells (10, 17, 35, 36). Consequently, there is no information about the quantitative expression of TMEM16A relative to TMEM16B, the presence of the known splice variants (8, 14), and the nature of translation products of these genes in any smooth muscle. The aim of the present study was to perform a quantitative analysis of TMEM16A and TMEM16B gene expression in different murine blood vessels that exhibit robust IClCa and to ascertain the validity of these genes as candidates of native CACCs.
EXPERIMENTAL PROCEDURES
Tissue collection.
BALB/c mice (6–8 wk of age) were killed by an overdose of pentobarbitone sodium in accordance with Schedule 1 of the United Kingdom Animals Act (1986) or sedated with isoflurane prior to cervical dislocation as approved by the local Institutional Animal Care and Use Committee in accordance with the Animal Welfare Act of the United States.
Cell dissociation.
Single smooth muscle cells were isolated from murine portal vein, as described previously (4, 31, 32, 34). Murine thoracic aorta and carotid artery were digested in a physiological salt solution containing 50 μM Ca2+, 1.5 mg/ml collagenase XI (Sigma), and 1 mg/ml protease XIV (Sigma) for 27 min and triturated gently for 7 and 20 min. Cells were kept at 4°C before use.
Patch-clamp electrophysiology.
IClCa were elicited using the conventional whole cell configuration of the patch-clamp technique with a pipette solution containing 3 mM ATP. The pipette solution also contained 10 mM BAPTA as the Ca2+ buffer, and free Ca2+ concentration was set to 500 nM by the addition of 7.08 mM CaCl2, estimated by the Ca2+ chelator program MaxC (version 2.50; http://www.stanford.edu/≃cpatton/downloads.htm). Contamination of IClCa from other types of current was minimized by the use of CsCl and tetraethylammonium chloride in the pipette solution and tetraethylammonium chloride in the external solution (2–4, 11, 12, 29, 32). IClCa was evoked immediately upon rupture of the cell membrane, and the voltage-dependent properties were monitored every 15 s by stepping from a holding potential (HP) of −50 mV to +90 mV for 1 s followed by repolarization to −80 mV for 1 s. Current-voltage relationships were constructed by stepping in 20-mV increments from the HP to test potentials between −100 and +130 mV for 1 s after 20 min of dialysis. For current-voltage relationships, IClCa was expressed as current density (pA/pF) by dividing the amplitude of the current measured at the end of the voltage-clamp step by the cell capacitance. All chemicals were obtained from Sigma unless otherwise stated.
TMEM16A overexpression studies.
Murine TMEM16A was kindly supplied by Professor Lily Y. Jan (Howard Hughes Medical Institute, University of California San Francisco). The full-length open reading frame was subcloned into pcDNA3.1/CT-GFP upstream of a green fluorescent protein (GFP) tag. Endotoxin-free plasmid DNA was prepared using the Endo-Free kit (Qiagen, Valencia, CA). HEK 293 cells were transfected with DNA at 250 ng/cm2 surface area using TransIT-LT1 reagent (Mirus, Madison, WI). After 18 h, these cells were used for immunofluorescence, Western blotting, or electrophysiology (see below).
RNA extraction and cDNA synthesis.
Total RNA extraction from various murine tissues was accomplished using the RNeasy Micro kit (Qiagen) or PureZOL RNA isolation reagent (Bio-Rad) according to the manufacturer's instruction and included an on-column DNase treatment step. RNA quality was measured using a Nanodrop spectrophotometer (Agilent Technologies) and reverse-transcribed with oligo(dT)(12–18) primers and Maloney's murine leukemia virus or SuperScript III reverse transcriptase (Invitrogen). Negative controls (RT−) were carried out in the absence of reverse transcriptase and used to check for genomic contamination. All samples were stored at −20°C prior to PCR amplification.
Primers.
The housekeeping genes β-actin and GAPDH served as references to which the gene of interest was normalized [quantitative PCR (qPCR) studies]. Primers for all genes were designed to allow amplification of exon-spanning amplicons, thereby allowing for identification of contaminating genomic DNA. Primers (Table 1) were synthesized by Invitrogen, Eurofins MWG Operon, or PrimerDesign (murine TMEM16B).
Table 1.
PCR primer pair sequences used to probe for TMEM16A and TMEM16B in various tissues
| Primer Sequence | GenBank Accession No. | Amplicon, bp | Region Spanned | |
|---|---|---|---|---|
| Conventional PCR primers | ||||
| TMEM16A | NM_178642 | 324 | 611–935 | |
| Sense | 5′-GGTGTCGGGTTTGTGAAGAT-3′ | |||
| Antisense | 5′-TGCACGTTGTTCTCTTCAGG-3′ | |||
| TMEM16B | NM_153589 | 480 | 1640–2120 | |
| Sense | 5′-GCAACAGCAGTCATCATCA-3′ | |||
| Antisense | 5′-TCAGCCCAGTGTAAGGCTCT-3′ | |||
| β-Actin | NM_007393 | 573 | 225–797 | |
| Sense | 5′-TGTTACCAACTGGGACGACA-3′ | |||
| Antisense | 5′-AAGGAAGGCTGGAAAAGAGC-3′ | |||
| Splice variant PCR primers | ||||
| Exon a annealing | NM_178642.4 | 413 | 222–635 | |
| Sense | 5′-GCGCCATCGAGGACCTGGGC-3′ | |||
| Antisense | 5′-GCCCAAACACTTCTAGGTACGC-3′ | |||
| Exon b spanning | NM_178642.4 | 741 | 613–1354 | |
| Sense | 5′-CGGGTTTGTGAAGATCCATGCG-3′ | |||
| Antisense | 5′-CGGACACGGTGTCGGGCACGG-3′ | |||
| Exon c annealing | NM_178642.4 | 421 | 1485–1906 | |
| Sense | 5′-GGAGGAGGAGGAAGCTGTCAAG-3′ | |||
| Antisense | 5′-CCTCTCCGATTGGAAGTTCCGG-3′ | |||
| Exon c spanning 1 | NM_178642.4 | 395 | 1140–1535 | |
| Sense | 5′-GCCTGTGCCACAGCCCGTGCC-3′ | |||
| Antisense | 5′-GGGTCTCGTCTCATACTTCGG-3′ | |||
| Exon c spanning 2 | NM_178642.4 | 200 | 1334–1534 | |
| Sense | 5′-GCCTGTACTTTGCCTGGCTTGG-3′ | |||
| Antisense | 5′-GGGTCTCGTCTCATACTTCGG-3′ | |||
| Exon d spanning | NM_178642.4 | 393 | 1513–1906 | |
| Sense | 5′-CCCAGAGCAGAGTATGAAGCC-3′ | |||
| Antisense | 5′-CCTCTCCGATTGGAAGTTCCGG-3′ | |||
| Exon d annealing | NM_178642.4 | 298 | 1608–1906 | |
| Sense | 5′-CCGATTCCCAGCCTATTTCACC-3′ | |||
| Antisense | 5′-CCTCTCCGATTGGAAGTTCCGG-3′ | |||
| qPCR primers | ||||
| TMEM16A | NM_178642 | 180 | 756–935 | |
| Sense | 5′-AGATCACAGACCCCATCCAG-3′ | |||
| Antisense | 5′-GGACTTCTCTTGTTGCACGT-3′ | |||
| TMEM16B | NM_153589 | 97 | 1776–1872 | |
| Sense | 5′-CCAAAGTACGAGAGAAACTGTTAAA-3′ | |||
| Antisense | 5′-TTATCTTCATCATCCTCATCATCCT-3′ | |||
| β-Actin | NM_007393 | 277 | 335–612 | |
| Sense | 5′-CTAAGGCCAACCGTGAAAAG-3′ | |||
| Antisense | 5′-ATGTCGAAGTGGTGGTGTCG-3′ | |||
| GAPDH | NM_008084 | 129 | 36–165 | |
| Sense | 5′-CGTCCCGTAGACAAAATGGT-3′ | |||
| Antisense | 5′-GAGGTCAATGAAGGGGTCG-3′ |
TMEM16A and TMEM16B, transmembrane proteins 16A and 16B; qPCR, quantitative PCR.
