Summary
Background
Glanzmann thrombasthenia (GT) is an inherited autosomal recessive platelet disorder characterized by a complete or partial lack, or mutation, of the GPIIb/IIIa complex (integrin αIIbβ3) on the thrombocytes’ surface, leading to a severe bleeding syndrome.
Material and Methods
Molecular genetic analysis was performed in patients with suspected GT. The aim of the present study was the identification of new natural variants, their impact on platelet function, and their relation to the risk of bleeding.
Results
Expression of the platelet integrin αIIbβ3 was determined by flow cytometry. Mutations were identified through sequencing of cDNA and genomic DNA. In addition, platelet function studies (PAC-binding, aggregations) were implemented. The study included 25 patients revealing 13 mutations (GPIIb: n = 9; GPIIIa: n = 4). Two of the 13 mutations were previously described (T207I; L214P). The remaining mutations have not been published yet, whereas 1 mutation in 2 unrelated families was identical (3062 T→C).
Conclusion
All patients with less than 25% of present αIIbβ3 have a medical history of bleeding.
Keywords: Glanzmann thrombasthenia, αIIbβ3, Molecular genetic analysis, Platelet function studies
Zusammenfassung
Hintergrund
Die Thrombasthenie Glanzmann (GT) ist eine angeborene, autosomal rezessiv vererbte Störung der Plättchenfunktion, die durch einen völligen oder partiellen Mangel bzw. eine Mutation des GPIIb/IIIa-Komplexes (Integrin αIIbβ3) auf der Thrombozytenmembran verursacht wird und zu einer schweren Blutungsneigung führt.
Material und Methoden
In der vorliegenden Studie wurden bei Patienten mit Verdacht auf GT molekulargenetische Analysen durchgeführt. Ziel der Studie war die Aufklärung neuer natürlicher Varianten, deren Einfluss auf die Funktionsfähigkeit der Thrombozyten und ihre Relation zum Blutungsrisiko. Die Expression des GPIIb/IIIa-Komplexes auf Thrombozyten wurde mittels Durchflusszytometrie überprüft. Die Identifikation der Mutationen erfolgte über die Sequenzierung von cDNA und genomischer DNA. Weiterhin wurden Plättchenfunktionsuntersuchungen (PAC-Messung, Aggregationen) durchgeführt.
Ergebnisse
Es wurden 25 Patienten untersucht, insgesamt wurden 13 Mutationen identifiziert (GPIIb: n = 9; GPIIIa: n = 4). Zwei dieser Mutationen wurden bisher beschrieben (T207I; L214P). Die übrigen Mutationen waren bisher noch nicht bekannt, wobei eine Mutation bei zwei nicht verwandten Patienten detektiert wurde (3062 T→C).
Schlussfolgerung
Patienten mit einem Rezeptorbesatz unter 25% hatten positive Blutungsanamnesen.
Introduction
Glanzmann thrombasthenia (GT) is a rare, recessively inherited, bleeding syndrome affecting the megakaryocyte lineage [1, 2]. Affected patients exhibit a lifelong moderate to severe bleeding tendency with mainly mucocutaneous bleeding. The thrombasthenic phenotype is linked to quantitative and/ or qualitative abnormalities in the platelet fibrinogen receptor, the αIIbβ3 integrin (glycoprotein (GP) IIb/IIIa, CD41/ CD61), which mediates the incorporation of platelets into an aggregate or thrombus at sites of vessel injury [3]. Laboratory parameters in severe GT show normal platelet count, prolonged bleeding time, absence of platelet aggregation in response to multiple physiologic agonists, and impaired adhesion to siliconized glass. The more common heterozygous GT patients show almost normal or slightly diminished platelet aggregation.
The αIIb (ITGA2B) and β3 (ITGB3) genes are located on chromosome 17 (q21–22). ITGA2B spans 17 kb and comprises 30 exons, ITGB3 spans 46 kb and has 15 exons. The fibrinogen binding site of the receptor complex seems to be a discontinuous surface made up of various regions of both subunits. Fibrinogen binding to activated αIIbβ3 induces further sequential conformational changes in the receptor complex, inducing exposition of cryptic ligand-binding sites with outside-in signal transduction processes and cytoskeletal rearrangement [4]. The exact course of events is not known but various regions in the αIIb and β3 subunits are known to be involved in fibrinogen binding [5,6,7,8,9,10,11,12].
