Abstract
Global protein identification through current proteomics methods typically depends on the availability of sequenced genomes. In spite of increasingly high throughput sequencing technologies, this information is not available for every microorganism and rarely available for entire microbial communities. Nevertheless, the protein-level homology that exists between related bacteria makes it possible to extract biological information from the proteome of an organism or microbial community by using the genomic sequences of a near neighbor organism. Here, we demonstrate a trans-organism search strategy for determining the extent to which near-neighbor genome sequences can be applied to identify proteins in unsequenced environmental isolates. In proof of concept testing, we found that within a CLUSTAL W distance of 0.089, near-neighbor genomes successfully identified a high percentage of proteins within an organism. Application of this strategy to characterize environmental bacterial isolates lacking sequenced genomes, but having 16S rDNA sequence similarity to Shewanella resulted in the identification of 300–500 proteins in each strain. The majority of identified pathways mapped to core processes, as well as to processes unique to the Shewanellae, in particular to the presence of c-type cytochromes. Examples of core functional categories include energy metabolism, protein and nucleotide synthesis and cofactor biosynthesis, allowing classification of bacteria by observation of conserved processes. Additionally, within these core functionalities, we observed proteins involved in the alternative lactate utilization pathway, recently described in Shewanella.
Introduction
Protein identification from peptide centric liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomics is currently limited to those organisms for which a genome or metagenome sequence is available. In the absence of sequence information, methods for identifying peptides include the use of de novo computational tools, as well as the use of trans-species comparisons or near neighbor genome sequences [1], [2]. Although interpretation of mass spectra using de novo tools has made considerable progress, the approach remains challenged by the shear number of possible amino acid sequence interpretations for measured fragmentation mass spectrum [3], [4]. Additionally, within any automated LC-MS/MS proteomics run, a large number of common contaminants are present [5]. Typically, masses derived from peptides belonging to these background proteins do not affect conventional searches. However, many of the proteins associated with contaminants, such as the keratins, contain large stretches of low complexity searches, which hit many other unrelated proteins in a sequence database search. Deconvolution and assignation of these low complexity regions to a single protein is difficult, if not impossible.
Recently, the UStags de novo approach [6] was published. As with other sequence tag identification strategies, UStags makes the assumption that ambiguous amino acids are located near the N- or C- terminus of a protein, regions that are usually more conserved[7], [8], [9], [10], [11]. Stretches of amino acids as small as 4 residues can be unique, allowing identification of a protein, using a peptide with ambiguous amino acids. However, as an error tolerant search, resulting candidate lists are large and require manual curation, though development of statistical models and automated filtering methodologies is underway [12], [13], [14].
An alternate approach involves using the genome from one organism to investigate the proteome of an unsequenced organism, which has been computationally investigated and experimentally demonstrated [1], [4], [12], [15], [16]. However, this approach has been constrained to bivariate comparisons and to comparisons within different strains of the same species. The majority of these investigations employed the “MS BLAST” homology searching protocol developed by Shevchenko, et al. [2]. MS BLAST is a sequence-based search strategy that involves de novo peptide sequencing, followed by a BLAST search to identify candidate proteins from these sequences. However, none of these studies addressed the question of how closely related an organism needs to be to generate meaningful data, especially, when multiple near neighbor (multiple species, strains, etc.) genome sequences exist.
In this study, we employed a systematic peptide identification strategy in which spectra derived from one organism were searched against the genome sequences of progressively more genetically distant neighbor organisms to measure the extent to which proteomic information could be obtained about one species when using the genomic sequence of another. Multiple genome sequences for Shewanellae were selected for proof of concept, not only because of the large number of publicly available genome sequences, but also because of the potential environmental importance of these organisms [17], [18], [19], [20], [21]. We also included sequences from two bacteria that are relatively distant from Shewanellae, i.e., Deinococcus radiodurans R1 and Salmonella enterica subsp. enterica serotype Typhimurium LT2 (S. Typhimurium) [22], [23], [24]. In an initial demonstration, we applied the strategy to identify proteins in four environment isolates of Shewanella obtained from sediments along the Columbia River in Washington state that lacked sequenced genomes [25]. These isolates had been identified as Shewanella by partial 16S rDNA sequencing. Depending upon the isolate, we identified 300–500 proteins from ∼4300 open reading frames based on sequenced Shewanellae. Note that species and strain designations are as in [26], except Shewanella putrefaciens CN32, which was originally described in [27].
Similar to most high throughput, mass spectrometry driven proteomic experiments, millions of unique spectra were generated for this empirical study, then analyzed using software tools that match measured spectra to a database of in silico spectra derived from genomic information. Ultimately, these tools allow for the identification of peptides and their parent proteins. Application of these tools to organisms without genome sequences (the approach demonstrated in this empirical study) is relatively new. In the future, emerging technologies, using a combination of de novo sequencing or unique sequence tags (UStags) may help expand the number of identified proteins, allowing further exploration of uncharacterized organisms.
Results and Discussion
Proof of concept
Global proteomics analysis
Spectra derived from previous studies of 11 Shewanella species, D. radiodurans, and S. Typhimurium were searched against their own genome sequences using the open source software tool X!Tandem [28], [29]. A total of 2,502,088 unique and fully tryptic peptide sequences containing at least six amino acid residues were identified and then filtered according to an X!Tandem calculated E-value of ≤5.01×10−09 to generate a list of the top 10% identified peptides. From these peptides, 30,528 proteins were identified by at least two unique peptides, and 26,539 of these proteins were observed in at least two organisms. The high degree of expressed protein homologs among the Shewanella organisms was expected because all were cultured aerobically in tryptic soy broth at 30°C. Tryptic soy broth represents a “universal medium” without going through an extended optimization process to develop a defined medium. Given the range of habitats the environmental isolates came from, tryptic soy broth was used minimize growth medium-related effects. The number of peptides/proteins identified for each organism was assumed to represent the maximum observable proteome for the particular growth and LC-MS/MS instrument conditions employed in this study.
Relationship between proteome and evolutionary distance of neighbor organisms
Spectra derived from a single condition for each organism were searched against the genome sequences of progressively more genetically distant (based on 16S-rDNA sequences) neighboring organisms. Normalized peptide/protein observation ratios were calculated by dividing the number of peptides/proteins identified (not observation count) for a particular organism when using the neighbor genome sequence into the number of peptides/proteins identified when using its own genome sequence. For example, spectra obtained for Shewanella sp. MR-7 that were searched against the Shewanella sp. MR-7 genome sequence yielded 4594 peptides. A search of the same spectra against the genome of near neighbor Shewanella oneidensis MR-1 yielded 3067 peptide identifications for a normalized peptide observation ratio of 0.67 (3067/4594). The normalized peptide ratios were plotted against evolutionary distances determined by CLUSTAL W [30], [31] (Table S1) and 16S rDNA (Figure 1) to examine the extent to which the genomic sequence of one organism can be used to identify proteins in another. Plots of the number of peptide (Figure S1) and protein (Figure S2) observations prior to normalization versus neighbor organism evolutionary distance also were generated for comparison.