PCRs.
Non- or semi-qPCR was performed using Platinum Taq DNA polymerase (Invitrogen) or GoTaq Green Mastermix (Promega). The two Taq polymerases have slightly different amplification profiles: an initial denaturation step at 94°C followed by 35 cycles of 94°C for 30 s and an annealing temperature specific for each primer set for 30 s and 72°C for 1 min (Invitrogen) or an initial denaturation step at 95°C followed by 35 cycles of 95°C for 30 s and an annealing temperature specific for each primer set for 30 s and 72°C for 30 s; the reaction was completed with a 5-min extension step. β-Actin or GAPDH served as internal control. PCR product amplification was confirmed with subsequent 2% agarose gel electrophoresis and sequence analysis (University of Dundee Sequencing Facility or Nevada Genomics Center) and checked using the National Center for Biotechnology Information Basic Local Alignment Search Tool (BLAST) program.
qPCR.
Quantitative analysis of mRNA expression was determined using Brilliant SYBR Green qPCR Master Mix (Stratagene) with the Mx3000 system (Stratagene). Duplicate reactions were performed in 25-μl volumes including 1.5 μl of cDNA, 12.5 μl of SYBR Green qPCR Master Mix, and 0.75 μl of the passive reference 5-carboxy-X-rhodamine (ROX) dye (supplied with Master Mix). The following cycling conditions were used: initial denaturation at 95°C for 10 min (segment 1) followed by 50 cycles of 95°C for 30 s, annealing at 60°C for 1 min, and extension at 72°C for 30 s (segment 2). A final segment (segment 3) of 95°C for 1 min, 55°C for 30 s, and 95°C for 30 s completed the protocol. Fluorescence data were collected and used for analysis from data acquired at segment 2. Data (means ± SE) are expressed as 2−ΔCt. All quantitative data were accrued from three different animals. Melt curve analysis was performed to ensure that each primer set amplified a single product; because SYBR Green will bind any double-stranded product, any amplification of nonspecific material will artificially increase the level of fluorescence. A single peak confirmed that a single product had been amplified and that the Ct (cycle threshold) values generated were accurate. Ct values were determined using MxPro qPCR software (Stratagene), and RNA abundance relative to β-actin or GAPDH was calculated using 2−ΔCt. Standard curves were plotted using 10-fold serial dilutions of cDNA from murine brain (deemed a suitable positive control from previous semi-qPCR) to ascertain the efficiency of amplification of each gene used in this study. After a logarithmic conversion, a linear response was generated, and only R2 values ≥0.99 were accepted. Additionally, it was ensured that efficiencies were comparable between each primer set in each individual experiment.
Western blot analysis.
Protein was prepared by homogenization of tissue in lysis buffer [20 mM Tris base, 137 mM NaCl, 2 mM EDTA, 1% NP-40, 10% glycerol (pH 8), and 10 μl/ml protease inhibitor cocktail; Sigma] and subsequent incubation on ice for 15 min prior to centrifugation to remove cell debris. HEK 293 cells were lysed by incubation in lysis buffer with two 1-min bursts of vortexing followed by centrifugation to remove cell debris. H441 cell protein was a kind gift from Dr. Baines (St. George's, University of London). All supernatants were short-term stored at −20°C until required. Equal concentrations of all proteins were denatured at 95°C for 5 min in the presence of sample buffer and reducing agent (Invitrogen); then the proteins were separated on SDS-polyacrylamide gels (Invitrogen) and transferred onto polyvinylidene difluoride (PVDF) membranes. TMEM16A expression products were immunodetected using a rabbit polyclonal anti-TMEM16A antibody [1:500 dilution; ab53213, an undiluted form used previously (8, 18)] and visualized with ECL (ThermoFisher Scientific) and Hyperfilm (Amersham). No control peptide was supplied for this antibody, and the sequence of the epitope targeted by this antibody is not available. Quantifiable differences in antibody binding between different TMEM16A isoforms were not previously reported (8), suggesting that the epitope for ab53213 does not lie over spliced sites. Protein lysates were interrogated with serum from a nonimmunogenic rabbit (negative control for ab53213).
Immunohistochemistry.
After dissection, murine portal vein, thoracic aorta, and carotid artery were flash-frozen in chilled isopentane, embedded in Tissue-Tek optimal cutting temperature compound (Sakura Finetek, Zoeterwoude, The Netherlands), and frozen at −20°C. Sections (4 μm) were prepared and mounted onto poly-l-lysine-coated slides prior to endoperoxidase and serum blocking steps, which were performed according to the manufacturer's instructions (ImmunoCruz Staining System, Santa Cruz Biotechnology). Primary antibodies (ab53213 and ab72984, Abcam) were applied to the sections for 18 h at 4°C, and the biotinylated secondary antibody was applied for 30 min. Streptavidin-conjugated horseradish peroxidase (HRP) was applied to the sections for 30 min; then a final diaminobenzidine-HRP reaction was performed until immunolocalization was apparent. Negative controls were performed by incubation of sections without primary antibody and run in parallel for each experiment, i.e., time matched to the experiment. Epitope sites for both antibodies used for immunohistochemistry were not provided by the manufacturer and are unknown (see Western blot analysis for further detail regarding ab53213).
Immunocytochemistry.
Enzymatically isolated smooth muscle and H441 cells were fixed in 4% paraformaldehyde solution for 1 min at room temperature and then incubated for 5 min with antibody diluent (PBS containing 0.1% Triton X-100 and 0.5% BSA). Cells were then incubated with primary antibody (ab53213, Abcam; see Western blot analysis for further details) in antibody diluent overnight in a humidified chamber at 4°C. For control experiments, the primary antibody was omitted. Cells were subsequently incubated with a donkey anti-rabbit secondary antibody conjugated with the fluorescent tetramethylrhodamine isothiocyanate (TRITC) probe (1:100 dilution; Jackson Immuno Research Laboratories) or biotinylated goat anti-rabbit antibody (1:2,000 dilution) with streptavidin (1:1,000 dilution) for 1 h at room temperature. Unbound secondary antibody was removed, and cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 5 min at room temperature. Single cells were imaged using a laser scanning confocal microscope (model LSM 510, Zeiss) with an excitation of 488 nm (TRITC) or 345 nm (DAPI). Images for each cell type were gain-matched to ensure accuracy between samples. A cross section of the cell was selected for display purposes.
Statistical analysis.
Use of statistics is highlighted throughout the manuscript.