GT is classified into 3 subtypes, depending on the level of present αIIbβ3. Patients with type I or classical GT are homozygous for the disease and have a virtual absence of αIIbβ3 (<5% of normal level). Type II GT patients can have up to 25% of the normal level. In the variant type, αIIbβ3 levels are near normal but functionally impaired, leading to defective binding of fibrinogen [13]. Heterozygotes for type I GT have approximately 50% of the normal level of platelet αIIbβ3, without bleeding problems. Since the cloning of the αIIb and β3 genes, a number of mutations have been found to be associated with GT. A continually updated database is available on the Internet (sinaicentral.mssm.edu/intranet/research/glanzmann); it currently lists 103 records of mutations in the αIIb and 68 records of mutations in the β3 gene. The types of mutations identified in both genes include minor and major deletions, insertions, inversions, and mostly point mutations. The molecular and functional characterization of many of them has provided important information about the biosynthesis and structure-function relationships of the αIIbβ3 complex as well as the biology of other molecules of the integrin family [14, 15].
Most of the documented single nucleotide substitutions are located in the coding sequence and cause missense or nonsense substitutions at the amino acid level, producing either normal-sized non-functional or truncated proteins [16,17,18,19,20].
Splice site defects are also widespread, and mutations that alter mRNA splicing are frequently nonsense mutations [21, 22] or mutations directly affecting the standard consensus splicing signals, and typically lead to skipping of the neighboring exon [23]. By and large, mutations are specific for each family. GT occurs in high frequency in certain ethnic populations with an increased incidence of consanguinity, such as Indians, Iranians, Iraqi Jews, Palestinian and Jordanian Arabs, and French Gypsies [24,25,26,27]. To elucidate the molecular basis of GT, we investigated 25 GT patients for gene mutations within αIIb and β3.
Material and Methods
Study Subjects
25 Patients, belonging to 12 unrelated families, with a diagnosis of GT, and their first-degree relatives if available were the subjects of the study (table 1). The investigated patients had different genetic backgrounds (Caucasian: n = 13; Asian: n = 9; African: n = 3). GT was diagnosed on the basis of clinical and hematologic parameters. The patients’ phenotypes and genotypes were studied to perform carrier studies in their families. Bleeding symptoms were evaluated by examining available hospital records. Mild bleeders were defined as those who had minor symptoms, such as epistaxis or gum bleeds, and moderate bleeders suffered, in addition, from bleeding complications after surgery and trauma. Severe bleeders were defined as those with a history of spontaneous or life-threatening hemorrhages, such as gastrointestinal bleeding, or who had repeated episodes requiring platelet transfusion. Blood samples for the studies described below were taken from the patient after informed consent was obtained. The study was approved by the Ethics Committee of the Landesärztekammer Hessen.
Table 1.
