Abstract
Protein L-isoaspartyl (D-aspartyl) O-methyltransferases (EC 2.1.1.77; PIMT or PCMT) are enzymes that initiate the full or partial repair of damaged L-aspartyl and L-asparaginyl residues, respectively. These enzymes are found in most organisms and maintain a high degree of sequence conservation. Arabidopsis thaliana (Arabidopsis L. Heynh.) is unique among eukaryotes in that it contains two genes, rather than one, that encode PIMT isozymes. We describe a novel Arabidopsis PIMT isozyme, designated AtPIMT2αω, encoded by the PIMT2 gene (At5g50240). We characterized the enzymatic activity of the recombinant AtPIMT2αω in comparison to the other AtPIMT2 isozymes, AtPIMT1, and to the human PCMT ortholog, to better understand its role in Arabidopsis. All Arabidopsis PIMT isozymes are active over a relatively wide pH range. For AtPIMT2αω maximal activity is observed at 50 °C (a lethal temperature for Arabidopsis); this activity is almost ten times greater than the activity at the growth temperature of 25 °C. Interestingly, enzyme activity decreases after pre-incubation at temperatures above 30°C. A similar situation is found for the recombinant AtPIMT2ψ and the AtPIMT2ω isozymes, as well as for the AtPIMT1 and human PCMT1 enzymes. These results suggest that the short-term ability of these methyltransferases to initiate repair under extreme temperature conditions may be a common feature of both the plant and animal species.
Introduction
Protein L-isoaspartate (D-aspartate) O-methyltransferases (PIMT or PCMT; EC 2.1.1.77) recognize the isomerized and racemized products from the spontaneous damage of L-aspartyl and L-asparaginyl residues. The predominant product of such damage, L-isoaspartyl (L-isoAsp) residues, is methyl esterified at the α-carboxyl by these enzymes in an S-adenosyl-L-methionine (AdoMet)-dependent reaction that is followed by non-enzymatic reactions that result in its conversion to a normal L-Asp residue approximately 25% of the time (Brennan et al. 1994; Clarke 2003). Since the accumulation of L-isoAsp residues can deleteriously affect protein function, these methyltransferases are thought to function in vivo to repair protein damage (Reissner and Aswad 2003; Doyle et al. 2003; Lanthier and Desrosiers 2004). This widespread enzyme maintains a high degree of sequence conservation in organisms such as bacteria, nematodes, flies, plants, and mammals, including humans (Kagan et al. 1997).
PIMT is present in some lower plants and in higher plants, especially in seed tissues, and has been associated with ageing processes (Mudgett et al. 1997). For example, PIMT activity increases with age in potato tubers (Kumar et al. 1999) and barley seeds (Mudgett et al. 1997), and is associated with the robustness of polycentenarian Nelumbo nucifera (sacred lotus) seeds (Shen-Miller et al. 1995). It has also been suggested that plant PIMT may play a role in protein repair in photo-damaged leaves (Thapar et al. 2001). Plant PIMT activity has been characterized in winter wheat (Mudgett and Clarke 1993), corn (Thapar et al. 2001), sacred lotus (Shen-Miller et al. 1995), tomato (Kester et al. 1997), and in Arabidopsis thaliana (Arabidopsis thaliana L. Heynh.) (Mudgett and Clarke 1996; Thapar and Clarke 2000; Xu et al. 2004). Considering that seed embryos often experience long periods of quiescence/dormancy prior to completing germination, it is essential to maintain seed proteins in their functional forms (or rapidly repair them upon imbibition) to ensure efficient germination. In mature seeds, metabolism becomes more energetically conservative (Borisjuk et al. 2004), so repair rather than degradation of proteins may become even more important. A notable case is the successful germination of lotus seeds up to 1300 years old that were recovered from a dry lake bed at Pulantien, China. When tested for methyltransferase activity and L-isoAsp residue content, two “old” lotus specimens had comparable levels to their progeny, consistent with a role in seed longevity (Shen-Miller et al. 1995).
Arabidopsis is unique amongst eukaryotes in that it contains not one but two genes (AtPIMT1 At3g48330 and AtPIMT2 At5g50240) that encode active protein repair methyltransferases (Xu et al. 2004). The AtPIMT2 gene has been shown to encode at least two active isozymes (the ψ and ω forms) via alternative splicing of the first intron where a QFQ sequence is either present or not in the amino terminal region (Xu et al. 2004). The human PCMT gene also encodes alternatively spliced forms but at the C-terminus, with either an RK or DEL terminal sequence (MacLaren et al. 1992; Ota et al. 1988). Additional variation in the human enzyme is provided by a polymorphism (Val or Ile) at position 119 (DeVry and Clarke 1999).
In this work, we characterize a third isozyme produced by alternative start site selection of the AtPIMT2 gene. To identify its optimal conditions for protein repair activity we examined the methyltransferase activity of this isozyme under conditions of pH and temperature that can lead to more rapid formation of L-isoAsp residues in proteins and thus a greater potential for damage accumulation. Temperature and pH extremes can lead to polypeptide denaturation and disruption of conformation, which makes proteins even more vulnerable to covalent damage such as deamidation and racemization (Aswad 1995; Slonczewski and Foster 1996). Peptides and proteins aged in vitro at alkaline pH form L-isoAsp linkages at an increased rate (Ota et al. 1987; Bhatt et al. 1990; Patel and Borchardt 1990a,b; Brennan and Clarke 1995). In vivo L-isoAsp accumulation in Escherichia coli is pH-dependent (Hicks et al. 2005). Having a means to repair polypeptide damage induced by pH and high temperature may be necessary to help organisms maintain the integrity of their proteome. In this work, we compare the enzymatic activity of the three known isozymes from AtPIMT2, as well as the AtPIMT1 species and the human PCMT species (MacLaren and Clarke 1995). Most significantly, we find that the activity of all of these enzymes is maximal under temperature conditions that are lethal to the organism, suggesting a role for this enzyme in short term responses to extremes of heat.
MATERIALS AND METHODS
Arabidopsis (Arabidopsis thaliana L. Heynh.) L-isoAsp Methyltransferase Overexpressing Strains
The construction of Nus-tag-AtPIMT2ω and Nus-tag-AtPIMT2ψ in pET43.1, and AtPIMT1-hexahistadyl-tag in pET23d was described previously (Xu et al., 2004). An Escherichia coli expression strain (based on BL21(DE3)RIL) was constructed that contained the plasmid AtPIMTαω-histag-pET23d. For this the PIMT2αω coding region was amplified from the longer Nus-tag-AtPIMT2ω in pET43.1 and cloned into the Nco I and Xho I sites of pET23d (Novagen Inc.) using BspH I and Sal I sites introduced into the amplicon by the primers. The construct was then introduced into the E. coli strain BL21(DE3)RIL (Stratagene, La Jolla, CA, USA).