RESULTS
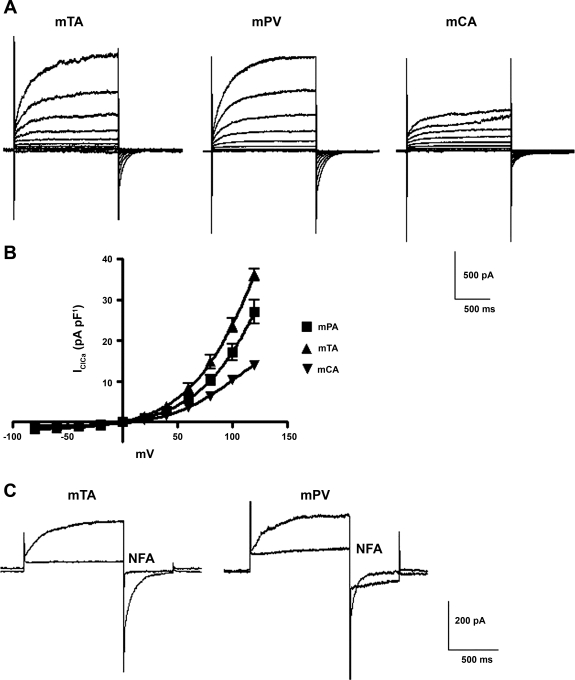
The purpose of the study was to ascertain whether TMEM16A is expressed in smooth muscle cells that exhibit IClCa. Most of our studies have focused on the rabbit pulmonary artery (2, 3, 11, 12, 34) and murine portal vein (4, 29, 31, 32). Figure 1A shows that, in addition to myocytes from murine portal vein, robust IClCa with distinctive voltage-dependent kinetics were also evoked in myocytes isolated from murine thoracic aorta and carotid artery with pipette solutions containing 500 nM Ca2+. In each cell type, the currents were qualitatively and kinetically similar. For instance, the current at +70 mV developed monoexponentially with mean time constants of 251 ± 32, 180 ± 24, and 173 ± 16 ms for myocytes from portal vein, thoracic aorta, and carotid artery, respectively (n = 6–8). Figure 1B shows that although current density for IClCa was slightly different in the three murine cell types (thoracic aorta > portal vein > carotid artery), all reversed near the predicted equilibrium potential for Cl− under these conditions (∼0 mV) and exhibited strong outwardly rectifying properties attributable to voltage-dependent gating. In addition to the biophysical properties, IClCa in each cell type showed the characteristic bimodal response to 100 μM niflumic acid (Fig. 1C), as described previously in murine portal vein (31, 32) and rabbit pulmonary artery smooth muscle cells (26, 34). These data showed that myocytes from different murine blood vessels exhibit IClCa with biophysical and pharmacological properties that are similar and consistent with findings in rabbit vascular myocytes, suggesting that a common molecular structure may underlie the current in each cell type.
Fig. 1.
Characteristics of Ca2+-dependent Cl− channels (IClCa) in murine vascular smooth muscle cells. A: representative families of IClCa recorded at test potentials from −100 to +120 mV from a holding potential (HP) of −50 mV in myocytes isolated from mouse thoracic aorta (mTA), portal vein (mPV), and carotid artery (mCA). IClCa was evoked by a pipette solution containing 500 nM free Ca2+. B: mean current-voltage (I-V) relationships for amplitude of IClCa recorded at the end of each test pulse normalized to cell capacitance in each of the 3 different vascular myocytes. Each point is mean ± SE of 6–15 cells from ≥3 different animals. C: typical superimposed IClCa recordings from a murine thoracic aorta (left) and a murine portal vein (right) myocyte obtained in the absence or presence of 100 μM niflumic acid (NFA). Voltage-clamp protocol consisted of 1-s test steps to +70 mV followed by a 500-ms return step to −80 mV (HP = −50 mV). Traces represent results from >4 cells from ≥3 animals.
Expression of TMEM16A and TMEM16B in vascular smooth muscle.
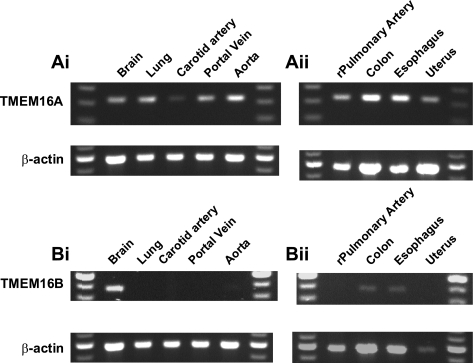
mRNA for TMEM16A has been identified in a number of different tissues, including brain, heart, lung, and kidney (5, 8, 30, 35), but not in vascular smooth muscle. Consequently, the suitability of TMEM16A as a molecular correlate for native CACCs in smooth muscle is unknown. Figure 2 shows that TMEM16A and TMEM16B transcripts were identifiable after 35 PCR cycles in tissues shown previously to express these genes [i.e., TMEM16A in brain and lung (30) and TMEM16B in brain (33)]. In addition, TMEM16A message was also detectable in murine portal vein, thoracic aorta, and carotid artery (Fig. 2Ai) as well as rat pulmonary artery. TMEM16A message was also detected in nonvascular murine smooth muscle samples (Fig. 2Aii). In contrast, expression of the TMEM16B gene product was undetectable in murine portal vein, thoracic aorta, or carotid artery (Fig. 2Bi) but was detected in murine esophagus and colon (Fig. 2Bii).
Fig. 2.
RT-PCR expression analysis of transmembrane proteins 16A and 16B (TMEM16A and TMEM16B) in smooth muscle tissues. mRNA was extracted from whole tissues and subjected to reverse transcription, and semiquantitative PCR was performed. After a standard 35 cycles, sequence-verified TMEM16A product (Ai and Aii) was amplified in a range of tissues, including those previously shown to express this gene. In contrast, amplification of TMEM16B (Bi and Bii) product showed a more restricted expression profile. The housekeeping gene β-actin acted as internal positive control for all PCRs. Images represent results from >3 animals.
Identification of TMEM16A splice variants.
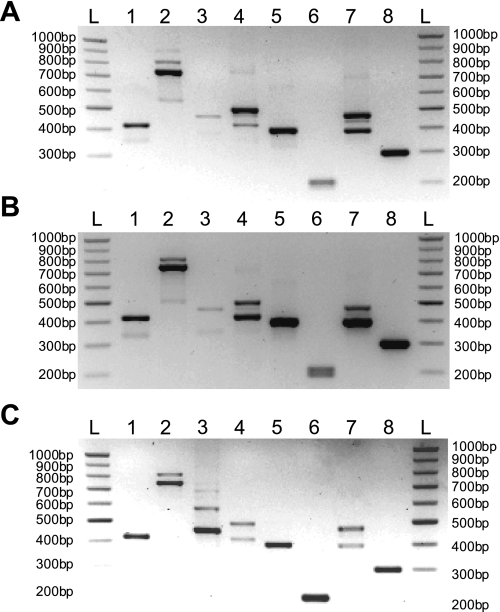
At least four alternatively spliced exons (labeled a, b, c, and d) were predicted by sequence analysis for human TMEM16A (6, 8). Using RT-PCR, we examined the expression pattern of these hypothetical transcripts in mouse by designing sets of primers that would span or anneal at one end to the target exon sequence (Table 1). Lane 1 in Fig. 3 shows a PCR product from a reaction using a primer pair designed with one primer annealing to part of the sequence corresponding to the exon encoding for exon splice variant “a” (mouse exon 2). This yielded a single band (413 bp) in all tissues probed, including carotid artery, thoracic aorta, and portal vein samples. These results indicate that these tissues express this variant, but our analysis cannot exclude the possibility that transcripts lacking this sequence are also expressed. Double bands were obtained in thoracic aorta, carotid artery, and portal vein tissues when a primer pair spanning the presumed exon encoding for variant b (lane 2) was used. Sequencing confirmed that the expected 741-bp amplicon corresponded to TMEM16A lacking the b variant in all three tissues. In the portal vein, we were also able to show that the upper band contained a 66-bp fragment that was not present in the lower band. When this sequence was translated, it was shown to be a good match (95%) to the protein sequences described as corresponding to exon b by Hartzell et al. (14). However, we were unable to confirm, by direct sequencing of gel extracts or by subcloning PCR products, that the upper band corresponded to a transcript that includes this spliced sequence, although the molecular weight on the gel was consistent with the predicted 66-bp transcript reported by Hwang et al. (17) in mouse colon. Thus the double bands indicate that these vascular smooth muscles contain TMEM16A transcripts that may include or exclude exon b. A second pair of primers, designed such that one of the pair annealed to part of exon b, was expected to produce only one band if the upper band of the doublet was the +b transcript (including the extra 66 bp). Single bands were indeed observed at the predicted 455 bp (lane 3), indicating the presence of exon b in the thoracic aorta and carotid artery. Reamplification of the PCR product derived from the portal vein cDNA was necessary before an appropriately sized band could be observed, suggesting that while exon b is present in this tissue, it is likely to be in lower quantities than in the thoracic aorta or portal vein.