Clinical characteristics and flow cytometry analysis in GT patients
| Patient (sex) | Bleeding severity | Bleeding time, min | CD41 anti-αllb, % | CD42a anti-GPMX, % | CD42b anti-GPIbα, % |
|---|---|---|---|---|---|
| TA (M)+ | severe | 33 | <1 | 99 | 105 |
| TG (M)+ | severe | >30 | <1 | 107 | 111 |
| TN (F)° | mild | 9 | 90 | 109 | 110 |
| TF (M)° | none | 6.5 | 78 | 94 | 93 |
| GE (F) | severe | 21 | <1 | 108 | 87 |
| CM (F)° | mild | 58 | 99 | 93 | |
| SB (F) | moderate | >15 | 23 | 120 | 125 |
| YF (M) | moderate | 3 | n.a. | 73 | |
| TE (M) | severe | <1 | 110 | 122 | |
| GI (F)° | mild | 97 | 102 | 103 | |
| TA (M)° | mild | 90 | 99 | 109 | |
| DZ (F) | 0 | 103 | 110 | ||
| LN (F) | <1 | 98 | 96 | ||
| LH (F)° | 67 | 130 | 129 | ||
| LB (M)° | 82 | 101 | 104 | ||
| HM (M) | moderate | 0 | normal | normal | |
| HS (F)° | n.a. | n.a. | n.a. | ||
| HA (M)° | n.a. | n.a. | n.a. | ||
| OK (F) | severe | >30 | 4 | 101 | 114 |
| MM (M) | severe | <1 | 98 | 97 | |
| MU (F)° | 82 | 94 | 86 | ||
| WC (M) | severe | 0 | 119 | 113 | |
| WV (F)° | 68 | 121 | 113 | ||
| WA (M)° | 78 | 101 | 84 | ||
| PJ (M) | severe | <1 | normal | normal |
F = Female; M = male; n.a. = not available
= siblings
= parents.
Blood Sampling and Processing
Blood was taken in citrated buffer or in ethylenediaminetetraacetic acid (EDTA) from the patients. Citrated platelet-rich plasma (PRP) was obtained by centrifugation at 180 × g for 10 min, platelet-poor plasma (PPP) by centrifugation at 2,500 × g for 20 min. PRP was used for flow cytometry, platelet aggregation, and binding experiments. DNA extraction from white blood cells was carried out in blood samples containing EDTA.
Flow Cytometric Analysis of Platelets
These studies were carried out as recently reported with some modifications [28]. Monoclonal antibodies (mabs) against CD42a (clone SZ1), CD42b (clone SZ2), αIIbβ3 (clone P2), CD36 (clone FAG.152), and mouse IgG isotypes were purchased from Beckman Coulter (Krefeld, Germany). Mab PAC-1, specific for activated αIIbβ3, was obtained from BD Bioscience (Heidelberg, Germany). To obtain information relating to the receptor count, 200 μl of PRP were fixed using 2% formaldehyde in PBS at room temperature for at least 1 h. The cells were sedimented by centrifugation at 600 × g for 3 min and resuspended in 200 μl HEPES buffer (20 mol/l HEPES, 150 mol/l NaCl with 60 mg/ml bovine serum albumin (Sigma-Aldrich, Steinheim, Germany); pH 7.5). Aliquots of platelet suspension were then incubated for 30 min in the dark with appropriate fluorescence-marked monoclonal antibodies. The samples were washed (2,400 rpm, 3 min) using CellWash (BD Bioscience), and resuspended in 400 μl of CellWash. Fluorescence-labeled isotype matched IgG antibodies were used as negative control. Fluorescence intensity was measured with a FACScan flow cytometer and analyzed with CellQuest 3.1 software (BD Bioscience) which allowed the parallel measurement of 3 different fluorescences. Quantum fluorescence microbeads (Calibrite beads; BD Bioscience) were used each day for standardization of the instrument settings. To study platelet GPIIb/IIIa activation and fibrinogen binding by flow cytometry, platelets in PRP were activated with either adenosine diphosphate (ADP, 10 μmol/l) or thrombin receptor-activating peptide (TRAP-6, 10 μmol/l) in the presence of saturating concentrations of FITC-conjugated PAC-1 (directed against activated αIIbβ3) or anti-fibrinogen mab. Following incubation for 15 min at room temperature, platelets were fixed for 15 min, washed with CellWash, and analyzed in a final volume of 400 μl for cell-bound fluorescence in the flow cytometer. The background was measured using FITC-conjugated anti-mouse IgG.
Light Transmission Aggregometry
Light transmission aggregometry (LTA) was performed on the APACT 4S Plus aggregometer (LABiTec, Ahrensburg, Germany). Platelet counts were not adjusted, and the baseline optical density was set with PPP. The used inducers were ADP (final concentrations: 2.5, 5, 10 μmol/l), collagen (0.5, 1, and 2 μg/ml; Horm® Nycomed Austria GmbH, Linz, Austria), epinephrine (10 μmol/l final concentration), and arachidonic acid (0.75 mol/l final concentration). Optical density changes were recorded photoelectrically for 7 min as platelets began to aggregate. The aggregation response is given as percentage of maximal light transmission (Amax).