Recombinant Protein Expression and Purification
Recombinant human isozyme II (rhPCMT) was overexpressed and purified based on a previous report (MacLaren and Clarke 1995) with some modifications. The pellets of previously frozen E. coli cells, which express rhPCMT in the presence of isopropylthio-β-galactoside, were resuspended in chilled purification buffer (5 mM sodium phosphate, 5 mM sodium EDTA, 0.1 mM dithiothreitol, 10% glycerol, 25 µM phenylmethylsulfonylfluoride and 1 tablet of Complete Mini EDTA-free (Roche, Indianapolis, IN) per 10 mL of buffer). The resuspended cells were pulse sonicated at 50% power for 30 sec bursts, six-times, with incubation on ice for 3 minutes between rounds. The supernatant was collected after the sonicated cells were centrifuged for 20 min at 13,000g. The following steps were all performed at 4 °C or on ice. With stirring, 4% protamine sulfate, in purification buffer, was slowly added to the supernatant over a 15 min interval, to give a final protamine sulfate concentration of 0.8%, and the mixture was then stirred for an additional 45 min. Subsequently, the rhPCMT supernatant/ protamine sulfate mixture was added to 1.3 volumes of DE-53 resin that had been saturated with the purification buffer, and this was stirred for 30 min at 4 °C. After removing the DE-53 by vacuum filtration through Whatman type 4 filter paper, ammonium sulfate was added to the filtrate to a 20% final concentration. The ammonium sulfate filtrate solution was loaded onto a phenyl Sepharose column that had been pre-equilibrated in 20 % ammonium sulfate in purification buffer. The column was washed with Buffer A (purification buffer with 20% ammonium sulfate) for 70 min at 2 mL/min. Then the column was washed with a linear buffer gradient that started with 100% Buffer A and ended with 100% of purification buffer for 180 min at 2mL/min, and fractions were collected. The OD280 of the fractions averaged 0.45, or about 0.45 mg protein/mL. To enhance protein precipitation with ammonium sulfate, bovine serum albumin was added to the pooled fractions until the total protein concentration was 1 mg/mL. This was followed by the slow addition of ammonium sulfate, with stirring, to a final concentration of 55% over a 15 min interval, and stirring was continued for another 30 min. The precipitated protein solution was then centrifuged at 13,000 × g for 20 min, the pellets were collected, and then these were combined and resuspended in 1 mL of purification buffer per 100 mg of wet weight. The resuspended pellet mixture was then fractionated on a Superdex 75 column with Buffer A at 0.4 mL/min. Every fraction was assayed for rhPCMT activity, and the most active fractions were pooled and used as rhPCMT stock.
Recombinant AtPIMT2ω and AtPIMT2ψ (rAtPIMT2ω and rAtPIMT2ψ) proteins were prepared as described previously (Xu et al., 2004). rAtPIMT1 and rAtPIMT2αω proteins were prepared from cleared E. coli lysates under native conditions using Ni-affinity chromatography. In brief, rAtPIMT1 and rAtPIMT2αω clones were cultured in 500 mL of Luria-Bertani broth to an OD600 of 0.5. At this point, the cultures were induced with 0.1 mM isopropylthio-β-galactoside for 18 h at 25 °C with shaking. The cell pellets were collected after spinning for 5 min at 5800g and 4 °C, and then the pellets were washed in 5 mL of lysis buffer (50 mM sodium phosphate, 300 mM NaCl, 20 mM imidazole, 10% glycerol, adjusted to a final pH of 8.0) per g of wet pellet, and spun as before. The washed pellets were stored overnight at −20 °C. About 5.7 g and 6.7 g of thawed rAtPIMT1 and rAtPIMT2αω cell pellet, respectively, were resuspended in chilled lysis buffer supplemented with 20 mM β-mercaptoethanol, 1 mM phenylmethylsulfonylfluoride (prepared as a 100 mM solution in dimethylsulfoxide), and 1 µM leupeptin (prepared as a 10 mg/ml solution in ethanol) at 5 ml/g wet mass immediately prior to use. Lysozyme was added to the resuspended cells to a final concentration of 1 mg/ml and the mixture was incubated for 30 min on ice. Phenylmethylsulfonylfluoride was added again to 1 mM and the cells were immediately sonicated on ice using a Model W-350 Sonifier at 40% duty cycle at an output setting of 4 with a microtip in six 10 s bursts with a 10 s cooling period between bursts. Subsequently, sonicated cells were centrifuged at 10,000g for 30 min at 4 °C and the cleared lysates were collected, filtered through two layers of Miracloth (Calbiochem, LaJolla, CA), and then kept on ice.
Approximately 25 mL of each cleared lysate was added to 1 mL of a chilled 50% (v/v) suspension of Ni-NTA agarose beads (Qiagen, Valencia, CA) and mixed at 4 °C for 1 h. The slurry was then added to a 10 cm × 1.5 cm borosilicate column at 4 °C, drained, washed with 8 mL of chilled lysis buffer, and His-tagged proteins were then eluted with 1 mL of chilled elution buffer (50 mM sodium phosphate, 300 mM NaCl, 250 mM imidazole, 10% (v/v) glycerol; pH 8.0). The purified proteins were stored at −80 °C in 10 µL aliquots, each meant for single usage.
Thermostability of Recombinant Methyltransferase Activity
Purified rAtPIMT and rhPCMT proteins were tested on different days in either elution buffer or phosphate buffer (250 mM sodium phosphate) resulting in a final pH of 6.9 or 8.0, respectively. A 2 µL or 6 µL aliquot of each dilution per reaction was transferred to a room temperature 1.6 mL ultra-clear polypropylene microcentrifuge tube (Neptune catalog #3445, San Diego, CA). These were pretreated for 10 min at the indicated temperature immediately before being assayed for methyltransferase activity.
Seed Methyltransferase Activity
Two-hundred mg of mature, dry Arabidopsis (Wassilewskija ecotype) seeds (stored at room temperature) were incubated in the dark for 4 h at either between 38 °C to 40 °C or at room temperature. The seeds were then immediately transferred to dry ice and the crude cytosol was prepared. In brief, seeds were powdered in liquid N2 with a chilled mortar and pestle. The seed powder was mixed with 1.2 mL of freshly-prepared chilled extraction buffer (100 mM HEPES, pH 7.5, 10 mM β-mercaptoethanol, 10 mM sodium hydrosulfite, 10 mM sodium metabisulfite, 10 % glycerol (v/v), 1 µM leupeptin (prepared as a 0.01 mg/ml solution in dimethylsulfoxide), and 25 µM phenylmethylsulfonylfluoride (prepared as a 1 mM solution in dimethylsulfoxide)). The seed suspensions were spun at 14,000g for 10 min at 4 °C and the supernatant was collected. Additional extraction buffer was added as before to the seed pellet and this was spun again as before. The supernatants, which are referred to as crude soluble protein solution, were pooled and stored at −80 °C.
Measurement of Protein Content
The protein content of each purified recombinant protein fraction and crude soluble protein solution was determined by precipitating protein with 1 mL of 10% (w/v) trichloroacetic acid followed by a modification of the Lowry method (Lowry et al. 1951). A standard curve was created using bovine serum albumin (Sigma Chemical Co., St. Louis, MO). The concentrations varied from 3.4 to 4.2 mg/ml and 7.4 to 8.6 mg/ml in the purified recombinant protein fractions and the crude soluble protein, respectively.
Methyltransferase Activity Determination
A vapor diffusion assay was performed, which involves the transfer of a radiolabeled methyl group from S-adenosyl-L-[methyl-14C]methionine ([14C]AdoMet) to a synthetic isoaspartyl-containing methyl-accepting peptide. The reaction mixture contained 10 µM [14C]AdoMet (60 mCi/mmol, Amersham Pharmacia Biotech, UK), 625 µM of ValTyrPro-(L-isoAsp)-HisAla (VYP-(L-isoAsp)-HA, California Peptide Research Inc., Napa, CA), and citrate buffer (unless otherwise indicated; see Fig. 3) at a final reaction pH of 6.9. The reaction buffer was equilibrated to the reaction temperature for 10 min prior to addition of an appropriate dilution of purified recombinant protein in elution buffer or 10 µL of crude soluble protein solution in a final volume of 40 µL. The reaction mixture was then incubated at the indicated temperature for 10 min and then immediately moved to dry ice except in the reaction temperature assay where wet ice was used. This was then quenched with 40 µL of 0.2 N NaOH/1% (w/v) SDS, which results in the hydrolysis of methyl esters to methanol. Sixty µL of quenched reaction mixture was then spotted onto a triple-pleated 1.5 × 8 cm thick filter paper (No. 1650962, Bio-Rad Laboratories, Hercules, CA) and placed in the neck of a 20 mL scintillation vial containing 5 mL of counting fluor (Safety Solve High Flashpoint Cocktail, Research Products International, Mt. Prospect, IL). The vial was capped and left at room temperature for 2 h. During this time, volatile [14C]methanol diffuses into the fluor and the unreacted [14C]AdoMet remains on the filter paper. After removal of the filter paper, vials were counted by liquid scintillation. Peptide specific activity was calculated by subtracting the radioactivity from a control reaction incubated in the absence of the peptide.