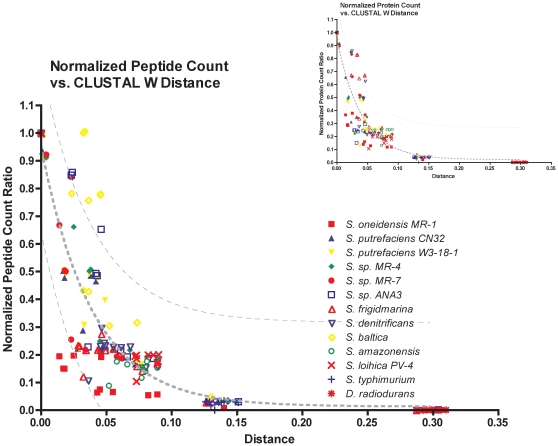
Figure 1. Peptide conservation (inset: protein conservation) was examined across the different species by graphing the normalized number of observed peptides (proteins) with respect to evolutionary distance.
As the distance increases, the number of successfully identified features decreases. Data were fit to a one-phase exponential decay; 95% confidence interval for the observed features is shown with a hashed line. Each point represents the reference proteome peptide count relative to the near neighbor peptide count.
Figure 1 shows that the numbers of observed peptides decrease as the evolutionary distance between an organism and its neighbors increases. The most rapid decrease appears between evolutionary distances of 0 to 0.05. This trend also is conserved across all organisms at the protein level (Figure 1 inset). Note that S. putrefaciens CN32 appears most closely related to S. oneidensis MR-1 (evolutionary distance of 0.016) and shares 4394 peptides observed in common. At approximate mid evolutionary distance (0.038), S. frigidmarina NCIMB400 shares only 1302 peptides with S. oneidensis. Between the two most genetically distant Shewanellae, i.e., S. oneidensis MR-1 and S. amazonensis SB2B (relative evolutionary distance 0.089), the number of peptides observed in common is 575, which means only 6% of the S. oneidensis MR-1 peptides are identified when searching S. oneidensis MR-1 spectra against the neighbor S. amazonensis SB2B genome. Furthermore, only 94 (0.9%) Shewanella peptides are identified when the S. Typhimurium LT2 (considered an outlier at an evolutionary distance of 0.11) genome is used to search the Shewanella spectra. Doubling the evolutionary distance to 0.299 (D. radiodurans R1) further decreases the number of identifications to a single peptide, i.e., insufficient peptide sequences for protein identification at these evolutionary distances (Figure 1 inset).
Comparison of protein functions assigned to observed orthologs
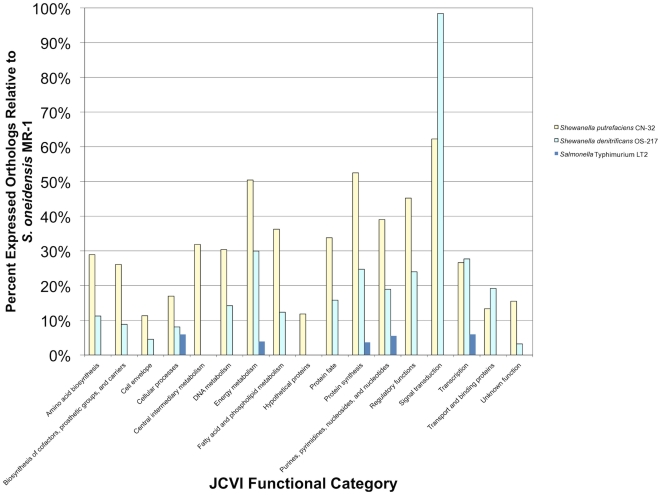
Using the proteins identified from searching the S. oneidensis MR-1 spectra against the genomes of S. putrefaciens CN32, S. denitrificans OS217, and S. Typhimurium LT2, orthologs were mapped to functional categories to determine the level of conservation of protein function among the organisms. The latter three organisms represent near, mid, and remote evolutionary distances relative to S. oneidensis MR-1. Figure 2 attests to the genetic similarity between S. oneidensis MR-1 and S. putrefaciens CN32 relative to the similarity between S. oneidensis MR-1 and the other two organisms. Note that 50% of orthologs within energy metabolism and protein synthesis functional categories were observed when S. oneidensis MR-1 spectra were searched against the S. putrefaciens CN32 genome sequence. After searching S. oneidensis MR-1 spectra against the mid distant neighbor S. denitrificans OS217 genome sequence, only 30% of orthologs were observed in the energy metabolism category and only 25% were observed in the protein synthesis category. When the S. Typhimurium LT2 genome sequence was used to identify peptides from S. oneidensis MR-1, only 15% of the total orthologs (not within a specific JCVI functional category) were observed. This low percentage of observed orthologs is due to the lack of genomic or proteomic sequence homology between the two organisms and highlights the fact that for a surrogate genome to be used for peptide/protein identification, the two organisms must be phylogentically close. For instance, MS/MS spectra may have been generated for a peptide in S. oneidensis that comes from an ortholog between S. oneidensis and S. Typhimurium, yet a lack of sequence conservation for this peptide explains why that MS/MS spectra was not conserved between the two organisms. Similarly, a high percentage of observed orthologs may occur between organisms with few predicted orthologs. For instance, between S. denitrificans and S. oneidensis, 31 predicted orthologous proteins fall within the signal transduction category, whereas S. putrefaciens has 49 predicted S. oneidensis orthologs. Because of the lower number of predicted orthologs between S. oneidensis and S. denitrificans, within this functional category the observed result appears somewhat anomalous.
Figure 2. Conservation of functional orthologs across four of the species in the study is displayed, using normalized protein observations.
Normalized protein observations were derived by dividing observed proteins for a single species within a category by the total number of proteins observed in the study. S. oneidensis orthologous groups from the JCVI Comprehensive Microbial Resource were employed to examine conservation of function.
Application to environmental Shewanella isolates
Following proof of concept, we applied the empirical strategy for characterizing bacterial proteomes across species in the absence of genomic sequences to identify peptides and proteins in four environmental Shewanella isolates from the Hanford Reach region of the Columbia River in Washington state. Although these isolates lacked sequenced genomes, two have 16S ribosomal DNA sequences indicative of phylogenetic affiliation with S. oneidensis MR-1, and two others have 16S ribosomal sequences indicative of an affiliation with S. putrefaciens CN32 [27] (Table S1). LC-MS/MS spectra were obtained for the four isolates, which were then systematically searched against the genome sequence of each Shewanella to identify proteins. The four isolates (HRCR-1, -2, -4 and -5) were cultured under the same conditions used in previous studies performed with sequenced Shewanella to allow for comparison of proteomes.
Extent of proteome information available for the isolates
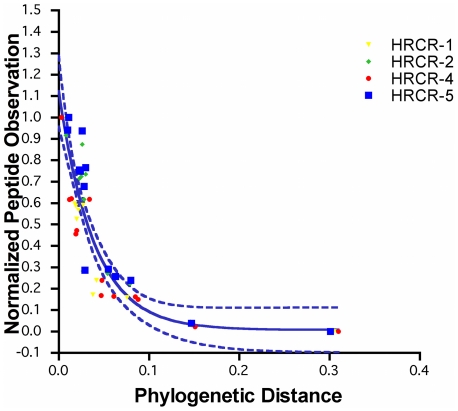
The number of peptides identified from each isolate was normalized to the number of near neighbor peptide identifications for each Shewanella and plotted against the neighbor evolutionary distance (Figure 3). Note that the resulting normalized data exhibit a sigmoidal regression line similar to the trans-organism comparison performed using Shewanellae with sequenced genomes, and peptide data points fall within the 95% prediction index. These results suggest that for these unsequenced Shewanella isolates, the sigmoidal regression curve can be used to predict the extent to which proteome information can be obtained from a sequenced near neighbor organism.