Fig. 3.
TMEM16A splice variant expression in murine vascular tissues. A–C: end-point PCR gels for thoracic aorta (A), carotid artery (B), and portal vein (C). Primers are listed in Table 1. Lane markers are as follows: ladder (L), exon a annealing (lane 1), exon b spanning (lane 2), exon b annealing (lane 3), exon c annealing (lane 4), exon c spanning (lanes 5 and 6), exon d spanning (lane 7), and exon d annealing (lane 8). Double bands in lanes 2 and 7 represent presence and absence of that exon product.
Lane 4 provides unequivocal evidence that the 12-bp sequence consistent with the c variant is present in all vascular tissues, since the forward primer of the pair used to form this amplicon partially anneals to exon c. If exon c was routinely absent from these tissues, no bands could have been observed, since the forward primer would have failed to anneal. The double band is explained by the fact that this primer set also spans exon d. Two further sets of primers were also used; both span exon 14, which encodes for exon c. One pair had an expected product size of ∼395 bp, while the other had a smaller expected product size of 200 bp. In the portal vein and thoracic aorta, the existence of a single transcript was revealed using both primer pairs (lanes 5 and 6), suggesting that only a +c transcript exists in these tissues. However, while the primers that amplified the larger region also showed a single band in the carotid artery (lane 5; predicted product size of 395 bp), when the smaller region was amplified (lane 6; predicted product size of 200 bp), greater resolution was achieved, and, unlike the portal vein and aorta, there was a double band. This shows likely alternative splicing of exon c in the carotid artery. As previously demonstrated by the double bands in lane 5, there was alternative splicing of the d variant isoform, which was confirmed by primer pairs designed to span exon d (lane 7; predicted product size of 393 bp). The doublet displayed was indicative of two transcripts, one +d and one −d form. Primers designed such that one annealed to the exon encoding for this variant showed a single band in all tissues at the expected size (298 bp; lane 8), confirming that the larger of the two transcripts observed in lane 7 was, in fact, the +d isoform. This experiment demonstrates that transcripts that include or exclude exon d may occur in the portal vein, thoracic aorta, and carotid artery.
Quantification of TMEM16A transcripts.
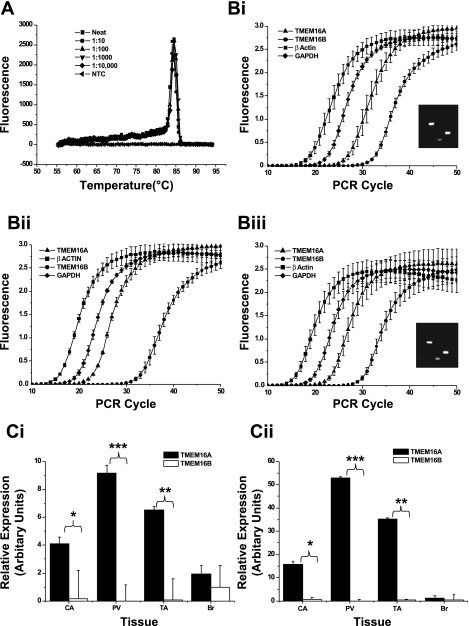
qPCR using SYBR Green technology was undertaken on mRNA isolated from murine brain, portal vein, thoracic aorta, and carotid artery to assess the relative abundance of TMEM16A and TMEM16B in these tissues. To confirm the specificity of all primers used in the reactions, melt-curve and agarose gel electrophoresis analyses were performed (see example of melt-curve analysis of primers specific for TMEM16A in Fig. 4A). In addition, efficiency of the primers in each experiment was established by generation of a standard curve from serial dilutions of cDNA from brain (positive control tissue for all genes examined; Fig. 4Bi–iii). The relative abundance of mRNA encoding the TMEM16A and TMEM16B genes was quantified on the basis of the expression ratio of the target gene vs. two reference genes, β-actin and GADPH. Figure 4, Ci and Cii, shows that TMEM16A was expressed in all three blood vessels, with considerably less expression of TMEM16B than TMEM16A in portal vein, thoracic aorta, and carotid artery. There was no significant difference in expression between TMEM16A and TMEM16B genes in brain (Fig. 4, Ci and Cii).
Fig. 4.
Quantitative PCR (qPCR) expression analysis of TMEM16A and TMEM16B. A: representative dissociation profile for TMEM16A primers in serially diluted concentrations of murine brain cDNA. B: amplification plots of each PCR primer set tested in murine carotid artery (i), portal vein (ii), and thoracic aorta (iii), including analysis of end-point PCR products on agarose gels for representative qPCR experiment (see insets). In insets, lane 1 is 25-bp ladder, lane 2 is β-actin RT+, lane 3 is GAPDH RT+, lane 4 is TMEM16A RT+, and lane 5 is TMEM16A RT−. C: relative expression of primer sets, normalized to β-actin (Ci) and GAPDH (Cii). Significance was determined by Student's 1-tailed paired t-test: *P = 0.01–0.05; **P = 0.01–0.001; ***P < 0.001. Values are means ± SE of tissues from 3 different animals, each with duplicate PCRs.
Post hoc analysis revealed no significant difference in TMEM16A expression levels between blood vessels, irrespective of the housekeeping gene against which the data were normalized (Student's t-test). However, expression of TMEM16A was significantly greater in murine portal vein than murine brain (P = 0.014 for data normalized to β-actin, P = 0.002 for data normalized to GAPDH). Taken together, analysis of TMEM16A gene expression yields data with the same trends, regardless of the housekeeping gene to which the Ct values have been normalized for this primer set. For TMEM16B, there were no differences in abundance between blood vessels when normalized to β-actin, but expression of TMEM16B was significantly lower in portal vein than carotid artery and thoracic aorta (P = 0.005 and 0.003, respectively) when normalized to GAPDH. Regardless of the housekeeping gene, TMEM16B was lower in blood vessels than brain (P < 0.05). These data provide a quantified analysis of TMEM16A and TMEM16B expression in vascular smooth muscle, which reveals considerably greater expression of TMEM16A than TMEM16B in murine blood vessels.
Protein translation of TMEM16A message.