Figure 3.
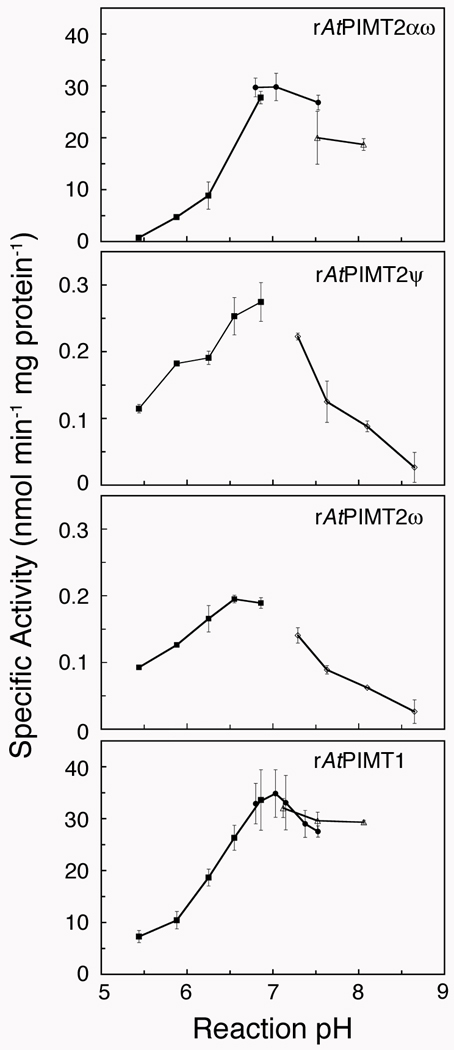
The effect of pH on the activity of recombinant Arabidopsis L-isoAsp methyltransferases. Methyltransferase activity at different pH reaction conditions was assayed as described under “Materials and Methods” with rAtPIMT2αω (120 ng protein), rAtPIMT2ψ (1.7 µg protein), rAtPIMT2ω (1.6 µg protein), and rAtPIMT1 (70 ng protein) at 40 °C for 10 min. In preliminary trials, the pHs of reactions at 40 °C were measured after dilution of each buffer, and each buffer was then used to adjust the reaction pH. The citrate-phosphate buffer (■; reaction pHs 5.4 to 7.0) was a 1 M citric acid solution mixed with 2 M sodium dibasic phosphate and diluted 5-fold into the final assay mixture. Additionally, 1 M buffer solutions of sodium phosphate (●; reaction pHs 6.9 to 7.6), HEPES (△; reaction pHs 7.1 to 8.1), and Tris-HCl (◊; reaction pHs 7.4 to 8.9) were diluted to a final concentration of 200 mM. The rAtPIMT2ψ and rAtPIMT2ω isozymes were purified in parallel and assayed in at least duplicate, as were rAtPIMT2αω and rAtPIMT1 except at a later date. Error bars represent the standard deviation from the mean.
RESULTS AND DISCUSSION
Identification of a Third Isozyme of the PIMT2 gene in Arabidopsis
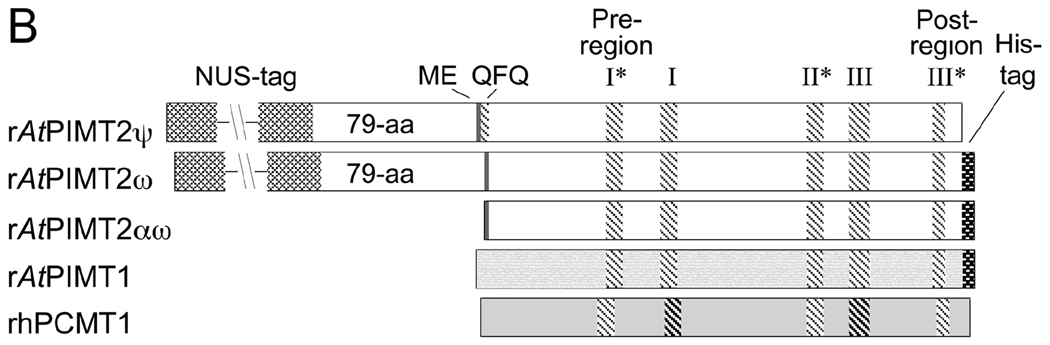
Two AtPIMT2 isozymes, AtPIMT2ψ and AtPIMT2ω, have been described as products of two alternatively-spliced mRNAs of the AtPIMT2 gene (GenBank accession no. AY496702) (Xu et al. 2004). Further analysis of GenBank provided evidence for a third mRNA encoded by AtPIMT2 (GenBank accession no. AK118104; Seki et al. 2002) (Fig. 1A). Here, a downstream transcriptional/translational start site results in an N-terminal truncated form of the enzyme. We now designate this enzyme AtPIMT2αω because it is identical to AtPIMT2ω except that the 79 amino acid N-terminal extension is absent (Xu et al., 2004). The mRNA corresponding to the AK118104 cDNA commences 154 nucleotides downstream of those encoding the AtPIMT2ψ and AtPIMT2ω isozymes and includes a 5’ untranslated region of 108 nucleotides. The resulting protein, AtPIMT2αω (Fig. 1A), is predicted to be an AtPIMT2 isozyme that more closely resembles the structure of the AtPIMT1 isozyme than the previously studied AtPIMT2 isozymes (Fig. 1B; Xu et al. 2004). AtPIMT2αω lacks the QFQ sequence that is a product of alternative splicing in AtPIMT2ψ (Fig. 1B). AtPIMT2αω is predicted to be 227 amino acids long, identical to the length of the PCMT1 isozyme I and similar to that of isozyme II (228 amino acids; Ingrosso et al. 1991), and has a predicted molecular mass of 24.3 kDa. Based on its structural similarity to the cytosolic isoform AtPIMT1 and the absence of any apparent nuclear localization signal (Xu et al. 2004), AtPIMT2αω may also be localized to the cytoplasm. AtPIMT2αω is 51% identical to the human isozymes, indicating a high degree of sequence conservation.
Figure 1.
Recombinant Arabidopsis protein repair methyltransferase constructs. A) The nucleotide and predicted amino acid sequence for AtPIMT2αω encoded by the GenBank AK118104 cDNA sequence is depicted. The apparent transcription initiation site (TIS) is marked with an arrow. Nucleotide positions are indicated on the left, amino acid positions on the right. Pre-region I and post-region III, which are characteristic amino acid sequence motifs of L-isoaspartyl methyltransferases, and motifs I, II, and III, which are characteristic of seven-beta strand methyltransferases, are highlighted by brackets. B) Representation of the recombinant protein structures (top to bottom): rAtPIMTψ, rAtPIMTω, rAtPIMT2αω, and rhPCMT isozyme II. The N-terminal ME sequence, encoded by exon 1, is indicated by shading, and the adjacent QFQ sequence, resulting from alternative 3’-splicing of intron 1 in AtPIMTψ, is indicated by cross-hatching (Xu et al., 2004). rAtPIMTψ and rAtPIMTω are both expressed with an Nus-tag at the N-terminus and both have an additional N-terminal 79 amino acid residues resulting from an upstream transcriptional and translational start site (Xu et al., 2004). rAtPIMT2αω and rAtPIMT1 are each expressed with a C-terminal His-tag sequence, -WVE(H)6 and -WDP(H)6, respectively. Motifs that share 100% identity in all five structures are marked with an asterisk. The rAtPIMT isozymes and the rhPCMT share 88% identity in Region I and 80% identity in Region III and these regions are highlighted with dark hatching in rhPCMT.