Figure 3. The number of peptides observed from the Columbia River Shewanella isolates graphed against evolutionary distance.
The resulting trend agrees with the trend observed from characterized Shewanella species.
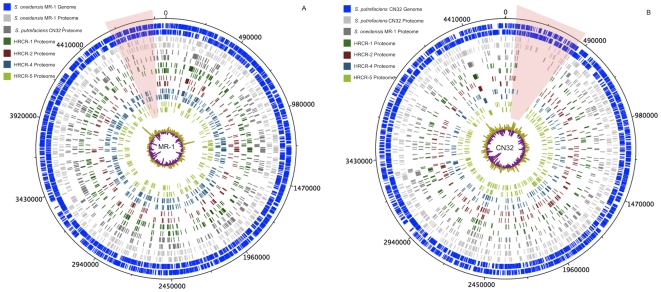
The greatest number of proteins for the environmental isolates was observed when the genome sequences of either S. oneidensis MR-1 or S. putrefaciens CN32, i.e., the nearest phylogenetic neighbors of the isolates were utilized for protein identification. The extent of proteome similarity was revealed when proteins from the isolates were mapped to the genomes of S. oneidensis MR-1 and S. putrefaciens CN32 (Figure 4). Isolates HRCR-1 (457 proteins) and HRCR-4 (534 proteins) were observed most similar to the proteome of S. oneidensis, whereas the proteomes of HRCR-2 (276 proteins) and HRCR-5 (301 proteins) most similar to the proteome of S. putrefaciens (Table 1).
Figure 4. Protein identifications from the Columbia River isolates are mapped to the reference genomes S. oneidensis MR-1 (panel A) and S. putrefaciens CN32 (B).
While all organisms were grown under the same conditions, observation of no protein expression compared to the reference proteome reveals that these organisms have undergone evolutionary divergence, which is reflected in protein expression. Also shown are the protein identifications for each of the Shewanella species mapped onto their respective genomes, as well as the protein orthologs across species. Two regions of ‘missing’ proteome information from the Hanford Reach isolates are highlighted.
Table 1. Conservation of peptides amongst Shewanella isolates from the Hanford Reach of the Columbia River.
| Closest Relative | Furthest Relative | |||||
| Isolate | Strain | CLUSTAL distance | Conserved Peptides/Proteins | Strain | CLUSTAL distance | Conserved Peptides/Proteins |
| HRCR-1 | S. oneidensis MR-1 | 0.009 | 2302/457 | S. loihica PV-4 | 0.087 | 344/72 |
| HRCR-2 | S. putrefaciens CN32 | 0.008 | 1268/276 | S. loihica PV-4 | 0.079 | 310/66 |
| HRCR-4 | S. oneidensis MR-1 | 0.003 | 2504/534 | S. loihica PV-4 | 0.088 | 378/81 |
| HRCR-5 | S. putrefaciens CN32 | 0.010 | 1407/301 | S. loihica PV-4 | 0.080 | 357/78 |
In Figure 4, the proteins mapped to the S. oneidensis MR-1 and S. putrefaciens CN32 genome sequences show distinct regions where proteins from the isolates were either lacking or not observed (Table S2 and Table S3). Figure 4A highlights a representative slice from the S. oneidensis genome in which no proteins were observed for HRCR 2, while Figure 4B shows that no proteins from any of the isolates were observed over a 30,000 base pair region (274 genes). In both maps, gene GC content and protein hydrophobicity (plotted in the center of Figure 4A and B) provided no insight into why these proteins were not observed.
Within the shaded region of the map in Figure 4A (proteins mapped to the S. oneidensis MR-1 genome) are genes that have predicted functions for formate metabolism, including formate dehydrogenase (locus tags SO4507–SO4515), as well as cytochrome c oxidase (SO4606–SO4609). S. oneidensis MR-1 contains two described determinants encoding metal efflux proteins, i.e., the Czc heavy metal and the Cus copper/silver efflux families. Although within the general region of the genome, metal efflux proteins were not observed in any of the isolates. Previous studies have demonstrated tight regulatory control of copper response elements in both Shewanella and other Gram-negative bacteria[32], [33]. Proteins responding to copper stress are only observed under stress-inducing growth conditions. Members of the Czc family of proteins are less well characterized, but also appear to be regulated as tightly as the Cus efflux protein family [34]. The shaded region in Figure 4B (proteins mapped to the S. putrefaciens CN32 genome) also contains several genes that encode proteins associated with formate metabolism and metal efflux protein families. Other proteins in this region are linked to fumarate metabolism and an additional two proteins contain putative 4Fe-4S ferredoxin iron-sulfur binding domains (locus tags CN32_0332, CN32_0336).
The absence of observed proteins in these regions could be due to ecoparalogy, where nucleotide substitutions in genes lead to differential regulation under the influence of a mutant regulator [35]. Ecoparalogy can result in an underestimation of the amount of protein information available when using a near neighbor organism genome sequence. Another plausible explanation for the absence of observed proteins in these regions may be linked to the growth of the organisms under study in highly aerated, rich growth medium. It is possible that a low nutrient, defined minimal medium may be more representative of the environment (i.e., Columbia River water/sediments) from which these bacteria were isolated. Growth of the Columbia River isolates under different nutritional conditions may result in a different complement of proteins expressed by the isolates, allowing investigation of alternate pathways, regulation, and protein expression within these regions. Alternatively, the lack of proteins in this region may simply be due to the absence of genes encoding these proteins in the isolate strains.
Proteome characterization of the isolates
Shewanellae are capable of using a vast respiratory network to reduce various organic and non-organic electron acceptors[1]. The utilization of a wide array of electron acceptors can be attributed to a large number of c-type cytochromes [1], [36], which have been shown to function as terminal reductases of metals [37], [38], [39] and radionuclides [40]. Within S. oneidensis MR-1 there are 42 putative c-type cytochromes that are expressed under a variety of conditions [41]. Under the nutrient rich, aerobic growth conditions used for this experiment, nine of the predicted c-type cytochromes were observed from the S. oneidensis MR-1 cultures (SO0970, SO1127, SO1778, SO1779, SO2178, SO2361, SO2363, SO2785, SO3420, SO4048, SO4666), while only two were detected in the S. putrefaciens CN32 cultures (CN32_0905, CN32_1958) (Table 2). The tetraheme cytochrome, fumarate reductase (SO0970 and CN32_0905) was observed in all isolates, suggesting that these isolates should be capable of fumarate respiration [42].
Table 2. Shewanella isolates were identified from the Columbia River, based on 16S rDNA sequencing.
| Number of identified peptides | ||||||||||
| S. oneidensis genome comparison | S. putrefaciens genome comparison | |||||||||
| MR-1 locus | CN32 locus | HRCR-1 | HRCR-2 | HRCR-4 | HRCR-5 | HRCR-1 | HRCR-2 | HRCR-4 | HRCR-5 | |
| SO0970 | CN32_0905 | 13 | 8 | 11 | 8 | 7 | 12 | 6 | 9 | |
| SO1777 | CN32_1477 | |||||||||
| SO1778 | CN32_1478 | 11 | 3 | 8 | 4 | |||||
| SO1779 | CN32_1479 | 5 | ||||||||
| SO2178 | CN32_2287 | 2 | 2 | |||||||
| SO2361 | CN32_1956 | 6 | 4 | 2 | 2 | 3 | ||||
| SO2363 | CN32_1958 | 3 | 5 | 3 | 5 | |||||
| SO3420 | CN32_2738 | 4 | 3 | 4 | 3 | |||||
| SO3980 | CN32_0685 | 3 | 2 | 2 | ||||||
| SO4666 | CN32_3908 | 6 | ||||||||
Peptides from these isolates were compared to 11 sequenced Shewanella genomes, with the result that the most peptides were identified when S. oneidensis MR-1 or S. putrefaciens CN32 was used as the reference genome. Data below shows the number of identified peptides from the c-type cytochromes using either S. oneidensis MR -1 or S. putrefaciens CN32 genomes.