DNA and RNA Extraction
DNA isolation from whole blood was carried out by using the QIAamp DNA Blood mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Total RNA in platelets was isolated from PRP as previously described [28].
Amplification of Genomic DNA by PCR and Synthesis of cDNA
Amplification of all individual exons of the αIIb and β3 genes, including splicing sites, was performed by using primers located in the flanking intron regions. Table 2 provides information about the length and location of all used primers. According to the different primers, polymerase chain reactions (PCRs) were performed using different annealing temperatures varying from 50 to 62 °C for 30 s to optimize the amplification products. Extension was performed at 72 °C for 60 s using Hot Start Taq Plus polymerase (Qiagen) for all exons. The amplification contained 35 cycles and was followed by a 5-min final extension step at 72 °C. Amplified DNA fragments were purified using QIAquick PCR Purification kit (Qiagen) according to the manufacturer's instructions. Platelet mRNA was reverse transcribed using the cDNA Synthesis System Plus (Amersham International, Little Chalfont, UK) as previously described [28].
Table 2.
List of primers used in the present study
| Exon | Sense location, bp | Sequence | Antisense location, bp | Sequence |
|---|---|---|---|---|
| allb | ||||
| 1 | −79-58 | GCAAGTTACTTGGGGTTCCAG | 307–287 | CGCAAATGGGAAACTCGAAGC |
| 2 | 3324–3343 | GCCTGGGATACGCTGGAATC | 3547–3526 | GCAGTCCACGTCCCTCTGAC |
| 3 | 3519–3538 | AAGGTTGGGGTCAGAGGGAC | 3731–3710 | AGTGTCCCTGCCCCCGATTG |
| 4 | 3739–3758 | AGGAGGAGCCCAAGTCTCGC | 4054–4033 | CAGATCCAAAGCAAGGGCTG |
| 5 | 4127–4145 | CTCCCTTCACCCCTGGGCTG | 4294–4273 | GGAAGGGAAGTCCTGAGGGT |
| 6 | 4220–4239 | GGCGAGTAGGGAGCAAAAGC | 4378–4359 | GCGAGAAGGGAGGGAGGTGT |
| 7 | 4387–4406 | GAGACCGCTTTGGGCTTCAC | 4598–4579 | TGACCCTCGGGGTGCTGGAA |
| 8 | 4886–4905 | CCCTGGAAAAGACTAATTTG | 5034–5015 | AGATTTCAGGAAGGCGCTCC |
| 9 | 5097–5166 | AAGTGGGTAGGTTCTAAGGC | 5243–5224 | AGAACTGGGATAAGGGGCTT |
| 10 | 5307–5326 | CCTGGAGTGGGAGGTTGCTT | 5481–5461 | TTATTCTGAAGTCTCAGTTCC |
| 11 | 5506–5525 | CTTAAAGAGGATGCTTGTCC | 5656–5637 | ATTTGGGACCCAACTGGGTA |
| 12 | 5767–5786 | GTCCAGTCCCATGTAACCAC | 6048–6029 | CCTCTGCAGCAAGTAGGGCT |
| 13 | 8409–8428 | AACAATCAGCCACTTCCTTT | 8662–8643 | CTTGGGCATTTCTAGCTGGA |
| 14 | 8826–8845 | ATCGCCAATTCTGACCCATT | 8962–8943 | GCCTCCTCTTGATGGCACAG |
| 15 | 8981–9000 | AATGGCAAGCCTACCCCATC | 9163–9144 | GAGGTCCCAGATCCTTTAAG |
| 16 | 9145–9183 | AGAAAGGCTCCAACCCCTGA | 9300–9281 | TCCAAGCCCACCTCCCTCCT |
| 17 | 9317–9336 | CCAGGGAGGTCCTGACTCTT | 9559–9540 | GTCCCAGTGGGTAAGTTCTAC |
| 18 | 9658–9677 | TAGCAAGATGGCCTGACTCT | 9869–9849 | ATTTGCTATCAGGGGTCCTG |
| 19 | 10730–10750 | CCAACTTACCCGCACACCCC | 10918–10899 | GCAGGTATGATAGGCAGAAA |
| 20 | 10962–10981 | CCAGGCTCCCTGGCTTCACT | 11179–11160 | GGTCCCGGGTACTGTTCCCA |
| 21 | 11681–11700 | TCTGTAATTTCTTTCTTGGA | 11844–11825 | TGGTGGAGACCCGGTACCAC |