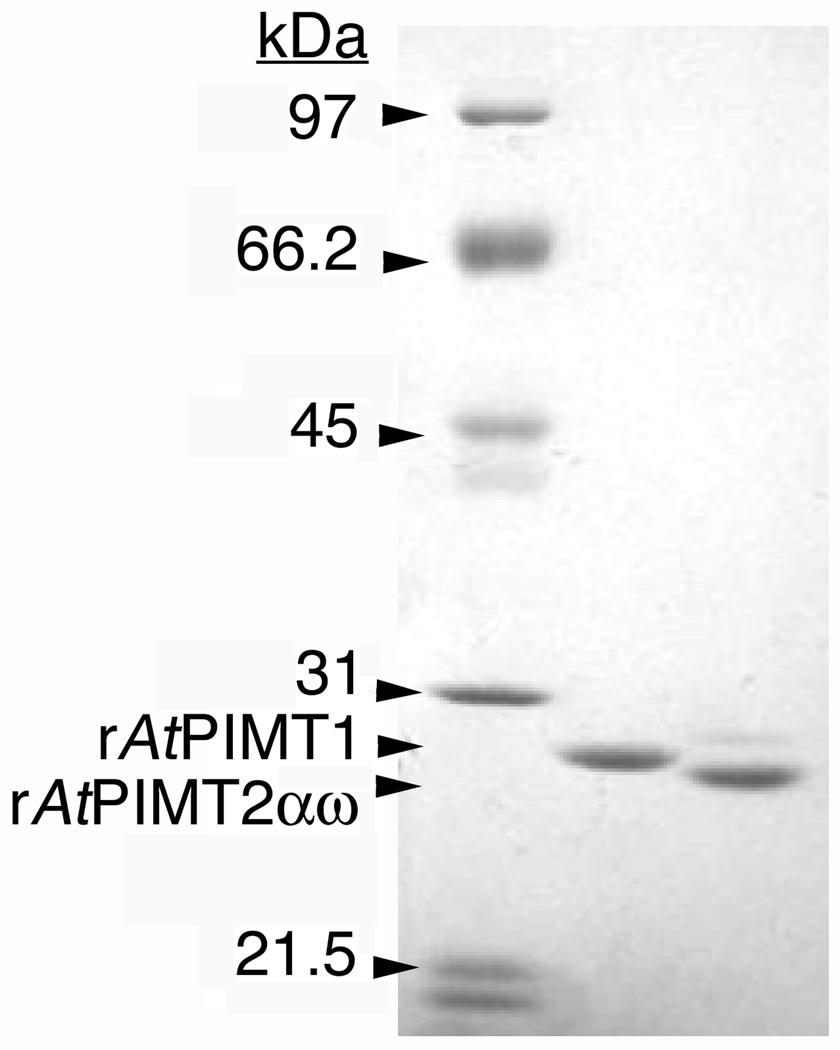
We inserted the DNA sequence corresponding to AtPIMT2αω into a pET23.1a expression plasmid and overexpressed its protein product in Escherichia coli as a carboxy-terminal, hexahistidine tagged recombinant protein (see "Materials and Methods"). We sequenced the plasmid insert DNA in both directions to verify that the sequence matched that of AK118104 (data not shown). The recombinant form (rAtPIMT2αω) is 236 amino acids long and has a predicted mass of 25.5 kDa. This enzyme was purified as described in “Materials and Methods” and analyzed by SDS gel electrophoresis. A single polypeptide mass corresponding to the predicted product was observed at 28.1 kDa (Fig. 2).
Figure 2.
Expression of recombinant protein repair methyltransfereases from Arabidopsis thaliana in E. coli. SDS-PAGE analysis of 2 µg of purified rAtPIMT1 protein (center lane) and 2 µg of purified rAtPIMT2αω protein (right lane) is shown. Gels contained 9.67 % acrylamide and 0.33 % N,N-methylene-bisacrylamide. Low range molecular mass standards (left lane; Bio-Rad, Hercules, CA) included phosphorylase a (97.4 kDa), bovine serum albumin (66.2 kDa), ovalbumin (45 kDa), and carbonic anhydrase (31 kDa). The rAtPIMT1 and rAtPIMT2αω polypeptide bands migrated to positions corresponding to molecular masses of approximately 28.8 kDa and 28.1 kDa, respectively.
Amino-terminal Nus-tagged rAtPIMT2ψ and rAtPIMT2ω, and carboxy-terminal hexahistadyl-tagged rAtPIMT1 were also purified in parallel (see "Materials and Methods") by Ni-column chromatography. SDS gel analysis of our rAtPIMT1 product indicated a single polypeptide band of 28.8 kDa (Fig. 2). For the rAtPIMT2ψ and rAtPIMT2ω enzymes, a Nus-tag construction was found to be necessary for protein solubility (Xu et al. 2004). SDS gel electrophoresis of these preparations demonstrated a band of the expected size, as well as some contaminant bands, accounting for the lower specific activity of these preparations (see below). We found that the purified recombinant proteins could be stored without loss of activity for at least three months in 10% glycerol at −80 °C.
pH Dependence of the activity of rAtPIMT1 and rAtPIMT2 Proteins
The average cytosolic pH of Arabidopsis leaf cells is 7.1 (Song et al. 2004), but this value can vary depending on environmental cues (Felle 2001). Neutral to alkaline pH conditions favor L-isoAsp formation from asparagine residues in proteins in vitro (Brennan and Clarke 1995) and in vivo (Hicks et al. 2005). The highest level of Arabidopsis methyltransferase activity was detected in seed yet a modest level was detected in other organs as well (Mudgett and Clarke 1996). So if we relate leaf cytosolic pH to seed cytosolic pH then understanding the pH dependence of AtPIMT is relevant to the role AtPIMT isozymes may play in protein repair. Therefore, we were interested in the pH dependence of the activity of the Arabidopsis PIMT2 isozymes. We thus compared the effect of pH on the ability of rAtPIMT1 and the rAtPIMT2 family of isozymes to utilize [14C]-AdoMet for methyl esterification of Val-Tyr-Pro-(L-isoAsp)-His-Ala, an L-isoAsp containing hexapeptide that serves as an excellent PIMT substrate (Lowenson and Clarke 1991; Fig. 3). We studied activity at 40 °C with each purified rAtPIMT by adjusting the reaction pH with one of the following buffers: citrate-phosphate (pH 5.4 to 6.8), phosphate (pH 6.8 to 7.5), HEPES (pH 7.5 to 8.1), or Tris-HCl (pH 7.3 to 8.7).
All of the proteins had optimal activity around pH 7 and all had similar pH dependence profiles where activity was seen over a broad range of pH values (Fig. 3). Specifically, rAtPIMT2αω maintained 13% and 63% of its optimal activity at pH values 6 and 8, respectively (Fig. 3). rAtPIMT2ψ preserved 67% and 33% of its optimal activity at pH values 6 and 8; similarly rAtPIMT2ω preserved 65% and 30% of its optimal activity at these respective values (Fig. 3). In addition, rAtPIMT1 maintained 30% and 84% of its optimal activity at pH values 6 and 8 (Fig. 3). The rAtPIMT1 profile over the pH range of 5.5 to 8.1 was similar to that reported previously for the pNT2 recombinant version except that this protein had lower activity overall and the amount of activity remaining at pH values of 6 and 8 was about 60% (Thapar and Clarke 2000). These differences may result from the modification of the second amino acid residue from lysine to glutamate and the lack of a carboxy-terminal -WVE(H)6 sequence in the pNT2 protein (Thapar and Clarke 2000).
Reaction Temperature Dependence of rAtPIMT1, rAtPIMT2s, and rhPCMT
A link between an increase in temperature and an increase in the presence of L-isoAsp has been previously shown (Sharma et al. 1993) and the PIMT protein repair system may be involved in reversing protein damage due to temperature stress. It has been shown that maize seed PIMT retains high levels of activity at relatively high reaction temperatures (up to 65 °C), and purified wheat seed PIMT and rice seed PIMT activity profiles are comparable (Thapar et al. Clarke 2001). Interestingly, a previous report of a purified recombinant Arabidopsis PIMT1 enzyme was shown to not be as robust under similar temperature conditions (Thapar and Clarke 2000).