Two other cytochromes (SO1778 and SO3420) were identified in all isolates when the S. oneidensis genome was employed for protein identification (Table 2). SO1778 is a decaheme cytochrome c, MtrC (OmcB) that has been implicated in metal and radionuclide reduction by S. oneidensis MR-1 [43], [44], [45], [46], [47]. In both S. oneidensis MR-1 and S. putrefaciens CN32, omcB is part of a metal reductase-containting locus that is typically co-expressed with omcA (SO1779), mtrA (SO1777) and mtrB (SO1776) [1], [36], so it is surprising that an OmcA homolog was observed in just one of the isolates, i.e., HRCR-4 (Table 2). A plausible explanation is that these cytochromes were not observed because of the high variability of mass spectrometry based proteomics. The second cytochrome detected, SO3420 is a cytochrome c' with little functional characterization and previously predicted to be a cytochrome solely through comparative genomic studies [48], [49].
Shewanellae's promiscuity for terminal electron acceptors is matched by a variety of pathways available for assimilating carbon beyond central metabolism [50]. For example, lactate is a common carbon and energy substrate for Shewanella that is oxidized completely under aerobic conditions and oxidized incompletely to acetate under anaerobic growth conditions. Similar to 2-oxoglutarate, the enzyme lactate dehydrogenase (dld; SO0968) was only observed in HRCR-1 and HRCR-4 when the S. oneidensis MR-1 genome sequence was used to identify proteins in the isolates. When the S. putrefaciens CN32 genome sequence was used, only the lactate dehydrogenase in HRCR-5 was observed. Pinchuk, et al. demonstrated the presence of an alternative lactate utilization pathway in S. oneidensis MR-1 [51], and we observed protein components (LldF, SO1519 and Lld-II, SO1521) of this second pathway in all isolates. While orthologs featuring similar topology for this second lactate utilization pathway exist in S. putrefaciens CN32, we only observed these orthologs in HRCR-1, -2, and -5, with HRCR-1 exhibiting two of the three proteins from L-lactate dehydrogenase and the entire D-lactate dehydrogenase. Differential observation of the components of these two lactate pathways across the proteomes is likely due to sequence divergence between S. oneidensis MR-1 and S. putrefaciens CN32.
Characterization of proteins associated with the glycolytic and TCA metabolic pathways in the isolates revealed little difference in the number of observed proteins within these pathways, regardless of the Shewanella genome sequence used for identification (Table 3). For example, with the exception of a few proteins, representation of glycolysis and the TCA cycle was complete, which implies that the proteins making up these pathways are part of the core proteome [52] associated with Shewanella. The exception encompassed four proteins in the glycolytic pathways (SO2486–SO2489 or CN32_1866- CN32_1869) involved in the conversion of glucose-6-phosphate to glyceraldehyde-3-phosphate (the pentose phosphate pathway). Across all of the environmental Shewanella isolates, only one enzyme in the pentose phosphate pathway, phosphogluconate dehydratase (Edd, SO2487 and CN32_1868) was observed. When the S. oneidensis MR-1 genome sequence was used to identify proteins expressed by the isolates, phosphogluconate dehydratase was observed in those strains that were more closely related to S. oneidensis MR-1, i.e., HRCR-1 and HRCR-4. This pattern was retained when the S. putrefaciens CN32 genome sequence was used for protein identification, i.e., phosphogluconate dehydratase was only observed in HRCR-2 and HRCR-5, which are the two strains most similar to S. putrefaciens CN32 (Table 4).
Table 3. S. oneidensis MR-1 peptide fragmentation patterns where mapped to theoretical spectra from organisms representing near, mid, and distant phylogenetic neighbors.
| Unique Peptides Identified | |||||
| Locus | Gene | S. oneidensis MR-1 | S. putrefaciens CN32 | S. denitrificans OS217 | S. Typhimurium |
| Glycolysis/Entner-Doudoroff Pathway | |||||
| SO0049 | gpmA | 5 | |||
| SO0932 | pgk | 8 | 2 | ||
| SO0933 | fba | 10 | 4 | 4 | |
| SO1200 | tpiA | 3 | |||
| SO2345 | gapA-2 | 7 | 3 | ||
| SO2347 | gapA-3 | 4 | |||
| SO2486 | eda | 3 | 2 | ||
| SO2487 | edd | 5 | |||
| SO2488 | pgl | 2 | |||
| SO2489 | zwf | 4 | |||
| SO2491 | pykA | 10 | 4 | ||
| SO2644 | ppsA | 15 | 6 | 3 | |
| SO3440 | eno | 8 | 3 | 2 | 2 |
| SO3547 | pgi | 3 | |||
| SO3991 | fbp | 4 | 3 | 2 | |
| TCA Cycle | |||||
| SO0343 | acnA | 5 | 3 | ||
| SO0344 | prpC | 4 | |||
| SO0432 | acnB | 30 | 10 | 3 | |
| SO0770 | mdh | 10 | 7 | 5 | 2 |
| SO0970 | SO0970 | 11 | 4 | ||
| SO1484 | aceA | 2 | |||
| SO1926 | gltA | 6 | 4 | 2 | |
| SO1928 | sdhA | 5 | 4 | 2 | |
| SO1929 | sdhB | ||||
| SO1930 | sucA | ||||
| SO1931 | sucB | 9 | |||
| SO1932 | sucC | 5 | 2 | ||
| SO1933 | sucD | 6 | 4 | ||
| SO2222 | SO2222 | 12 | 2 | ||
| SO2629 | icd | 50 | 12 | 4 | |
| SO4118 | SO4118 | 4 | |||
Data represented here are from highly conserved central metabolic pathways, as unique peptide count – the number of peptides identified belonging to a protein with an ortholog in the S. oneidensis genome.