| 22 | 12382–12401 | ACTTGGGCAGTGACCTTGGC | 12537–12518 | GGGGAAGGGTGGTGGGTAGG |
| 23 | 13084–13103 | CTGGCCCTGTTTCTCCTCAT | 13233–13214 | AATGCCATCTCCCTTCTCCA |
| 24 | 13287–13306 | ATCACACTCTCTCTGGGGGT | 13471–13452 | AGTTCTGAGGACCCGCTCAC |
| 25 | 13484–13504 | TTAAGCTCCCCACACCCTGCC | 13707–13688 | GCCTTCCCAGGTCTTTCTTC |
| 26 (1) | 13719–13738 | AACCACCGGGGCACCTCTGT | 13974–13955 | ATCCCCTCTGCCCCGTGGGT |
| 26 (2) | 13755–13774 | GAGACCTGGGCCTGACCACT | 13975–13955 | ATCCCCTCTGCCCCGTGGGT |
| 27 | 14336–14355 | TGGGGGATGATGGGGTGATG | 14561–14542 | AAAGTGGTCCCCGCCCAGAA |
| 28 | 14711–14730 | TGGGTGTAAACGGCTTTCAA | 14881–14862 | AGGACTGGTCTCTGCTCCAT |
| 29 | 14988–15007 | AAGGCAGGTGTCAAGGTGA | 15207–15188 | AGGGCAGCGCCAAGCCTGTG |
| 30 (1) | 17023–17042 | TGTGCTCTGGGGCCAGCAAA | 17385–17366 | TCCAAAACATTCCTTCGGTC |
| 30 (2) | 16975–16995 | TTCTGCGCTGGTCCAGGGAGG | 17333–17313 | GCTCAGCATGAGGGCTCAGTC |
| β3 | ||||
| 1 | −113-93 | CGTTGCGTCCCACCCACCGCG | 151–131 | CGCGGTGGGTGGGACGCAACG |
| 2 | 20490–20510 | AGGATGAGGCAGGCAAGTACC | 29752–20694 | CTTCATGGGTCTGTTTCTGCAC |
| 3 | 29427–29447 | CTGCAGGAGGTAGAGAGTCGC | 29752–29732 | CCTCCACCTTGTGCTCTATGC |
| 4 | 30523–30543 | CTGCTTATTCAATCTTGGTGG | 30898–30878 | GAACCAGGACTTGGACCTTCC |
| 5 | 32344–32363 | CATGCTGCCTTTTCCATGAAG | 32637–32617 | CCCAGAGTTGAGTCACTTCCT |
| 6 | 33141–33161 | TCTCTACCAGTGACATGGCTG | 33431–33411 | CTAGATTAGGGCAACCCTCAC |
| 7 | 35760–35780 | CTTTAGACCCTAGAGATGTAC | 35989–35969 | TGCTAGCACAGGCTGGCTACC |
| 8 | 36232–36252 | CATCTCAGGACTCTCAGTGGG | 36471–36451 | AAGCCAAATGAGGTGGACTCTCCGC |
| 9 | 37035–37059 | GCATTTCCCGTTTCCTTTCAGTTCA | 37269–37246 | CACAGATGCTCCAGGACAAAGGCC |
| 10 | 38227–38252 | AGTGTTAACTGGGCCCAACTGTGTCT | 38773–38749 | TCCCAGTGGTTGCAGGTATATGAGGG |
| 10 (intra) | 38408–38432 | AGGCCCAAGCTGAACCTAATAGCC | − | |
| 11 | 45387–45410 | GCTGCCATGGAGTGGAGCTCTCG | 45720–45698 | GATCCTCTCCTACCTCCCAGCCT |
| 12 | 46536–46556 | TGCATGGAGATCAGAGCTGGA | 46758–46736 | GGTTCTGGGATTGAATTAGAAAC |
| 13 | 48793–48815 | GGTTTGCAGTGGTCCCATCTTCC | 49047–49025 | ATTCCTGCCTAACATGGTTCTCC |
| 14 | 53552–53573 | AGGTTCAAGTGACTCCTGCTTCA | 53839–53816 | TGCTGCTTCCACTTAAGAGTCTC |
| 15 | 56218–56240 | TTCAGGGTAGGGAAGGACTTAAG | 56410–56389 | ACATGATGGCAGGGACTCCTGC |
Nucleotide Sequence Analysis
Cycle sequencing of the entire αIIb and β3 DNA was accomplished by using the primers from table 2 and the BigDye® Terminator v1.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. cDNA sequencing of αIIb and β3 was performed with overlapping primers (sequences not shown). The cycle sequencing products were purified by using DyeEx 2.0 Spin kit (Qiagen) pursuant to the manufacturer's instructions. Analysis of the sequencing products was executed in ABI Prism 310 (Applied Biosystems) sequencer utilizing Performance Optimized Polymer 6 (Applied Biosystems) and ABI Prism Genescan software (Applied Biosystems). The regions containing the mutation were sequenced 3 times in both directions.