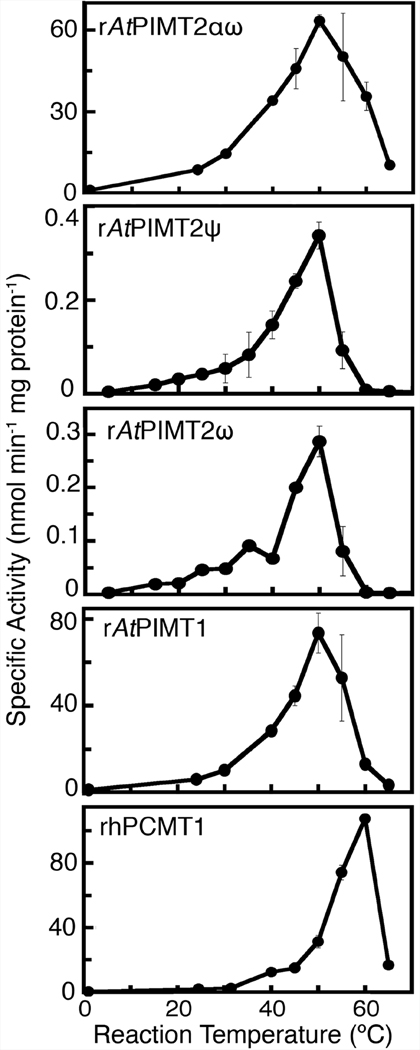
To better understand the role AtPIMT2 isozymes may play in the repair of temperature induced L-isoAsp formation, we tested the reaction temperature dependence of each purified recombinant Arabidopsis isozyme for the methyl-esterification of synthetic L-isoAsp containing hexapeptide (Fig. 4). The temperature dependence activity profiles were similar in that each had an exponential-like increase in activity from low temperatures to the optimal temperature (50 °C), followed by a rapid decline at higher temperatures. In addition, the temperature of optimal activity did not appear to depend on the structure of the recombinant protein. The optimum activity of rAtPIMT2αω was 63.4 nmol min−1 mg−1 (Fig. 4); activity declined by 50% to 60% at temperatures ten-degrees above and below the optimum. rAtPIMT2ψ and rAtPIMT2ω had much lower activity overall (Fig. 4; due to the presence of the Nus fusion protein and the presence of contaminating proteins), but shared the same value for the temperature of optimal activity with rAtPIMT2αω. rAtPIMT2ψ and rAtPIMT2ω activity were more sensitive to temperature: activity dropped below 45% at 40 °C and was not detected at 60 °C. We show that rAtPIMT1 has an optimal activity of 73.6 nmol min−1 mg−1, which declined to 40% at 40 °C and 20% at 60 °C (Fig. 4). In comparison, the pNT2 version of the recombinant Arabidopsis PIMT1 protein was also active across the temperature range 25 °C and 45 °C. However, the overall activity was several fold lower and the activity was barely detectable at 55 °C. These differences may also reflect the amino acid differences in these two PIMT1 proteins (see above).
Figure 4.
The effect of reaction temperature on the activity of recombinant Arabidopsis L-isoAsp methyltransferases. Methyltransferase activity assays were performed as described under “Materials and Methods” at various temperatures over the range of 0 °C to 65 °C with rAtPIMT2αω (120 ng protein), rAtPIMT2ψ (1.7 µg protein), rAtPIMT2ω (1.6 µg), and rAtPIMT1 (70 ng protein), and rhPCMT (50 ng protein). The rAtPIMT2ψ and rAtPIMT2ω isozymes were purified in parallel and assayed in at least duplicate, as were rAtPIMT2αω and rAtPIMT1 except at a later date. rhPCMT was purified and studied separately. Error bars represent the standard deviation from the mean.
We also compared the activity of the recombinant human PCMT1 enzyme under these conditions (MacLaren and Clarke 1995). Surprisingly, rhPCMT was most active at 60 °C with a specific activity of 108 nmol min−1 mg−1 (Fig. 4). rhPCMT was able to maintain 20% activity at 65 °C, similar to rAtPIMT2αω, the only rAtPIMT noticeably active at this temperature. At the physiological temperature of 37°C, rhPCMT was only about 15% as active as at 60 °C.
Thermostability of rAtPIMT1, rAtPIMT2αω, and rhPCMT
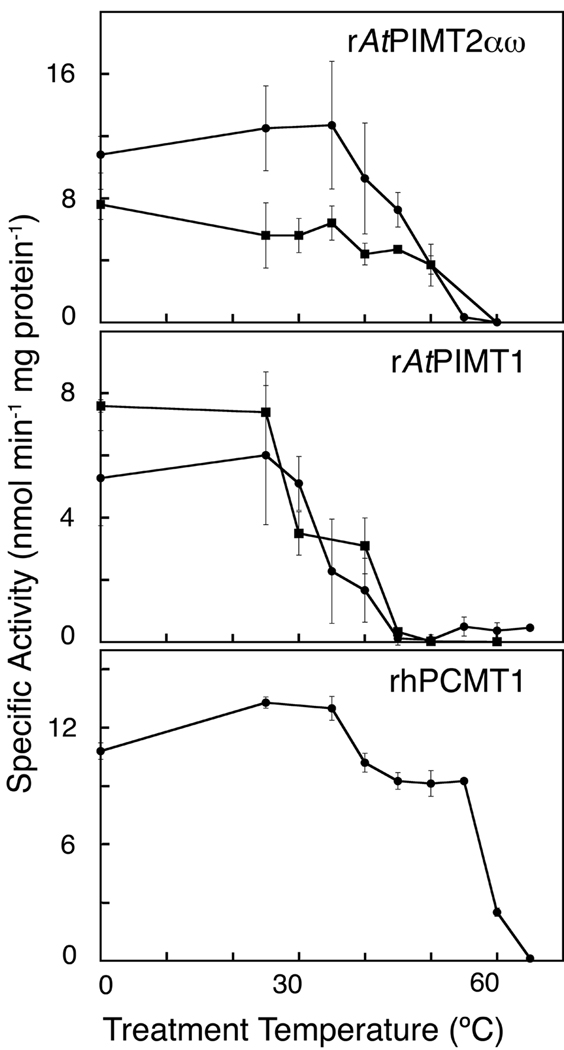
To further understand the role of rAtPIMT isozymes in temperature stress, we focused on the thermostability of rAtPIMT1 and rAtPIMT2αω when treated over a range of temperature conditions for 10 min at pH values of 6.9 and 8.0 (Fig. 5). In comparison to rAtPIMT1, rAtPIMT2αω better maintains its ability to methyl-esterify the L-isoAsp containing hexapeptide after exposure to relatively high temperatures. rAtPIMT2αω maintained essentially all its methyltransferase activity after being exposed to temperatures between 0 °C and 35 °C (Fig. 5). However, its activity declined when pretreated at 40 °C and this decline continued at higher temperatures. Little or no activity remained following exposure to temperatures greater than 55 °C. In comparison, rAtPIMT1 was not as thermostable as rAtPIMT2αω (Fig. 5). Maximum activity was seen in rAtPIMT1 when pretreated between 0 °C and 25 °C. Activity declined markedly by 35 °C and was nearly lost at temperatures greater than 45 °C.
Figure 5.
Thermostability of recombinant Arabidopsis L-isoAsp methyltransferase. Purified recombinant enzyme was pretreated in elution buffer (λ; final pH of 8.0), or sodium phosphate buffer (ν; final pH of 6.9) over the temperature range of 0 °C to 65 °C (see Materials and Methods) for 10 min prior to its addition to the methyltransferase assay reaction mixture. Methyltransferase activity assays were performed for 10 min at 40 °C with rAtPIMT2αω (60 or 120 ng protein), rAtPIMT1 (70 ng protein), and rhPCMT (130 ng protein). The rAtPIMT2αω and rAtPIMT1 isozymes were purified in parallel and assayed in at least duplicate. rhPCMT was purified and studied separately. Error bars represent the standard deviation from the mean.