Table 4. Shewanella isolates were identified from the Columbia River, based on 16S rDNA sequencing.
| Number of identified peptides | |||||||||||
| S. oneidensis genome comparison | S. putrefaciens genome comparison | ||||||||||
| MR-1 locus | CN32 locus | Gene | HRCR-1 | HRCR-2 | HRCR-4 | HRCR-5 | HRCR-1 | HRCR-2 | HRCR-4 | HRCR-5 | |
| Glycolysis/Entner-Doudoroff Pathway | |||||||||||
| SO0049 | CN32_0040 | gpmA | 3 | 2 | |||||||
| SO0932 | CN32_0874 | pgk | 7 | 5 | 7 | 5 | 5 | 3 | 4 | 4 | |
| SO0933 | CN32_0875 | fba | 6 | 6 | 5 | 6 | 4 | 5 | 4 | 5 | |
| SO1200 | CN32_2838 | tpiA | 3 | 3 | 4 | 3 | 3 | 3 | |||
| SO2345 | CN32_1889 | gapA-2 | 4 | 3 | 2 | 3 | 3 | 3 | 2 | 2 | |
| SO2347 | CN32_1891 | gapA-3 | 6 | 3 | 5 | 3 | 3 | 2 | 3 | 4 | |
| SO2486 | CN32_1869 | eda | |||||||||
| SO2487 | CN32_1868 | edd | 2 | 2 | 2 | 3 | |||||
| SO2488 | CN32_1867 | pgl | |||||||||
| SO2489 | CN32_1866 | zwf | |||||||||
| SO2491 | CN32_1864 | pykA | 3 | 8 | 4 | 6 | 5 | 3 | 5 | ||
| SO2644 | CN32_2243 | ppsA | 11 | 7 | 9 | 8 | 8 | 4 | 5 | 3 | |
| SO3440 | CN32_2757 | eno | 6 | 7 | 6 | 7 | 3 | 6 | 2 | 5 | |
| SO3547 | CN32_1048 | pgi | 2 | 2 | |||||||
| SO3991 | CN32_0676 | fbp | 5 | 3 | 5 | 4 | 4 | 3 | 3 | 4 | |
| TCA Cycle | |||||||||||
| SO0343 | CN32_3646 | acnA | 4 | 4 | 5 | 3 | 4 | 4 | 4 | 5 | |
| SO0344 | CN32_3645 | prpC | 2 | 2 | 3 | 3 | 2 | 3 | 2 | 3 | |
| SO0432 | CN32_3409 | acnB | 19 | 13 | 19 | 12 | 14 | 12 | 11 | 12 | |
| SO0770 | CN32_3219 | mdh | 6 | 5 | 7 | 5 | 5 | 4 | 6 | 3 | |
| SO0970 | CN32_0905 | SO0970 | 13 | 8 | 11 | 8 | 7 | 12 | 6 | 9 | |
| SO1484 | CN32_1239 | aceA | 3 | 2 | 2 | 2 | 2 | 2 | |||
| SO1926 | CN32_2274 | gltA | 4 | 4 | 5 | 6 | 5 | 3 | 3 | 4 | |
| SO1928 | CN32_2271 | sdhA | 7 | 5 | 3 | 3 | 5 | 5 | 2 | 2 | |
| SO1929 | CN32_2270 | sdhB | |||||||||
| SO1930 | CN32_2269 | sucA | 6 | 4 | 9 | 4 | 2 | 4 | 4 | 4 | |
| SO1931 | CN32_2268 | sucB | 4 | 4 | 6 | 4 | 4 | 3 | 3 | 4 | |
| SO1932 | CN32_2267 | sucC | 3 | 2 | 4 | 3 | 2 | 2 | 2 | ||
| SO1933 | CN32_2266 | sucD | 4 | 2 | 4 | 2 | 4 | 2 | 4 | ||
| SO2222 | CN32_1807 | SO2222 | 7 | 6 | 5 | 5 | 5 | 8 | 6 | 6 | |
| SO2629 | CN32_2230 | icd | 15 | 5 | 12 | 10 | 8 | 7 | 6 | 11 | |
| SO4118 | CN32_0554 | SO4118 | 2 | 2 | |||||||
Peptides from these isolates were compared to 11 sequenced Shewanella genomes, with the result that the most peptides were identified when S. oneidensis MR-1 or S. putrefaciens CN32 was used as the reference genome. Data below shows the number of identified peptides from central metabolic pathways using either S. oneidensis MR -1 or S. putrefaciens CN32 genomes.
A high percentage of the TCA cycle proteins were observed in all isolates (Table 3). For example, 2-oxoglutarate dehydrogenase, a member of a three-enzyme complex that converts alpha-glutarate to succinyl-CoA was observed in each of the isolates, but not observed in the proteomes of either S. oneidensis MR-1 or S. putrefaciens CN32. Observation of this protein in the isolates and the concomitant lack of observation in S. oneidensis MR-1 and S. putrefaciens CN32 may be due to a difference in growth stage or regulatory control, causing 2-oxoglutarate dehydrogenase to be present in greater abundance in the environmental Shewanella isolates.
We demonstrated a strategy for selecting and utilizing near neighbor organism genome sequences that enabled proteomics characterization of unsequenced environmental isolates lacking sequenced genomes. In spite of the fact that rapid microbial bacterial genome sequencing is becoming increasingly affordable, it is not yet practical to generate whole genome sequences for all organisms isolated from a complex environmental sample nor may it be warranted.
The proof of concept portion of this study revealed that the largest number of peptide identifications for an organism resulted when the evolutionary distance of the sequenced neighbor fell within 0–0.046, after which the extent of proteome characterization derived from a near neighbor genome decreased as evolutionary distance increased. Application of the strategy to characterize Columbia River Shewanella isolates revealed that the Shewanella were genetically related to either S. oneidensis MR-1 or Shewanella putrefaciens CN32. In the absence of whole genome sequences for these isolates, application of the strategy also resulted in the identification of 300–500 proteins, which represents the first proteome characterization of these isolates beyond partial 16S rDNA sequencing. As demonstrated here, there is a limit to how close a near-neighbor genome needs to be in order to make meaningful protein identification, within confidence limits. However, the proteome information generated provided a starting point for elucidating underlying metabolic networks that define adaptation to different environments and ultimately speciation [53], [54], [55]. Tandem mass spectrometry data for the isolates is available through the Biological MS Data and Software Distribution Center website at http://omics.pnl.gov.
With the careful application of error-tolerant search methodologies, such as de novo peptide sequencing, or the USTags approach [8], additional identifications of orthologous proteins that contain sequence polymorphisms may result. Additionally, the generation of high-resolution tandem mass spectra may improve quality and confidence scores associated with spectral matching and de novo tools, resulting in a larger number of proteins identified (see citations [56], [57]for reviews).
Materials and Methods
Bacterial growth conditions
In earlier studies, Shewanella sp. samples analyzed using LC-MS/MS to generate peptide reference databases for the Shewanella Federation were grown aerobically in tryptic soy broth without dextrose (BD Diagnostics, Sparks, MD, USA) at 30°C with shaking at 200 rpm to an OD600 ∼0.5. In other earlier studies, Salmonella serovar Typhimurium strain LT2 was grown in Luria-Bertani broth [58] at 37°C and Deinococcus radiodurans R1, in TGY medium at 30°C. Cells were harvested by centrifugation (8000× g for 10 min at 4°C), flash frozen in liquid nitrogen, and then stored at –80°C until processing. Environmental Shewanella isolates were obtained from samples of the water-sediment interface in the Hanford Reach region of the Columbia River near Richland, Washington [27].
Proteins were prepared as outlined in Lipton, et al. [59]. In brief, cells were lysed by bead beating in 100 mM NH4HCO3 buffer (pH ∼8). Proteins were eluted and denatured with 7M urea, 2M thiourea, and 5 mM DTT at 60°C for 30 min. For soluble and insoluble analyses, cell pellets were treated as above, and the lysate was centrifuged. The supernatant (soluble preparation) was transferred to a fresh tube, and the remaining pellet was resuspended in 7M urea, 2M thiourea, 1% CHAPS in 50 mM NH4HCO3, and 5 mM DTT at 60°C for 30 min (insoluble preparation). For all analyses, the denatured proteins were diluted with buffer to reduce the salt concentration and digested with trypsin for 3 h at 37°C. Cleanup was performed by passing the samples through a C18 SPE column [60]. The sample solutions were concentrated in a speed-vac to a final volume of ∼50–100 µL, quick frozen in liquid nitrogen, and stored at –80°C until needed for analysis.