Results
Molecular Analysis of αIIb and β3 Transcripts
To determine the molecular basis underlying the abnormalities of αIIbβ3 receptor in the patients, full-length αIIb and β3 (1–2 kb fragments) from the patients’ platelets were amplified using PCR and sequenced by a modified Sanger method. A list of mutations identified within the αIIb and β3 genes is shown in tables 3 and 4. We found 8 different mutations of the αIIb gene in 20 patients and 4 mutations of the β3 gene in 5 patients. Four missense mutations, 4 splicing site mutations, 2 insertions of a single nucleotide, 1 deletion of a single nucleotide, and a deletion of 2 nucleotides were found. Overall, 9 patients were homozygous for a mutation in the αIIb gene (n = 7) or in the β3 gene (n = 2), whereas only 1 patient was found to be compound heterozygous with both mutations located in the β3 gene (table 5). In 14 patients, only a mutation in the heterozygous state was found and no additional gene defect was identified. We assume that 3 out of those 14 hetero zygotes are compound heterozygous (GE, SB, WC), however the second mutations could not yet be located. To exclude that the mutations found in our patients represent common polymorphisms, 50 randomly chosen samples from different ethnic or geographic backgrounds were screened for all mutations. None of these subjects were found to carry one of the mutations.
Table 3.
Mutations in the am gene identified in GT patients
| N | Mutation | Location | Patient | Genotype | Phenotype | Effect |
|---|---|---|---|---|---|---|
| 1 | 620C→T | exon 5 | OK | homozygous | missense | T207I |
| 2 | 641T→C | exon 6 | MM | homozygous | missense | L214P |
| MU | heterozygous | missense | L214P | |||
| 3 | 1754T→C | exon 17 | DZ | homozygous | splicing site | alternative splicing |
| 4 | 1878G→C | exon 18 | HM | homozygous | splicing site | alternative splicing |
| HS | heterozygous | splicing site | alternative splicing | |||
| HA | heterozygous | splicing site | alternative splicing | |||
| 5 | 2051T→G | exon 20 | GE | heterozygous | missense | L684R |
| CM | heterozygous | missense | L684R | |||
| 6 | 2232Gins | exon 22 | WC | heterozygous | ins. out of frame | frameshift |
| WV | heterozygous | ins. out of frame | frameshift | |||
| 7 | 3060G→A | exon 29 | TA | homozygous | splicing site | alternative splicing |
| TG | homozygous | splicing site | alternative splicing | |||
| TN | heterozygous | splicing site | alternative splicing | |||
| TF | heterozygous | splicing site | alternative splicing | |||
| 8 | 3062T→C | exon 29 | LN | homozygous | splicing site | alternative splicing |
| LH | heterozygous | splicing site | alternative splicing | |||
| LB | heterozygous | splicing site | alternative splicing | |||
| 9 | 3062T→C | exon 29 | SB | heterozygous | splicing site | alternative splicing |
N = Consecutive numbering of the found mutations; ins. = insertion.