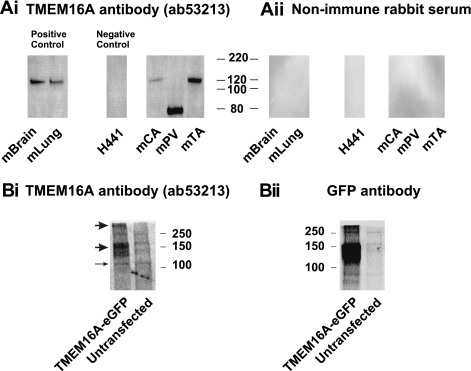
Immunodetection of TMEM16A expression products has mainly focused on overexpressed cells or sections of epithelial structures (1, 17, 19, 35); in no studies, have native tissues been subjected to Western blot analysis. These studies were controlled in a number of ways: the nonimmunogenic serum from the host animal was run; a tissue shown not to express TMEM16A (H441 cells) was used (19); tissues shown to express TMEM16A (brain and lung) were used; and HEK 293 cells overexpressing GFP-tagged TMEM16A were used. Figure 5A shows that bands (∼120 kDa) close to the theoretical molecular weight of TMEM16A (∼114 kDa for the mouse TMEM16A protein, including all 4 alternatively spliced variants) were identified in protein lysates from murine brain and lung. In contrast, no bands were detectable in H441 cells. Immunodetection of the various blood vessels showed an interesting pattern of bands. Murine carotid artery and thoracic aorta exhibited a band at approximately the same molecular weight (∼120 kDa) as that seen in positive control samples. However, portal vein protein exhibited a dominant band of ∼80 kDa (absent in all other samples) and no obvious band at ∼120 kDa. These bands were not due to nonspecific binding, as no bands were present when nonimmune serum was used (Fig. 5B). Overexpression of a fusion protein construct comprising mouse TMEM16A and GFP linked to its COOH-terminal end yielded a pronounced band at ∼147 kDa when probed with a specific anti-GFP antibody [predicted molecular weight is ∼138 kDa; this TMEM16A clone lacked the amino acid residues of the spliced exons corresponding to the b, c, and d variants and, thus, contained only the NH2-terminal sequence containing the a variant (6)], which was not apparent in untransfected cells (Fig. 5C, right). The same antibody (ab53213) used to probe vascular smooth muscle proteins (Fig. 5A, right) also detected a broad band within a range of molecular weights similar to that produced with the anti-GFP antibody, as well as a band that equates to the endogenous, GFP-free TMEM16A (∼120 kDa). The same antibody detected an additional band that exhibited a molecular weight approximately double that of the main band, perhaps suggesting that TMEM16A, at least under these artificial conditions, is minimally assembled as a dimeric protein. These data suggest that the posttranslational processing of TMEM16A may be complex and tissue specific.
Fig. 5.
Western blot analysis of murine protein lysates. TMEM16A protein expression in various murine tissues was determined by SDS-PAGE. H441 cells acted as a negative control for TMEM16A protein expression. Aii: no band was detected by Western blot analysis of the same protein lysates samples in Ai when rabbit nonimmune serum was used in place of primary antibody. B: immunodetection of TMEM16A and green fluorescent protein (GFP) in HEK 293 cells transfected with enhanced GFP (eGFP)-tagged TMEM16A. Blot performed using the anti-TMEM16A antibody (Ab 53213) shows molecular weight of the principal product, most likely corresponding to monomeric protein, is consistent with that expected for a TMEM16A-GFP construct (∼138 kDa). There also appears to be a band at approximately the predicted size for endogenous human TMEM16A (∼114 kDa; full-length would be predicted to lie near 117 kDa) in protein obtained from control and TMEM16A-GFP-transfected cells (thin arrow). A similar display of immunodetection is seen in protein samples processed from the same cells, but stained with an anti-GFP antibody (600-101-215, Rockland; 1:1,000 dilution). GFP staining confirms that the band observed at 147 kDa after staining with the anti-TMEM16A antibody is specific to this protein. Images are representative of results from >3 experiments.
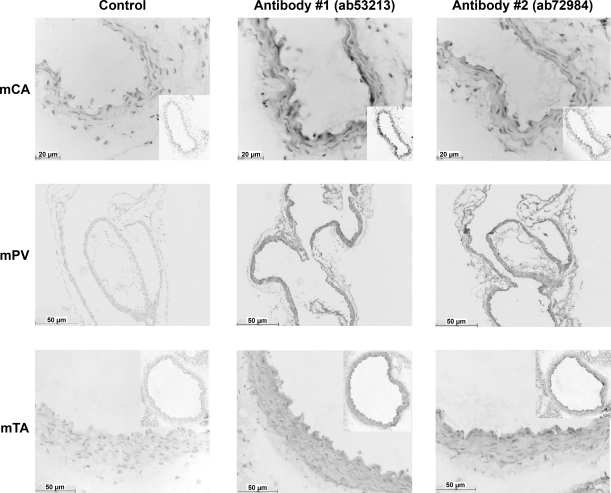
Experiments were undertaken to ascertain the localization of TMEM16A protein in the walls of murine blood vessels. Figure 6 shows representative transverse sections from murine thoracic aorta and murine carotid artery, as well as a longitudinal section of murine portal vein. Figure 6 shows control sections, i.e., sections not treated with primary antibody; sections treated with the primary antibody ab53213; and sections treated with a different primary antibody for TMEM16A, ab72984. In addition, all sections were counterstained for nuclei. In all tissues tested, the control sections (i.e., those not treated with TMEM16A antibody) showed minimal immunoreactivity compared with the obvious diaminobenzidine staining in the smooth muscle layer (Fig. 6, middle and right). As both primary antibodies show similar levels of immunoreactivity, we are confident of specificity of signal over background. These data show that TMEM16A-encoded protein is present in the smooth muscle layers of the murine blood vessels under investigation.
Fig. 6.
TMEM16A is expressed in the smooth muscle layer of murine vessels. Immunohistochemistry was performed on 4-μm-thick transverse (carotid artery and thoracic aorta) and longitudinal (portal vein) sections. Representative time-matched sections incubated in control serum (without primary antibody) reveal only very faint false-positive staining. Sections treated with the primary antibodies ab53213 and ab72984 show obvious diaminobenzidine staining compared with control. All sections were counterstained for nuclei. Images are representative of results from 3 experiments.
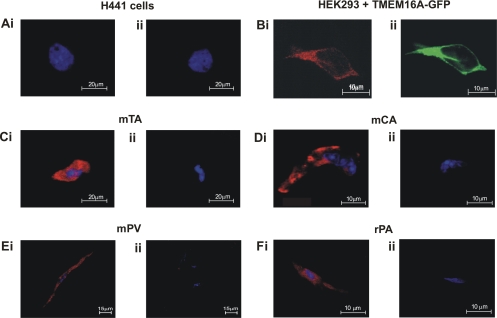
Figure 7 shows the results of immunocytochemical experiments on single smooth muscle cells isolated from murine carotid artery, thoracic aorta, and portal vein, as well as positive and negative control cell types (HEK 293 cells overexpressing TMEM16A and H441 cells, respectively). As expected from the Western blot data and mRNA analysis (19), no immunofluorescence was observed in any H441 cell with the TMEM16A antibody. In contrast, HEK 293 cells overexpressing GFP-tagged TMEM16A exhibited considerable immunofluorescence that overlapped completely with the GFP-based staining, again confirming the specificity of this antibody. TMEM16A-specific fluorescence was also detected in the cytoplasm and membrane regions of all smooth muscle cells tested compared with little or no fluorescence in myocytes incubated only with the secondary antibody (Fig. 7, C–Eii) or H441 cells (Fig. 7A). The pattern of staining was consistent, regardless of the vascular myocyte under study. Overall, these studies show that smooth muscle cells express TMEM16A and that the encoded protein is abundant in smooth muscle cells.
Fig. 7.