Samples were fractionated by strong cation exchange chromatography [59]. Approximately 25 fractions were collected from each sample, and each fraction was dried under vacuum and then dissolved in 30 µL of 25 mM NH4HCO3. Aliquots containing 10 µg of protein were analyzed by LC-MS/MS, using an LTQ ion trap mass spectrometer (ThermoFisher Scientific Corp., San Jose, CA) and previously defined parameters [61].
Peptide/protein identification using a trans-organism search strategy
The X!Tandem algorithm [28], [29] was employed to match MS/MS spectra with predicted tryptic peptides from a protein file. Our search strategy allowed for partial tryptic peptides to pass the first round of searching by X!Tandem. The scores produced by X!Tandem are probability-based scores similar to the E-value or bit score from BLAST. Genomic sequences for each bacterial species were obtained from publicly available databases.
Spectra for each of the bacterial samples were systematically searched relative to the translated genome sequences of all species to identify common peptides. Salmonella and Deinococcus were included as outliers, similar to the inclusion of distantly related organisms when constructing and calculating confidence of genetic trees [62], [63]. A total of 4261 X!Tandem searches were performed using the PRISM computing cluster (260 days of CPU time across 32 processing nodes) [64]. Percentages of observed orthologs were calculated as the number of orthologs observed from S. oneidensis MR-1 spectra when searched using one of the three neighboring genome sequences divided by the number of orthologs observed from the same S. oneidensis MR-1 spectra when searched against its own genome sequence.
Data analysis of X!Tandem results
For the Shewanella species in this study, distribution of X!Tandem log-transformed E-values was divided into five intervals. Intervals represented the 10th (Interval A; all values ≤−8.3), 25th (−8.3> Interval B≤−6.1), 50th (−6.1 > Interval C≤−3.7), 75th (−3.7> Interval D ≤−1.6) and 90th (Interval E; all values >−1.6) percentiles. Only fully tryptic peptides having a minimum amino acid length of 6 residues and a log E-value ≤−8.3 were used in this evaluation. A protein was considered positively observed when identified by at least two unique peptides.
Regression analysis was performed in GraphPad Prism (GraphPad Software, Inc., La Jolla, CA). Several nonlinear regression models were tested, including exponential decay and polynomial association. The simplest model with the largest R2 value and most significant F-test was selected as the model with the best fit (in this case a two sigmoidal dose-response model).
A 95% prediction index rather than 95% confidence interval was calculated using GraphPad Prism (GraphPad Software, Inc.). This index was used to predict the next Y value for a given X, which in this case was the number of peptides/proteins for a specified evolutionary distance from a neighboring strain or species. Unlike confidence intervals obtained for replicate data, a prediction interval was used in cases where there was only a single observation of Y. Because the uncertainty of each peptide identification was unknown, all observations were given the same weight.
Near neighbor evolutionary distance calculation
Because of the small amount of sequence data available for each isolate, a partial 16S rDNA sequence that represented the 5′ end of the 16S rDNA gene (850 bp) was used to generate the CLUSTAL W genetic distance matrix. Sequence alignment was accomplished using the CLUSTAL W alignment algorithm accessed from the San Diego Supercomputing Center [65], [66]. Near neighbor evolutionary distances were reported as CLUSTAL W distances.
Supporting Information
Sequences for 16S rRNA were used for determination of evolutionary distance between Shewanella strains and the outlier species, Salmonella Typhimurium LT2 and Deinococcus radiodurans R1. Distance calculations were carried out using CLUSTAL, hosted at the San Diego Supercomputer Center Biology Workbench (http://workbench.sdsc.edu/). Values are CLUSTAL distances.
(0.07 MB DOC)
S. oneidensis MR-1 loci with poor proteome coverage from analysis with the Columbia River Shewanella isolates. ND indicates Not Detected, P indicates Present.
(0.57 MB DOC)
S. putrefaciens CN32 loci with poor proteome coverage from analysis with the Columbia River Shewanella isolates. ND indicates Not Detected, P indicates Present.
(0.47 MB DOC)
Plot of the number of peptide observations prior to normalization versus neighbor organism evolutionary distance.
(0.11 MB TIF)
Plot of the number of protein observations prior to normalization versus neighbor organism evolutionary distance.
(0.10 MB TIF)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Department of Energy Office of Biological and Environmental Research (DOE/BER; ER63232-1018220-0007203), the National Institute of Allergy and Infectious Diseases (NIH/DHHS through interagency agreement Y1-AI-4894-01), and the NIH National Center for Research Resources (RR18522). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lo I, Denef VJ, Verberkmoes NC, Shah MB, Goltsman D, et al. Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature. 2007;446:537–541. doi: 10.1038/nature05624. [DOI] [PubMed] [Google Scholar]
- 2.Shevchenko A, Sunyaev S, Loboda A, Bork P, Ens W, et al. Charting the proteomes of organisms with unsequenced genomes by MALDI-quadrupole time-of-flight mass spectrometry and blast homology searching. Anal Chem. 2001;73:1917–1926. doi: 10.1021/ac0013709. [DOI] [PubMed] [Google Scholar]
- 3.Den Hartigh A, Sun Y, Sondervan D, Heuvelmans N, Reinders M, et al. Differential requirements for virB1 and virB2 during Brucella abortus infection. Infect Immun. 2004;72:5143–5149. doi: 10.1128/IAI.72.9.5143-5149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Habermann B, Oegema J, Sunyaev S, Shevchenko A. The power and the limitations of cross-species protein identification by mass spectrometry-driven sequence similarity searches. Mol Cell Proteomics. 2004;3:238–249. doi: 10.1074/mcp.M300073-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Craig R, Cortens JC, Fenyo D, Beavis RC. Using annotated peptide mass spectrum libraries for protein identification. J Proteome Res. 2006;5:1843–1849. doi: 10.1021/pr0602085. [DOI] [PubMed] [Google Scholar]
- 6.Shen Y, Tolić N, Hixson KK, Purvine SO, Paša-Tolić L, et al. Proteome-wide identification of proteins and their modifications with decreased ambiguities and improved false discovery rates using unique sequence tags. Anal Chem. 2008;80:1871–1882. doi: 10.1021/ac702328x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnesen T, Van Damme P, Polevoda B, Helsens K, Evjenth R, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci U S A. 2009;106:8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han SJ, Hu J, Pierce B, Weng Z, Renne R. Mutational analysis of the latency-associated nuclear antigen DNA binding domain of Kaposi's sarcoma-associated herpesvirus reveals structural conservation among {gamma}-herpesvirus origin binding proteins. J Gen Virol. doi: 10.1099/vir.0.020958-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu P, Kenney JM, Stiller JW, Greenleaf AL. Genetic organization, length conservation and evolution of RNA polymerase II carboxyl-terminal domain. Mol Biol Evol. doi: 10.1093/molbev/msq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villafane R, Costa S, Ahmed R, Salgado C. Conservation of the N-terminus of some phage tail proteins. Archives of Virology. 2005;150:2609–2621. doi: 10.1007/s00705-005-0597-7. [DOI] [PubMed] [Google Scholar]