Table 4.
Mutations in the p3 gene identified in GT patients
| N | Mutation | Location | Patient | Genotype | Phenotype | Effect |
|---|---|---|---|---|---|---|
| 1 | 856G→A | exon 6 | TE | heterozygous | missense | G286R |
| TA | heterozygous | missense | G286R | |||
| 2 | 1129Ains | exon 9 | TE | heterozygous | ins. out of frame | frameshift |
| GI | heterozygous | ins. out of frame | frameshift | |||
| 3 | 1550Gdel | exon 10 | YF | homozygous | del. out of frame | frameshift |
| 4 | 2068delGT | exon 13 | PJ | homozygous | del. out of frame | premature termination |
N = Consecutive numbering of the found mutations; ins. = insertion; del. = deletion.
Table 5.
Genetic findings in GT patients
| Patient (sex) | Genotype | Gene | First mutation |
Second mutation |
||
|---|---|---|---|---|---|---|
| location | effect | location | effect | |||
| OK (F) | homozygous | αIIb | exon 5 | T207I | − | − |
| MM (M) | homozygous | αIIb | exon 6 | L214P | − | − |
| MU (F) | heterozygous | αIIb | exon 6 | L214P | − | − |
| DZ (F) | homozygous | αIIb | exon 17 | splicing site | − | − |
| HM (M) | homozygous | αIIb | exon 18 | Q626H | − | − |
| HS (F) | heterozygous | αIIb | exon 18 | Q626H | − | − |
| HA (M) | heterozygous | αIIb | exon 18 | Q626H | − | − |
| TA (M) | homozygous | αIIb | exon 29 | splicing site | − | − |
| TG (M) | homozygous | αIIb | exon 29 | splicing site | − | − |
| TN (F) | heterozygous | αIIb | exon 29 | splicing site | − | − |
| TF (M) | heterozygous | αIIb | exon 29 | splicing site | − | − |
| LN (F) | homozygous | αIIb | exon 29 | splicing site | − | − |
| LH (F) | heterozygous | αIIb | exon 29 | splicing site | − | − |
| LB (M) | heterozygous | αIIb | exon 29 | splicing site | − | − |
| SB (F) | (compound) heterozygous | αIIb | exon 29 | splicing site | unknown | |
| GE (F) | (compound) heterozygous | αIIb | exon 20 | L684R | unknown | |
| CM (F) | heterozygous | αIIb | exon 20 | L684R | − | − |
| WC (M) | (compound) heterozygous | αIIb | exon 22 | ins. out of frame | unknown | |
| WV (F) | heterozygous | αIIb | exon 22 | ins. out of frame | − | − |
| WA (M) | heterozygous | αIIb | unknown | unknown | − | − |
| TE (M) | compound heterozygous | β3 | exon 6 | G286A | exon 9 | frameshift |
| TA (M) | heterozygous | β3 | exon 5 | G286A | − | − |
| GI (F) | heterozygous | β3 | exon 9 | frameshift | − | − |
| YF (M) | homozygous | β3 | exon 10 | G517DelGframeshift | − | − |
| PJ (M) | homozygous | β3 | exon 13 | premature termination | − | − |
F = Female; M = male; ins. = insertion.