TMEM16A protein localization in enzymatically isolated smooth muscle cells. A: lack of staining in H441 cells incubated with TMEM16A antibody (ab53213, Ai) compared with cells incubated with secondary antibody only (Aii). Experiments were run in parallel with thoracic aorta myocytes (represented in C). Bi: colocalization of TMEM16A staining (ab53213, 1:50 dilution) with GFP tagging visible. Pattern of TMEM16A immunostaining and GFP location show a clear correlation, indicating that the TMEM16A antibody specifically stains TMEM16A in these cells. Fluorescence immunocytochemistry revealed specific expression of TMEM16A protein in murine carotid artery, portal vein, and thoracic aorta cells (Ci, Di, Ei,) and rat pulmonary artery cells (Fi). Control (no primary antibody) cells showed minimal fluorescence, confirming specificity of the signals (Cii, Dii, Eii, and Fii).
Pharmacological properties of Cl− currents generated by TMEM16A overexpression.
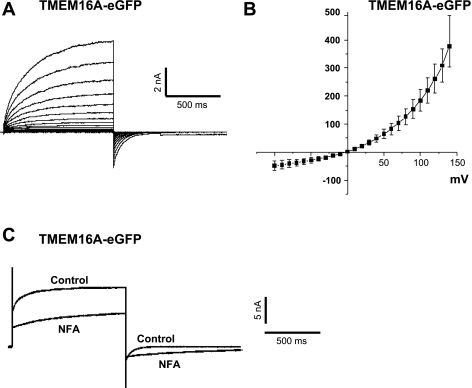
Cl− currents similar to previous studies of TMEM16A were recorded from HEK 293 cells transfected with GFP-tagged TMEM16A. All cells showing GFP fluorescence exhibited IClCa when dialyzed with a pipette solution containing 500 nM. These currents displayed considerable similarity to those recorded from vascular myocytes (Fig. 1), including time-dependent relaxation at positive potentials and slow deactivation kinetics on return to the HP (Fig. 8A), strong outward rectification and a reversal potential near the predicted equilibrium potential for Cl− (∼0 mV; Fig. 8B), and a similar pattern of blockade by niflumic acid characterized by a marked slowing of deactivation kinetics at negative potentials (Fig. 8C). Untransfected wild-type HEK 293 cells (n = 3) or HEK 293 cells transfected with a plasmid vector containing only GFP (n = 3) displayed small time- and voltage-dependent currents that were indistinguishable from leak when recorded under similar recording conditions (data not shown).
Fig. 8.
IClCa in HEK 293 cells transiently transfected with TMEM16A tagged with enhanced GFP. A: representative family of currents from a TMEM16A-transfected HEK 293 cell dialyzed with 500 nM Ca2+. Currents were evoked by stepping in 10-mV increments from HP of −50 mV to 1-s test potentials ranging from −100 to +140 mV following 10 min of dialysis. B: mean current-voltage relationship generated from families of currents similar to those described in A (n = 9). C: superimposed IClCa traces recorded before and after application of 100 μM niflumic acid (NFA). Current recorded in drug-free solution (control) was generated by stepping from HP of −50 mV to +140 mV for 1 s, then repolarizing to −80 mV for 1 s, following 10 min of dialysis with 5 mM ATP and 500 nM Ca2+. NFA (100 μM) was then applied for 10 min, and current trace labeled “NFA” was recorded in a manner similar to control current. Traces represent results from 6 experiments.
DISCUSSION
Since the initial identification of the TMEM16A expression product as a Cl− channel (6, 30, 35), an explosion of data has shown that this gene and paralogs contribute to native CACCs in epithelial (16, 19, 24, 28) and gastrointestinal pacemaker (10, 17, 36) cells. However, with the exception of one recent study in rat pulmonary artery myocytes (23), there has been no evidence that these genes are the molecular correlate of CACCs in vascular smooth muscle cells, where robust IClCa have been recorded (2–4, 11, 12, 31, 32). In the present study, we present a quantitative analysis of TMEM16A expression and protein translation in a number of vascular tissues. Our results have revealed that vascular smooth muscle cells, which exhibit IClCa with distinctive biophysical and pharmacological features, express a considerably greater abundance of TMEM16A mRNA than the closely related TMEM16B isoform when quantified relative to two different housekeeping genes. Moreover, the relative level of TMEM16A was higher in thoracic aorta, portal vein, and carotid artery than brain. This study also showed that the different NH2-terminal splice variants of TMEM16A (6, 8, 14, 16) are variably expressed in vascular smooth muscle. While the a variant was detected in all vessels, it is unknown whether transcripts lacking this variant are also expressed in these tissues, since primers could not be designed to span the target exon. The b and d variants were present and absent in all blood vessels studied, similar to other nonvascular tissues (8). Although the c variant was detected in all three blood vessels, only the carotid artery also expressed a transcript lacking the short 12-bp sequence. This is an interesting observation, since the b and c variants are known to affect Ca2+ sensitivity and voltage-dependent kinetics, respectively (8). Furthermore, inclusion or exclusion of the b variant has been suggested to determine the presence of the d variant (8). Although the precise effects of coexpression of different TMEM16A splice variants are not known, our data suggest that considerable variation in the functional properties and fine tuning CACCs in different vascular beds may be possible.
In addition to the RNA study, we identified TMEM16A expression products in smooth muscles. Similar to immunohistological studies on human and murine gastrointestinal tract (10, 16, 36), mouse and human airways (19, 24, 28), and male and female reproductive tracts (16), TMEM16A staining was detected using two different antibodies in 4-μm-thick sections of murine blood vessels. In every case, the staining was dominant in the smooth muscle layer. In addition, TMEM16A immunoreactivity was detected in isolated smooth muscle cells from mouse aorta, carotid artery, and portal vein. Similar to the staining pattern for other ion channels [e.g., Cav3.1 (20) and Kv1.5 (5)], the staining was not localized to the cell membrane but generally appeared throughout the cytosol, with some patchy staining occasionally observed at the membrane. A similar pattern of TMEM16A staining has been reported in murine and human tracheal epithelia (19) and gastrointestinal interstitial cells (10), whereas TMEM16A localizes predominantly to the cell membrane in heterologous expression systems (19, 24). These data reflect the normal posttranslational processing of native TMEM16A by the endoplasmic reticulum, Golgi apparatus, and trafficking machinery. Moreover, each of the smooth muscles expresses a differential pattern of NH2-terminal splice variants (a, b, c, and d isoforms) originally identified by Caputo et al. (6) and Ferrera et al. (8), and no distinction of splice variants was possible by the antibodies used in the present study. In addition to the rather diffuse intracellular staining, distinct areas of accumulated anti-TMEM16A fluorescence were visible in all cells tested. It has been proposed that TMEM16A-dependent fluorescence exhibits clustering when overexpressed in HEK 293 cells; this led to the speculation that TMEM16A may exist in lipid microdomains (19). It is intriguing that the amplitude and pharmacology of native IClCa in vascular smooth muscles cells are affected markedly by the cholesterol-depleting agent methyl β-cyclodextrin, which also causes translocation of raft markers (31). Consequently, if TMEM16A-encoded proteins contribute to the native CACC in smooth muscle cells, it may be possible that some of the native CACCs exist in localized microdomains. The Western blot experiments suggest that posttranslational processing of TMEM16A may differ between cell types. Thus immunoreactive bands were present in most lysates near the theoretical molecular weight (∼120 kDa for the full-length mouse protein comprising all 4 splice variants), but in the portal vein, an antibody-specific band was revealed with the same antibody at ∼80 kDa. It is unlikely that the discrepancy is due to selective exclusion of the b, c, and d splice variants, as the difference in molecular weights is too large to be accounted for by this known alternative splicing (8). This antibody also positively identified mouse TMEM16A-GFP expressed in HEK 293 cells that matched a similar band detected by a GFP-specific antibody, supporting the hypothesis that the bands revealed by Western blot analysis of protein lysates from blood vessels indeed corresponded to TMEM16A protein. This conclusion is further supported by the observations that this antibody successfully revealed endogenous human TMEM16A protein (∼110–114 kDa) in HEK 293 cells, which express TMEM16A mRNA, but failed to do so in H441 cells, which do not express this gene (19). These carefully controlled studies suggest that TMEM16A gene products exhibit tissue-specific differences in the molecular weight of the native protein but do not highlight whether these differences are due to alternative splicing and/or posttranslational modifications (e.g., N-linked glycosylation, proteolytic cleavage, and phosphorylation), which require additional investigation.