- 11.Maddocks SE, Oyston PC. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 12.Tabb DL, Saraf A, Yates JR., 3rd GutenTag: High-throughput sequence tagging via an empirically derived fragmentation model. Anal Chem. 2003;75:6415–6421. doi: 10.1021/ac0347462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liska AJ, Sunyaev S, Shilov IN, Schaeffer DA, Shevchenko A. Error-tolerant EST database searches by tandem mass spectrometry and MultiTag software. Proteomics. 2005;5:4118–4122. doi: 10.1002/pmic.200401262. [DOI] [PubMed] [Google Scholar]
- 14.Sunyaev S, Liska AJ, Golod A, Shevchenko A. MultiTag: Multiple error-tolerant sequence tag search for the sequence-similarity identification of proteins by mass spectrometry. Anal Chem. 2003;75:1307–1315. doi: 10.1021/ac026199a. [DOI] [PubMed] [Google Scholar]
- 15.Pandhal J, Snijders AP, Wright PC, Biggs CA. A cross-species quantitative proteomic study of salt adaptation in a halotolerant environmental isolate using 15N metabolic labelling. Proteomics. 2008;8:2266–2284. doi: 10.1002/pmic.200700398. [DOI] [PubMed] [Google Scholar]
- 16.Denef VJ, Shah MB, Verberkmoes NC, Hettich RL, Banfield JF. Implications of strain- and species-level sequence divergence for community and isolate shotgun proteomic analysis. J Proteome Res. 2007;6:3152–3161. doi: 10.1021/pr0701005. [DOI] [PubMed] [Google Scholar]
- 17.Saini G, Wood BD. Metabolic uncoupling of Shewanella oneidensis MR-1, under the influence of excess-substrate and 3, 3′, 4′, 5 tetrachlorosalicylanilide (TCS). Biotechnol Bioeng. 2007 doi: 10.1002/bit.21702. [DOI] [PubMed] [Google Scholar]
- 18.Mertens B, Blothe C, Windey K, De Windt W, Verstraete W. Biocatalytic dechlorination of lindane by nano-scale particles of Pd(0) deposited on Shewanella oneidensis. Chemosphere. 2007;66:99–105. doi: 10.1016/j.chemosphere.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 19.Guha H, Jayachandran K, Maurrasse F. Microbiological reduction of chromium(VI) in presence of pyrolusite-coated sand by Shewanella alga Simidu ATCC 55627 in laboratory column experiments. Chemosphere. 2003;52:175–183. doi: 10.1016/S0045-6535(03)00104-8. [DOI] [PubMed] [Google Scholar]
- 20.Dennis PC, Sleep BE, Fulthorpe RR, Liss SN. Phylogenetic analysis of bacterial populations in an anaerobic microbial consortium capable of degrading saturation concentrations of tetrachloroethylene. Can J Microbiol. 2003;49:15–27. doi: 10.1139/w03-008. [DOI] [PubMed] [Google Scholar]
- 21.Nealson KH, Belz A, McKee B. Breathing metals as a way of life: Geobiology in action. Antonie Van Leeuwenhoek. 2002;81:215–222. doi: 10.1023/a:1020518818647. [DOI] [PubMed] [Google Scholar]
- 22.Lipton MS, Paša-Tolić L, Anderson GA, Anderson DJ, Auberry DL, et al. From the cover: Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. PNAS. 2002;99:11049–11054. doi: 10.1073/pnas.172170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferguson PL, Smith RD. Proteome analysis by mass spectrometry. Annu Rev Biophys Biomol Struct. 2003;32:399–424. doi: 10.1146/annurev.biophys.32.110601.141854. [DOI] [PubMed] [Google Scholar]
- 24.Paša-Tolić L, Lipton MS, Masselon CD, Anderson GA, Shen Y, et al. Gene expression profiling using advanced mass spectrometric approaches. J Mass Spectrom. 2002;37:1185–1198. doi: 10.1002/jms.423. [DOI] [PubMed] [Google Scholar]
- 25.Popa R, Mashall MJ, Nguyen H, Tebo BM, Brauer S. Limitations and benefits of arisa intra-genomic diversity fingerprinting. J Microbiol Methods. 2009;78:111–118. doi: 10.1016/j.mimet.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, et al. Towards environmental systems biology of Shewanella. Nat Rev Microbiol. 2008;6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 27.Fredrickson JK, Zachara JM, Kennedy DW, Dong HL, Onstott TC, et al. Biogenic iron mineralization accompanying the dissimilatory reduction of hydrous ferric oxide by a groundwater bacterium. Geochimica Et Cosmochimica Acta. 1998;62:3239–3257. [Google Scholar]
- 28.Craig R, Beavis RC. Tandem: Matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 29.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res. 2004;3:1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 30.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, et al. Multiple sequence alignment with the CLUSTAL series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toes AC, Daleke MH, Kuenen JG, Muyzer G. Expression of copA and cusA in Shewanella during copper stress. Microbiology. 2008;154:2709–2718. doi: 10.1099/mic.0.2008/016857-0. [DOI] [PubMed] [Google Scholar]
- 33.Espariz M, Checa SK, Audero ME, Pontel LB, Soncini FC. Dissecting the Salmonella response to copper. Microbiology. 2007;153:2989–2997. doi: 10.1099/mic.0.2007/006536-0. [DOI] [PubMed] [Google Scholar]
- 34.Grosse C, Grass G, Anton A, Franke S, Santos AN, et al. Transcriptional organization of the czc heavy-metal homeostasis determinant from Alcaligenes eutrophus. J Bacteriol. 1999;181:2385–2393. doi: 10.1128/jb.181.8.2385-2393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez-Perez G, Mira A, Nyiro G, Pasic L, Rodriguez-Valera F. Adapting to environmental changes using specialized paralogs. Trends Genet. 2008;24:154–158. doi: 10.1016/j.tig.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Beliaev AS, Thompson DK, Khare T, Lim H, Brandt CC, et al. Gene and protein expression profiles of Shewanella oneidensis during anaerobic growth with different electron acceptors. OMICS: A Journal of Integrative Biology. 2002;6:39–60. doi: 10.1089/15362310252780834. [DOI] [PubMed] [Google Scholar]
- 37.Reardon CL, Dohnalkova AC, Nachimuthu P, Kennedy DW, Saffarini DA, et al. Role of outer-membrane cytochromes MtrC and OmcA in the biomineralization of ferrihydrite by Shewanella oneidensis MR-1. Geobiology. 8:56–68. doi: 10.1111/j.1472-4669.2009.00226.x. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Liu C, Wang X, Marshall MJ, Zachara JM, et al. Kinetics of reduction of fe(iii) complexes by outer membrane cytochromes MtrC and OmcA of Shewanella oneidensis MR-1. Appl Environ Microbiol. 2008;74:6746–6755. doi: 10.1128/AEM.01454-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beliaev AS, Saffarini DA. Shewanella putrefaciens mtrB encodes an outer membrane protein required for Fe(III) and Mn(IV) reduction. J Bacteriol. 1998;180:6292–6297. doi: 10.1128/jb.180.23.6292-6297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marshall MJ, Plymale AE, Kennedy DW, Shi L, Wang Z, et al. Hydrogenase- and outer membrane c-type cytochrome-facilitated reduction of technetium(VII) by Shewanella oneidensis MR-1. Environ Microbiol. 2008;10:125–136. doi: 10.1111/j.1462-2920.2007.01438.x. [DOI] [PubMed] [Google Scholar]