In comparison, the thermostability of rhPCMT was also studied (Fig. 5). Previous findings report that rhPCMT loses none of its activity after being heated for 30 min at 50 °C (MacLaren and Clarke 1995), so we were interested in testing its activity after pre-exposure to a range of temperatures. We observed that rhPCMT has maximal activity after being exposed to 30 °C and 35 °C for 10 min, and its level of activity was comparable to that of rAtPIMT2αω. In the range of 40 °C and 55 °C, activity was surprisingly conserved within 77% to 70% of the maximal activity observed at 30 °C for rhPCMT. Just 5 degrees above this range, the activity of rhPCMT sharply declined to 19%, but this was still remarkable when compared to the almost total loss of activity seen in both Arabidopsis enzymes when pre-exposed to 60 °C. At 65 °C, no activity was observed with rhPCMT.
Effect of Heat Stress on Induction of PIMT Activity in Seeds
Given the wide range of temperatures that we observed methyltransferase activity with the recombinant proteins in vitro, we then asked if the expression of PIMT might be enhanced in seeds under conditions leading to thermotolerance. Thermotolerance in Arabidopsis seedlings is acquired with exposure to sublethal heat stress temperatures of 35 °C to 40 °C (Burke et al. 2000; Hong and Vierling 2000; Hong et al. 2003). To see if L-isoAsp-peptide dependent methyltransferase activity is present under heat stress conditions we tested the activity in extracts from mature, dry seeds that were heated for 4 h between 38 °C and 40 °C, since activity is predominantly detected in Arabidopsis seeds, not vegetative tissue, under normal conditions (Mudgett and Clarke 1996; Xu et al. 2004). However, we found no detectable difference in peptide-dependent activity of unheated and heated seeds (1.1 pmol min−1 mg protein−1 +/− 0.069 and 1.1 pmol min−1 mg protein−1 +/− 0.034, respectively). Although this result indicates that enzyme activities are not increased under these conditions, it is interesting to note that we see no loss of activity, even though we see a significant decline in recombinant activity under similar conditions (Fig. 5). These results suggest that stabilizing factors exist in cells, although we cannot exclude the possibility that new AtPIMT is produced in seeds under these conditions. Consistent with our findings, corn seeds and wheat seedlings retain their repair methyltransferase activity after 10 h at 37 °C (Thapar et al. 2001; Mudgett and Clarke 1994).
Physiological Implications
It has been speculated that AtPIMT2 may be involved in plant stress response as AtPIMT2 transcripts are influenced by developmental stage, abscisic acid, and abiotic stress (Xu et al. 2004). Considering that alternative splicing, leading to a different protein length and/or amino acid composition, can play a role in stress responses (Kazan 2003; Iida 2004), we describe a third AtPIMT2 isoenzyme that arises via alternative transcriptional start site selection.
We find that the rAtPIMT isozymes are most active in vitro near neutral pH conditions, but retain activity over a broad range of pH values. Nearly 50% of the maximal activity of the rAtPIMT2ω and rAtPIMT2ψ isozymes is found at pH values near 5.5, whereas the loss of activity in the acidic range is sharper for rAtPIMT2αω and rAtPIMT1. Under acidic conditions, very little L-isoAsp formation from asparaginyl residues is expected, but such altered residues would continue to form from aspartyl residues (Ota et al. 1987). At least 50% of maximal activity of the rAtPIMT2αω and rAtPIMT1 isozymes is found near pH 8, whereas the loss of activity in the basic range is sharper for rAtPIMT2ω and rAtPIMT2ψ, but this may be a buffer specific effect. At basic pH values L-isoAsp formation from asparagine residues would be expected to occur at faster rates (Patel and Borchardt 1990a,b). These results suggest that these enzymes have been optimized to be active over a range of pH values where L-isoAsp formation occurs.
The rate of accumulation of altered aspartyl residues would be expected to increase with temperature as one part of the thermal stress-induced damage to the proteome. Significantly, our finding here that PIMT activity of the Arabidopsis isozymes is maximal at 50 °C suggests the importance of its repair activity as a component of plant defense mechanisms. Optimal Arabidopsis growth occurs between 16 °C and 25 °C (Weigel and Glazebrook 2002). While Arabidopsis vegetative tissue can withstand up to 2 h of exposure at 42 °C without damage and Arabidopsis seeds complete germination even after a 220 min heat treatment of 45 °C (Binelli and Mascarenhas 1990; Larkindale et al. 2005), temperatures of 45 °C and higher are considered lethal for seedlings (Burke et al. 2000; Hong et al. 2003). However, Arabidopsis can acquire some tolerance to temperatures of 45 °C to 50 °C after preexposure to high, but nonlethal, temperatures (Burke et al. 2000; Hong and Vierling 2000; Hong et al. 2003; Larkindale et al. 2005; Charng et al. 2006). Our in vitro data shows that while rAtPIMT enzyme activity is found up to at least 55 °C in a 10 min assay, the activity does decline with time at temperatures above 25 °C to 30 °C. These results suggest that the methyltransferase can function for brief exposures of plants to high temperatures, but not to extended exposures, especially above 40 °C. However, the stability of the enzyme in tissues may be greater, as shown here for seeds incubated at 38 °C to 40 °C. These results suggest a physiological role for the repair methyltransferase in response to the enhanced accumulation of L-isoAsp residues at elevated temperatures in addition to the normal accumulation that occurs under more optimal growth conditions.
We note that an optimal catalytic activity at lethal temperatures is also found for other enzymes in Arabidopsis. Ribulose-1,5-bisphosphate carboxylase/oxygenase and adenine phosphoribosyltransferase have optimal enzyme activity of at least 60 °C (Salvucci et al. 2001) and 65 °C (Lee and Moffat 1993), respectively. Additionally, it has been speculated that ribulose-1,5-bisphosphate caboxylase/oxygenase activase may be alternatively spliced as a result of environmental regulation since evidence shows a marked difference in temperature optima between its isoforms (Crafts-Brandner et al. 1997).
PIMT specialization to function under high temperatures is seen in other organisms as well. Genetic analyses show that the E. coli L-isoaspartyl methyltransferase confers to the organism the ability to withstand thermal stress (Li and Clarke 1992; Visick et al. 1998; Kindrachuck et al. 2003). In addition, an increase in PCMT activity is observed at 42 °C in HeLa cells (Ladino and O’Connor 1992). The ability to deploy a repair system that is predicted to function in the maintenance of the structural integrity at Asn and Asp sites in polypeptides under high temperature conditions is probably most compelling in thermophilic organisms. Recombinant Thermatoga maritima PIMT has a maximal activity at 85 °C and has been shown to resist denaturation at temperatures as high as 100 °C (Ichikawa and Clarke 1998). A similar situation is found in recombinant PIMT from Pyrococcus furiosus, although the thermal stability is limited above 75 °C (Thapar et al. 2002).
Plants and bacteria exist in environments where elevated temperatures generally cannot be avoided. However, warm blooded animals are adapted to maintain constant internal temperatures. Why then does recombinant human PCMT1 have maximal activity at 60 °C? One explanation may involve damage and repair to polypeptides associated with burns, particularly in cuticular/epidermal polypeptides such as collagen (Haley et al. 1966; Bornstein 1970; Fledelius et al. 1997; Cloos and Gledelius 2000). Although hPCMT1 is cytosolic, burn damage may release it into the extracellular matrix where it may act to repair damaged proteins.
Acknowledgements
We are very grateful to Dr. Jonathan Lowenson at UCLA for his advice and for providing recombinant human protein L-isoAsp methyltransferase. We also thank Dr. Jonathan Katz for his helpful advice and Sharon Niv for her assistance in recombinant Arabidopsis protein L-isoAsp methyltransferase purification.
This work was supported by awards from the National Institutes of Health (GM055052 to S.T.V., GM07185 to S.T.V., GM026020 to S.G.C., AG018000 to S.G.C.), the National Science Foundation (MCB-0448533 to S.G.C., MCB-0449646 to A.B.D), the Ford Foundation (S.T.V.), the U.S. Department of Education (S.T.V.), and the UCLA Graduate Division (S.T.V.)