TMEM16A protein is a constituent of native CACCs: caveats.
Unlike currents generated by overexpression of previous candidate genes for the CACC, i.e., CLCA and bestrophin, the currents generated by overexpression of TMEM16A/B resembled native IClCa recorded in acinar cells, neurons, and smooth muscle cells [compare Fig. 8 and Yang et al. (35) with Fig. 1 or Frings et al. (9) and Angermann et al. (2)]. The identification in the present study of robust TMEM16A, but not TMEM16B, expression in blood vessels that exhibit distinctive IClCa adds weight to the possibility that products of this gene contribute to native CACCs in these tissues. This view was further supported by a recent report, undertaken contemporaneously with our study, which showed that siRNA targeted against TMEM16A reduced the amplitude of IClCa recorded from smooth muscle cells from the rat pulmonary artery compared with scrambled siRNA (23). This recent report (23) also showed that transcripts of TMEM16A were far higher than transcripts of TMEM16B, similar to our findings on mouse blood vessels. In addition, splice variants of TMEM16A showed the same expression pattern as in the mouse blood vessels shown in the present study. Consequently, the similarity of TMEM16A-mediated IClCa and the siRNA study, the siRNA in rat pulmonary artery smooth muscle cells (23) provides considerable evidence that TMEM16A expression products contribute to the native CACC in blood vessels. However, some caution must be exercised in ascribing TMEM16A expression products as the only components of smooth muscle CACCs. 1) The single-channel conductance of native CACCs (1–3 pS) (15, 27) is closer to TMEM16B-encoded (1.2 pS) (25) than TMEM16A-encoded (8.3 pS) channels (35), although the unitary conductance of TMEM16A splice variants (8) is unknown. 2) There is no readily identifiable motif consistent with a Ca2+-binding domain within either terminal end of TMEM16A, so the mechanism by which a rise in Ca2+ gates channel opening is undefined. 3) IClCa in vascular smooth muscle cells are suppressed by CaMKII-dependent phosphorylation and augmented by Ca2+-dependent and Ca2+-independent phosphatase activity (2, 3, 11, 12). There are a number of kinase consensus sequences in the NH2 terminus of TMEM16A (14), but there is no information about how these channels are regulated by CaMKII or the effect of phosphatases. 4) Native IClCa in smooth muscle cells exhibit a bimodal response to Cl− channel blockers such as niflumic acid (25, 31, 33) and have considerable pharmacological overlap with KCa1.1 (13), being activated by NS1619 and isopimaric acid and blocked by paxilline, tamoxifen, penitrem A, and iberiotoxin (28, 31). The effect of the K+ channel modulators is lost following cholesterol depletion by methyl β-cyclodextrin (31). Pharmacological studies on TMEM16A-encoded currents show moderate sensitivity to standard Cl− channel blockers, but neither stimulation by niflumic acid or A-9-C nor any effect of KCa1.1 channel modulators has been reported. Moreover, there is no information as to how the three different TMEM16A splice variants (b, c, and d) (8) contribute to the native CACC and influence the pharmacology of these channels. Consequently, there is considerable evidence that TMEM16A-encoded proteins contribute to native CACCs in smooth muscle cells. However, it is likely that these proteins exist as part of a multimeric signaling complex that most likely exhibits a variable composition and architecture in different cell types appropriately tuned for their respective function.
GRANTS
A. J. Davis and W. R. Sones were funded by British Heart Foundation Grants PG/07/127/24235 and PG/05/038 to I. A. Greenwood. A. S. Forrest was supported by National Heart, Lung, and Blood Institute Grant 5 R01 HL-075477 to N. Leblanc. The publication was also made possible by National Institutes of Health National Center for Research Resources Grants 5 P20 RR-15581 (to N. Leblanc) and 5 P20 RR-15581 and 5 PPG HL-76611 (to M. L. Valencik) supporting a Center of Biomedical Research Excellence at the University of Nevada School of Medicine.
DISCLAIMER
The contents of the manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
ACKNOWLEDGMENTS
We thank Drs. Laura Pellatt, Suman Rice, and Ken Laing for qPCR advice; Kinga Szewczyk and Ray F. Moss (Image Resource Facility, Division of Biomedical Sciences, St. George's, University of London) for excellent technical support with the immunological experiments; and Vani Kilari for preparation of the TMEM16A-GFP plasmid and assistance in optimizing our transfection studies. We acknowledge the services of the Medical Biomics Centre, St. George's, University of London, and the Nevada Genomics Centre, University of Nevada.