- 41.Meyer TE, Tsapin AI, Vandenberghe I, de Smet L, Frishman D, et al. Identification of 42 possible cytochrome c genes in the Shewanella oneidensis genome and characterization of six soluble cytochromes. OMICS. 2004;8:57–77. doi: 10.1089/153623104773547499. [DOI] [PubMed] [Google Scholar]
- 42.Maier TM, Myers JM, Myers CR. Identification of the gene encoding the sole physiological fumarate reductase in Shewanella oneidensis MR-1. J Basic Microbiol. 2003;43:312–327. doi: 10.1002/jobm.200390034. [DOI] [PubMed] [Google Scholar]
- 43.Carpentier W, De Smet L, Van Beeumen J, Brige A. Respiration and growth of Shewanella oneidensis MR-1 using vanadate as the sole electron acceptor. J Bacteriol. 2005;187:3293–3301. doi: 10.1128/JB.187.10.3293-3301.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Myers CR, Myers JM. Cell surface exposure of the outer membrane cytochromes of Shewanella oneidensis MR-1. Lett Appl Microbiol. 2003;37:254–258. doi: 10.1046/j.1472-765x.2003.01389.x. [DOI] [PubMed] [Google Scholar]
- 45.Myers JM, Myers CR. Role for outer membrane cytochromes OmcA and OmcB of Shewanella putrefaciens MR-1 in reduction of manganese dioxide. Appl Environ Microbiol. 2001;67:260–269. doi: 10.1128/AEM.67.1.260-269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Myers JM, Myers CR. Overlapping role of the outer membrane cytochromes of Shewanella oneidensis MR-1 in the reduction of manganese(IV) oxide. Lett Appl Microbiol. 2003;37:21–25. doi: 10.1046/j.1472-765x.2003.01338.x. [DOI] [PubMed] [Google Scholar]
- 47.Beliaev AS, Saffarini DA, McLaughlin JL, Hunnicutt D. MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Mol Microbiol. 2001;39:722–730. doi: 10.1046/j.1365-2958.2001.02257.x. [DOI] [PubMed] [Google Scholar]
- 48.Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson WC, et al. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat Biotechnol. 2002;20:1118–1123. doi: 10.1038/nbt749. [DOI] [PubMed] [Google Scholar]
- 49.Daraselia N, Dernovoy D, Tian Y, Borodovsky M, Tatusov R, et al. Reannotation of Shewanella oneidensis genome. OMICS: A Journal of Integrative Biology. 2003;7:171–175. doi: 10.1089/153623103322246566. [DOI] [PubMed] [Google Scholar]
- 50.Serres MH, Riley M. Genomic analysis of carbon source metabolism of Shewanella oneidensis MR-1: Predictions versus experiments. J Bacteriol. 2006;188:4601–4609. doi: 10.1128/JB.01787-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinchuk GE, Rodionov DA, Yang C, Li X, Osterman AL, et al. Genomic reconstruction of Shewanella oneidensis MR-1 metabolism reveals a previously uncharacterized machinery for lactate utilization. Proc Natl Acad Sci U S A. 2009;106:2874–2879. doi: 10.1073/pnas.0806798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Callister SJ, McCue LA, Turse JE, Monroe ME, Auberry KJ, et al. Comparative bacterial proteomics: Analysis of the core genome concept. PLoS ONE. 2008;3:e1542. doi: 10.1371/journal.pone.0001542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chain PS, Comerci DJ, Tolmasky ME, Larimer FW, Malfatti SA, et al. Whole-genome analyses of speciation events in pathogenic Brucellae. Infect Immun. 2005;73:8353–8361. doi: 10.1128/IAI.73.12.8353-8361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jungblut PR, Holzhutter HG, Apweiler R, Schluter H. The speciation of the proteome. Chem Cent J. 2008;2:16. doi: 10.1186/1752-153X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilkins MJ, Verberkmoes NC, Williams KH, Callister SJ, Mouser PJ, et al. Proteogenomic monitoring of Geobacter physiology during stimulated uranium bioremediation. Appl Environ Microbiol. 2009;75:6591–6599. doi: 10.1128/AEM.01064-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hughes C, Ma B, Lajoie GA. De novo sequencing methods in proteomics. Methods Mol Biol. 604:105–121. doi: 10.1007/978-1-60761-444-9_8. [DOI] [PubMed] [Google Scholar]
- 57.Xu C, Ma B. Software for computational peptide identification from MS-MS data. Drug Discov Today. 2006;11:595–600. doi: 10.1016/j.drudis.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 58.Miller JH. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1972. Experiments in molecular genetics. [Google Scholar]
- 59.Lipton MS, Romine MF, Monroe ME, Elias DA, Paša-Tolić L, et al. AMT tag approach to proteomic characterization of Deinococcus radiodurans and Shewanella oneidensis. Methods Biochem Anal. 2006;49:113–134. [PubMed] [Google Scholar]
- 60.Callister SJ, Nicora CD, Zeng X, Roh JH, Dominguez MA, et al. Comparison of aerobic and photosynthetic Rhodobacter sphaeroides 2.4.1 proteomes. J Microbiol Methods. 2006;67:424–436. doi: 10.1016/j.mimet.2006.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masselon C, Paša-Tolić L, Tolić N, Anderson GA, Bogdanov B, et al. Targeted comparative proteomics by liquid chromatography-tandem fourier ion cyclotron resonance mass spectrometry. Anal Chem. 2005;77:400–406. doi: 10.1021/ac049043e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gascuel O, Steel M. Neighbor-joining revealed. Mol Biol Evol. 2006;23:1997–2000. doi: 10.1093/molbev/msl072. [DOI] [PubMed] [Google Scholar]
- 63.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 64.Kiebel GR, Auberry KJ, Jaitly N, Clark DA, Monroe ME, et al. PRISM: A data management system for high-throughput proteomics. Proteomics. 2006 doi: 10.1002/pmic.200500500. [DOI] [PubMed] [Google Scholar]
- 65.Sauro HM, Hucka M, Finney A, Wellock C, Bolouri H, et al. Next generation simulation tools: The systems biology workbench and biospice integration. OMICS. 2003;7:355–372. doi: 10.1089/153623103322637670. [DOI] [PubMed] [Google Scholar]
- 66.Subramaniam S. The biology workbench—a seamless database and analysis environment for the biologist. Proteins. 1998;32:1–2. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences for 16S rRNA were used for determination of evolutionary distance between Shewanella strains and the outlier species, Salmonella Typhimurium LT2 and Deinococcus radiodurans R1. Distance calculations were carried out using CLUSTAL, hosted at the San Diego Supercomputer Center Biology Workbench (http://workbench.sdsc.edu/). Values are CLUSTAL distances.
(0.07 MB DOC)
S. oneidensis MR-1 loci with poor proteome coverage from analysis with the Columbia River Shewanella isolates. ND indicates Not Detected, P indicates Present.
(0.57 MB DOC)
S. putrefaciens CN32 loci with poor proteome coverage from analysis with the Columbia River Shewanella isolates. ND indicates Not Detected, P indicates Present.
(0.47 MB DOC)
Plot of the number of peptide observations prior to normalization versus neighbor organism evolutionary distance.
(0.11 MB TIF)
Plot of the number of protein observations prior to normalization versus neighbor organism evolutionary distance.
(0.10 MB TIF)