Abbreviations
- [14C]AdoMet
S-adenosyl-L-[methyl-14C]methionine
- AdoMet
S-adenosyl-L-methionine
- AtPIMT
Arabidopsis thaliana protein L-isoaspartyl O-methyltransferase
- DE-53
Diethylaminoethylcellulose
- hPCMT
Human Protein L-isoaspartate (D-aspartate) O-methyltransferase
- Ni-NTA
Nickel nitrilotriacetic
- Nus-tag
NusA protein
- PCMT
Protein L-isoaspartyl (D-aspartyl) O-methyltransferase
- rAtPIMT
Recombinant Arabidopsis thaliana protein L-isoaspartyl O-methyltransferase
- rhPCMT
Recombinant human protein L-isoaspartate (D-aspartate) O-methyltransferase
- TIS
transcription initiation site
References
- Aswad D. Deamidation and Isoaspartate Formation in Peptides and Proteins. Boca Raton: CRC Press; 1995. Effects of incubation variable on rates of isoaspartate formation; pp. 105–109. [Google Scholar]
- Bhatt NP, Patel K, Borchardt RT. Chemical pathways of peptide degradation. I. Deamidation of adrenocorticotropic hormone. Pharm Res. 1990;7:593–599. doi: 10.1023/a:1015862026539. [DOI] [PubMed] [Google Scholar]
- Binelli G, Mascarenhas JP. Arabidopsis: Sensitivity of Growth to High Temperature. Dev Gen. 1990;11:294–298. [Google Scholar]
- Borisjuk L, Rolletschek R, Radchuk R, Weschke W, Wobus U, Weber H. Seed development and differentiation: A role for metabolic regulation. Plant Biol. 2004;6:375–386. doi: 10.1055/s-2004-817908. [DOI] [PubMed] [Google Scholar]
- Bornstein P. Structure of alpha-1-CG8, a large cyanogens bromide produced fragment form the alpha-1 chain of rat collagen. Biochem. 1970;9:2408–2421. doi: 10.1021/bi00814a004. [DOI] [PubMed] [Google Scholar]
- Brennan TV, Clarke S. Deamidation and isoaspartate formation in model synthetic peptides: The effects of sequence and solution environment. In: Aswad DW, editor. Deamidation and isoaspartate formation in peptides and proteins. Ann Arbor, Michigan: CRC Press; 1995. pp. 65–90. [Google Scholar]
- Brennan TV, Anderson JW, Jia Z, Waygood EB, Clarke S. Repair of spontaneously deamidated HPr phosphocarrier protein catalyzed by the L-isoaspartate-(D-aspartate) O-methyltransferase. J Biol Chem. 1994;269:24586–24595. [PubMed] [Google Scholar]
- Burke JJ, O’Mahony PJ, Oliver MJ. Isolation of Arabidopsis mutants lacking components of acquired thermotolerance. Plant Physiol. 2000;123:575–587. doi: 10.1104/pp.123.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng Y-Y, Liu H-C, Lie N-Y, Hsu F-C, Ko S-S. Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol. 2006;140:1297–1305. doi: 10.1104/pp.105.074898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S. Aging as war between chemical and biochemical processes: protein methylation and the recognition of age-damage proteins for repair. Ageing Res Rev. 2003;2:263–285. doi: 10.1016/s1568-1637(03)00011-4. [DOI] [PubMed] [Google Scholar]
- Cloos PA, Fledelius C. Collagen fragments in urine derived from bone resorption are highly racemized and isomerized: a biological clock of protein aging with clinical potential. Biochem J. 2000;345:473–480. [PMC free article] [PubMed] [Google Scholar]
- Crafts-Brandner SJ, van de Loo FJ, Salvucci ME. The two forms of ribulose-1,5-bisphosphate carboxylase/oxygenase activase differ in sensitivity to elevated temperature. Plant Physiol. 1997;114:439–444. doi: 10.1104/pp.114.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVry CG, Clarke S. Polymorphic forms of the protein L-isoaspartate (D-aspartate) O-methyltransferase involved in the repair of age-damaged proteins. J Hum Genet. 1999;44:275–288. doi: 10.1007/s100380050161. [DOI] [PubMed] [Google Scholar]
- Doyle HA, Gee RJ, Mamula MJ. A failure to repair self-proteins leads to T cell hyperproliferation and autoantibody production. J Immunol. 2003;171:2840–2847. doi: 10.4049/jimmunol.171.6.2840. [DOI] [PubMed] [Google Scholar]
- Felle HH. pH: Signal and messenger in plant cells. Plant Biol. 2001;3:577–591. [Google Scholar]
- Fledelius C, Johnsen AH, Cloos PA, Bonde M, Qvisdt P. Characterization of urinary degradation products derived from type I collagen. Identification of a beta-isomerized Asp-Gly sequence within the C-terminal telopeptide (alpha1) region. J Biol Chem. 1997;272:9755–9763. doi: 10.1074/jbc.272.15.9755. [DOI] [PubMed] [Google Scholar]
- Haley EE, Corcoran BJ, Dorer FE, Buchanan DL. Beta-aspartyl peptides in enzymatic hydrolysates of protein. Biochem. 1966;5:3229–3235. doi: 10.1021/bi00874a024. [DOI] [PubMed] [Google Scholar]
- Hicks WM, Kotlajich MV, Visick JE. Recovery from long-term stationary phase and stress survival in Escherichia coli require the L-isoaspartyl protein carboxyl methyltransferase at alkaline pH. Microbiol. 2005;151:2151–2158. doi: 10.1099/mic.0.27835-0. [DOI] [PubMed] [Google Scholar]
- Hong S-W, Lee U, Vierling E. Arabidopsis hot mutants define multiple functions required for acclimation to high temperature. Plant Physiol. 2003;132:757–767. doi: 10.1104/pp.102.017145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S-W, Vierling E. Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci. 2000;97:4392–4397. doi: 10.1073/pnas.97.8.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa JK, Clarke S. A highly active protein repair enzyme from an extreme thermophile: the L-isoaspartyl methyltransferase from Thermotoga maritima. Arch Biochem Biophys. 1998;358:222–231. doi: 10.1006/abbi.1998.0830. [DOI] [PubMed] [Google Scholar]
- Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, Konagaya A, Shinozaki K. Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nuc Acid Res. 2004;32:5096–5103. doi: 10.1093/nar/gkh845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingrosso D, Kagan RM, Clarke S. Distinct C-terminal sequences of isozymes I and II of the human erythrocyte L-isoaspartyl/D-aspartyl protein methyltransferase. Biochem Biophys Res Commun. 1991;175:351–358. doi: 10.1016/s0006-291x(05)81242-2. [DOI] [PubMed] [Google Scholar]
- Kagan RM, McFadden HJ, McFadden PN, O’Connor C, Clarke S. Molecular phylogenetics of a protein repair methyltransferase. Comp Biochem Physiol. 1997;117:379–385. doi: 10.1016/s0305-0491(96)00333-1. [DOI] [PubMed] [Google Scholar]
- Kazan K. Alternative splicing and proteome diversity in plants: the tip of the iceberg has just emerged. Trend Plant Sci. 2003;10:468–471. doi: 10.1016/j.tplants.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Kester ST, Geneve RL, Houtz RL. Priming and accelerated aging affect L-isoaspartyl methyltransferase activity in tomato (Lycopersicon esculentum Mill.) seed. J Exp Bot. 1997;309 949-949. [Google Scholar]
- Kindrachuk J, Parent J, Davies GF, Dinsmore M, Attah-Poku S, Napper S. Overexpression of L-isoaspartate O-Methyltransferase in Escherichia coli increases heat shock survival by a mechanism independent of methyltransferase activity. J Biol Chem. 2003;278:50880–50886. doi: 10.1074/jbc.M308423200. [DOI] [PubMed] [Google Scholar]
- Kumar GN, Houtz RL, Knowles NR. Age-induced protein modifications and increased proteolysis in potato seed-tubers. Plant Physiol. 1999;119:89–100. doi: 10.1104/pp.119.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladino CA, O’Connor CM. Methylation of atypical protein aspartyl residues during the stress response of HeLa cells. J Cell Physiol. 1992;153:297–304. doi: 10.1002/jcp.1041530209. [DOI] [PubMed] [Google Scholar]