REFERENCES
- 1.Almaca J, Tian Y, Aldehni F, Ousingsawat J, Kongsuphol P, Rock JR, Harfe BD, Schreiber R, Kunzelmann K. TMEM16 proteins produce volume regulated chloride currents that are reduced in mice lacking TMEM16A. J Biol Chem 284: 28571–28578, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angermann JE, Sanguinetti AR, Kenyon JL, Leblanc N, Greenwood IA. Mechanism of the inhibition of Ca2+-activated Cl− currents by phosphorylation in pulmonary arterial smooth muscle cells. J Gen Physiol 128: 73–87, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayon R, Sones W, Forrest AS, Wiwchar M, Valencik ML, Sanguinetti AR, Perrino BA, Greenwood IA, Leblanc N. Complex phosphatase regulation of Ca2+-activated Cl− currents in pulmonary arterial smooth muscle cells. J Biol Chem 284: 32507–32521, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britton FC, Ohya S, Horowitz B, Greenwood IA. Comparison of the properties of CLCA1 generated currents and ICl(Ca) in murine portal vein smooth muscle cells. J Physiol 539: 107–117, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burg ED, Platoshyn O, Tsigelny IF, Lozano-Ruizn B, Rana BK, Yuan JXJ. Tetramerization domain mutations in KCNA5 affect channel kinetics and cause abnormal trafficking patterns. Am J Physiol Cell Physiol 298: C496–C509, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caputo A, Caci E, Ferrera L, Pedemonte N, Barsanti C, Sondo E, Pfeffer U, Ravazzolo R, Zegarra-Moran O, Galietta LJV. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science 322: 590–594, 2008 [DOI] [PubMed] [Google Scholar]
- 7.Chipperfield AR, Harper AA. Chloride in smooth muscle. Prog Biophys Mol Biol 74: 175–221, 2000 [DOI] [PubMed] [Google Scholar]
- 8.Ferrera L, Caputo A, Ubby I, Bussani E, Zegarra-Moran O, Ravazzolo R, Pagani F, Galietta LJV. Regulation of TMEM16A chloride channel properties by alternative splicing. J Biol Chem 284: 33360–33368, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frings S, Reuter D, Kleene SJ. Neuronal Ca2+-activated Cl− channels—homing in on an elusive channel species. Prog Neurobiol 60: 247–289, 2000 [DOI] [PubMed] [Google Scholar]
- 10.Gomez-Pinilla PJ, Gibbons SJ, Bardsley MR, Lorincz A, Pozo MJ, Pasricha PJ, de Rijn MV, West RB, Sarr MG, Kendrick ML, Cima RR, Dozois EJ, Larson DW, Ordog T, Farrugia G. Ano1 is a selective marker of interstitial cells of Cajal in the human and mouse gastrointestinal tract. Am J Physiol Gastrointest Liver Physiol 296: G1370–G1381, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenwood IA, Ledoux J, Leblanc N. Differential regulation of Ca2+-activated Cl− currents in rabbit arterial and portal vein smooth muscle cells by Ca2+/calmodulin-dependent kinase. J Physiol 534: 395–408, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenwood IA, Ledoux J, Sanguinetti A, Perrino BA, Leblanc N. Calcineurin Aα but not Aβ augments IClCa in rabbit pulmonary artery smooth muscle cells. J Biol Chem 279: 38830–38837, 2004 [DOI] [PubMed] [Google Scholar]
- 13.Greenwood IA, Leblanc N. Overlapping pharmacology of Ca2+-activated Cl− and K+ channels. Trends Pharmacol Sci 28: 1–5, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Hartzell HC, Yu K, Xiao Q, Chien LT, Qu Z. Anoctamin/TMEM16 family members are Ca2+-activated chloride channels. J Physiol 587: 2127–2139, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirakawa Y, Gericke M, Cohen RA, Bolotina VM. Ca2+-dependent Cl− channels in mouse and rabbit aortic smooth muscle cells: regulation by intracellular Ca2+ and NO. Am J Physiol Heart Circ Physiol 277: H1732–H1744, 1999 [DOI] [PubMed] [Google Scholar]
- 16.Huang F, Rock JR, Harfe BD, Cheng T, Huang X, Jan YN, Jan LY. Studies on expression and function of the TMEM16A calcium-activated chloride channel. Proc Natl Acad Sci USA 106: 21413–21418, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang SJ, Blair PJA, Britton FC, O'Driscoll KE, Hennig G, Bayguinov YR, Rock JR, Harfe BD, Sanders KM, Ward SM. Expression of anoctamin 1/TMEM16A by interstitial cells of Cajal is fundamental for slow wave activity in gastrointestinal muscles. J Physiol 587: 4887–4904, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kashyap MK, Marimutha A, Kishore CJH, Peri S, Keerthikumar S, Prasad TSK, Mahmood R, Rao S, Ranganathan P, Sanjeeviah RC, Vijayakumar M, Kumar KVV, Montgomery E, Kumar RV, Pandey A. Genomewide mRNA profiling of esophageal squamous cell carcinoma for identification of cancer biomarkers. Cancer Biol Ther 8: 36–46, 2009 [DOI] [PubMed] [Google Scholar]
- 19.Kunzelmann K, Kongsuphol P, Aldehni F, Tian Y, Ousingsawat J, Warth R, Schreiber R. Bestrophin and TMEM16—Ca2+ activated Cl− channels with different functions. Cell Calcium 46: 233–241, 2009 [DOI] [PubMed] [Google Scholar]
- 20.Kuo IY, Ellis A, Seymour VAL, Sandpow SL, Hill CE. Dihydropyridine-insensitive calcium currents contribute to function of small cerebral arteries. J Cereb Blood Flow Metab 30: 1226–1239, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Large WA, Wang Q. Characteristics and physiological role of the Ca2+-activated Cl− conductance in smooth muscle. Am J Physiol Cell Physiol 271: C435–C454, 1996 [DOI] [PubMed] [Google Scholar]
- 22.Leblanc N, Ledoux J, Saleh S, Sanguinetti A, Angermann J, O'Driscoll K, Britton F, Perrino BA, Greenwood IA. Regulation of calcium-activated chloride channels in smooth muscle cells: a complex picture is emerging. Can J Physiol Pharmacol 83: 541–556, 2005 [DOI] [PubMed] [Google Scholar]
- 23.Manoury B, Tamuleviciute A, Tammaro P. TMEM16A/anoctamin 1 protein mediates calcium-activated chloride currents in pulmonary arterial smooth muscle cells. J Physiol 588: 2305–2314, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ousingsawat J, Martins JR, Schreiber R, Rock JR, Harfe BD, Kunzelmann K. Loss of TMEM16A causes a defect in epithelial Ca2+-dependent chloride transport. J Biol Chem 284: 28698–28703, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pifferi S, Dibattista M, Menini A. TMEM16B induces chloride currents activated by calcium in mammalian cells. Pflügers Arch 458: 1023–1038, 2009 [DOI] [PubMed] [Google Scholar]
- 26.Piper AS, Greenwood IA, Large WA. Dual effect of blocking agents on Ca2+-activated Cl− currents in rabbit pulmonary artery smooth muscle cells. J Physiol 539: 119–131, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piper AS, Large WA. Multiple conductance states of single Ca2+-activated Cl− channels in rabbit pulmonary artery smooth muscle cells. J Physiol 547: 181–196, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rock JR, O'Neal WK, Gabriel SE, Randell SH, Harfe BD, Boucher RC, Grubb BR. Transmembrane protein 16A (TMEM16A) is a Ca2+ regulated Cl− secretory channel in mouse airways. J Biol Chem 284: 14875–14880, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saleh SN, Angermann JE, Sones WR, Leblanc N, Greenwood IA. Stimulation of Ca2+-gated Cl− currents by the calcium-dependent K+ channel modulators NS1619 [1,3-dihydro-1-[2-hydroxy-5-(trifluoromethyl)phenyl]-5-(trifluoromethyl)-2H-benzimidazol-2-one] and isopimaric acid. J Pharmacol Exp Ther 321: 1075–1084, 2007 [DOI] [PubMed] [Google Scholar]
- 30.Schroeder BC, Cheng T, Jan YN, Jan LY. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell 134: 1019–1029, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sones WR, Davis AJ, Leblanc N, Greenwood IA. Cholesterol depletion alters amplitude and pharmacology of vascular calcium-activated chloride channels. Cardiovasc Res 87: 476–484, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sones WR, Leblanc N, Greenwood IA. Inhibition of vascular calcium-gated chloride currents by blockers of KCa1.1, but not by modulators of KCa2.1 or KCa2.3, channels. Br J Pharmacol 158: 521–531, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stohr H, Heisig JB, Benz PM, Schoberl S, Milenkovic VM, Strauss O, Aartsen WM, Wijnholds J, Weber BHF, Schulz HL. TMEM16B, a novel protein with calcium-dependent chloride channel activity, associates with a presynaptic protein complex in photoreceptor terminals. J Neurosci 29: 6809–6818, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiwchar M, Ayon R, Greenwood IA, Leblanc N. Phosphorylation alters the pharmacology of Ca2+-activated Cl− channels in rabbit pulmonary arterial smooth muscle cells. Br J Pharmacol 158: 1356–1365, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang YD, Cho H, Koo JY, Tak MH, Cho Y, Shim WS, Park SP, Lee J, Lee B, Kim BM, Raouf R, Shin YK, Oh U. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature 455: 1210–1215, 2008 [DOI] [PubMed] [Google Scholar]
- 36.Zhu MH, Kim TW, Ro S, Yan W, Ward SM, Koh SD, Sanders KM. A Ca2+-activated Cl− conductance in interstitial cells of Cajal linked to slow wave currents and pacemaker activity. J Physiol 587: 4905–4918, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]