- Lanthier J, Desrosiers RR. Protein L-isoaspartyl methyltransferase repairs abnormal aspartyl residues accumulated in vivo in type-I collagen and restores cell migration. Exp Cell Res. 2004;291:96–105. doi: 10.1016/j.yexcr.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Larkindale J, Hall JD, Knight MR, Vierling E. Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 2005;138:882–897. doi: 10.1104/pp.105.062257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Moffatt BA. Purification and characterization of adenine phosphoribosyltransferase from Arabidopsis thaliana. Physiol Plant. 1993;87:483–492. [Google Scholar]
- Li C, Clarke S. A protein methyltransferase specific for altered aspartyl residues is important in Escherichia coli stationary-phase survival and heat-shock resistance. Proc Natl Acad Sci. 1992;89:9885–9889. doi: 10.1073/pnas.89.20.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenson JD, Clarke S. Structural elements affecting the recognition of L-isoaspartyl residues by the L-isoaspartyl/D-aspartyl protein methyltransferase. Implications for the repair hypothesis. J Biol Chem. 1991;266:19396–19406. [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- MacLaren DC, Clarke S. Expression and purification of a human recombinant methyltransferase that repairs damaged proteins. Protein Expr Purif. 1995;6:99–108. doi: 10.1006/prep.1995.1013. [DOI] [PubMed] [Google Scholar]
- MacLaren DC, Kagan RM, Clarke S. Alternative splicing of the human isoaspartyl protein carboxyl methyltransferase RNA leads to the generation of a C-terminal –RDEL sequence in isozyme II. Biochem Biophys Res Commun. 1992;185:277–283. doi: 10.1016/s0006-291x(05)80987-8. [DOI] [PubMed] [Google Scholar]
- Mudgett MB, Clarke S. Characterization of plant L-isoaspartyl methyltransferases that may be involved in seed survival: Purification, cloning, and sequence analysis of the wheat germ enzyme. Biochem. 1993;32:11100–11111. doi: 10.1021/bi00092a020. [DOI] [PubMed] [Google Scholar]
- Mudgett MB, Clarke S. Hormonal and environmental responsiveness of a developmentally-regulated protein repair L-isoaspartyl methyltransferase in wheat. J Biol Chem. 1994;269:25605–25612. [PubMed] [Google Scholar]
- Mudgett MB, Clarke S. A distinctly regulated protein repair L-isoaspartylmethyltransferase form Arabidopsis thaliana. Plant Mol Biol. 1996;30:723–737. doi: 10.1007/BF00019007. [DOI] [PubMed] [Google Scholar]
- Mudgett MB, Lowenson JD, Clarke S. Protein repair L-isoaspartyl methyltransferase in plants. Phylogenetic distribution and the accumulation of substrate proteins in aged barley seeds. Plant Physiol. 1997;115:1481–1489. doi: 10.1104/pp.115.4.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota IM, Ding L, Clarke S. Methylation at specific altered aspartyl and asparaginyl residues in glucagon by the erythrocyte protein carboxyl methyltransferase. J Biol Chem. 1987;262:8522–8533. [PubMed] [Google Scholar]
- Ota IM, Gilbert JM, Clarke S. Two major isozymes of the protein D-aspartyl/L-isoaspartyl methyltransferase from human erythrocytes. Biochem Biophys Res Commun. 1988;151:1136–1143. doi: 10.1016/s0006-291x(88)80484-4. [DOI] [PubMed] [Google Scholar]
- Patel K, Borchardt RT. Chemical pathways of peptide degradation. II. Kinetics of deamidation of an asparaginyl residue in a model hexapeptide. Pharm Res. 1990a;7:703–711. doi: 10.1023/a:1015807303766. [DOI] [PubMed] [Google Scholar]
- Patel K, Borchardt RT. Chemical pathways of peptide degradation. III. Effect of primary sequence on the pathways of deamidation of asparaginyl residues in hexapeptides. Pharm Res. 1990b;7:787–793. doi: 10.1023/a:1015999012852. [DOI] [PubMed] [Google Scholar]
- Reissner KJ, Aswad DW. Deamidation and isoaspartate formation in proteins: unwanted alterations or surreptitious signals? Cell Mol Life Sci. 2003;60:1281–1295. doi: 10.1007/s00018-003-2287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvucci ME, Osteryoung KW, Crafts-Brandner SJ, Vierling E. Exceptional sensitivity of Rubisco activase to thermal denaturation in vitro and in vivo. Plant Physiol. 2001;127:1053–1064. [PMC free article] [PubMed] [Google Scholar]
- Seki M, Iida K, Satou M, Sakurai T, Akiyama K, Ishida J, Nakajima M, Enju A, Kamiya A, Narusaka M, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K. Direct Submission to NCBI database. 2002 [Google Scholar]
- Sharma S, Hammen PK, Anderson JW, Leung A, Georges F, Hengstenberg W, Klevit RE, Waygood EB. Deamidation of HPr, a phosphocarrier protein of the phosphoenolpyruvate:sugar phosphotransferase system, involves asparagines 38 (HPr-1) and asparagines 12 (HPr-2) in isoaspartyl acid formation. J Biol Chem. 1993;268:17696–17704. [PubMed] [Google Scholar]
- Shen-Miller J, Mudgett MB, Schopf JW, Clarke S, Berger R. Exceptional seed longevity and robust growth: ancient sacred lotus from China. Amer J Botany. 1995;82:1367–1380. [Google Scholar]
- Slonczewski JL, Foster JW. pH-regulated genes and survival at extreme pH. In: Neidhardt C, editor. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Washington DC: American Society for Microbiology Press; 1996. pp. 1539–1552. [Google Scholar]
- Song CP, Guo Y, Qiu Q, Lambert G, Galbraith DW, Jagendorf A, Zhu JK. A probable Na+(K+)/H+ exchanger on the chloroplast envelope functions in pH homeostasis and chloroplast development in Arabidopsis thaliana. Proc Natl Acad Sci. 2004;101:10211–10216. doi: 10.1073/pnas.0403709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapar N, Clarke S. Expression, purification, and characterization of the protein repair L-isoaspartyl methyltransferase from Arabidopsis thalianaM. Protein Expr Purif. 2000;20:237–251. doi: 10.1006/prep.2000.1311. [DOI] [PubMed] [Google Scholar]
- Thapar N, Griffith SC, Yeates TO, Clarke S. Protein repair methyltransferase from the hyperthermophilic archaeon Pyrococcus furiosus. Unusual methyl-accepting affinity for D-aspartyl and N-succinyl-containing peptides. J Biol Chem. 2002;277:1058–1065. doi: 10.1074/jbc.M108261200. [DOI] [PubMed] [Google Scholar]
- Thapar N, Kim A, Clarke S. Distinct patterns of expression but similar biochemical properties of protein L-isoaspartyl methyltransferase in higher plants. Plant Physiol. 2001;125:1023–1035. doi: 10.1104/pp.125.2.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick JE, Cai H, Clarke S. The L-isoaspartyl Protein Repair Methyltrasnferase Enhances Survival of Aging Escherichia coli subjected to secondary environmental stresses. J Bacteriol. 1998;1998:2623–2629. doi: 10.1128/jb.180.10.2623-2629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. Arabidopsis: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2002. p. 7. [Google Scholar]
- Xu Q, Belcastro MP, Villa ST, Dinkins RD, Clarke SG, Downie AB. A second protein L-isoaspartyl methyltransferase gene in Arabidopsis produces two transcripts whose products are sequestered in the nucleus. Plant Physiol. 2004;136:2652–2664. doi: 10.1104/pp.104.046094. [DOI] [PMC free article] [PubMed] [Google Scholar]