Missense Mutations (n = 4)
Two missense mutations were detected within the αIIb gene (table 3). In exon 5, a homozygous Thr 207 to Ile (ACT→ATT) substitution was identified, and in exon 6, a homozygous T-to-C transition leading to a Leu 214 to Pro (CTT→CCT) substitution. A heterozygous Gly 286 (GGA→AGA) to Arg mutation was detected within the β3 gene (table 4) of patient TE. Analysis of the parents’ DNA showed that the missense mutation was inherited from the father (TA). All detected missense mutations are non-conservative substitutions that involve strictly conserved residues among vertebrates in the αIIb as well as in the β3 gene. The Thr 207 to Ile substitution allows for a non-polar instead of a polar and aliphatic amino acid in the β-propeller domain of the N-terminus part of the mature protein (fig. 1). The Leu 214 to Pro substitution (Patient MM) occurs also in the β-propeller domain and involves substitution from an aliphatic amino acid to heterocyclic amino acid. In the β3 gene, the heterozygous missense mutation (Gly 286 Arg) lies, according to the known crystal structure [29], in a highly conserved position within a proposed intramolecular disulfide loop, between Cys 232 and Cys 273 [30]. Furthermore, a heterozygous Leu 684 (CTG→CGG) to Arginin substitution was found in 2 related patients (GE, CM).
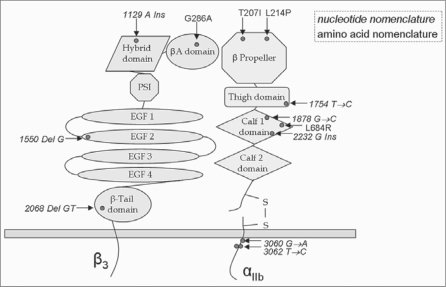
Fig. 1.
Schematic illustration of αIIbβ3. Shown is the distribution of all mutations (red dots) onto the different domains of αIIb and β3. Nucleotide nomenclature is italicized and refers to splicing site and frameshift mutations whereas the amino acid nomenclature is non-italicized referring to point mutations causing an amino acid exchange.
Deletions (n = 2)
Two mutations were detected within the β3 gene (table 3). A homozygous deletion at Gly 517 in exon 10 (1550delG) leads to changes of the corresponding amino acids of exon 10. In addition, a homozygous deletion of 2 nucleotides in exon 13 (2068delGT) is predicted to lead to changes of the corresponding amino acids of exon 13 and to a premature stop codon at position 696.
Insertions (n = 2)
Two mutations were detected within the αIb gene (table 3). A heterozygous insertion at Ala744 (2232Gins) in exon 22 is predicted to lead to changes in the corresponding amino acids, and was detected in 2 affected blood relatives (WC, WV). One insertion mutation was located within the β3 gene (table 3), and was found in the heterozygous state at Ile377 in exon 9 (1129Ains) in 2 related patients. This mutation would lead to changes in the corresponding amino acids encoded by the subsequent portion of exon 9.
Splicing Site Mutations (n = 4)
All splicing site mutations were found in the αIIb gene (table 3). Sequencing of exon/intron regions using genomic DNA revealed a homozygous splicing donor site mutation (3060G→A) in exon 29 in 2 siblings (TA, TG). Analysis of the entire cDNA showed a homozygous deletion of exon 29 in the αIIb subunit, the putative transmembrane region. As expected, both parents (TN, TF) were heterozygous carriers of that mutation. A further homozygous splicing donor site mutation (3062T→C) was found in exon 29 (LN) and as heterozygous condition in the patients’ parents (LH, LB). In addition, this splicing site mutation (3062T→C) was also detected, as heterozygous condition, in an additional patient (SB). Although no blood relationship was reported, these 2 patients were from the same region in Germany. Moreover, homozygous splicing donor site mutations in exon 17 (1754T→C) (DZ) or exon 18 (1878G→C) (HM) were identified.
Polymorphisms (n = 5)
Relating to GT, there are various single nucleotide polymorphisms known in αIIb and β3, which can lead to an amino acid exchange but have no impact on the function of the thrombocytes in terms of aggregation or adhesion ability. The investigation of all patients revealed previously described polymorphisms in homozygous and heterozygous forms concerning several patients (DZ, WC, TE, YF, PJ). The polymorphism Ile843Ser in the αIIb subunit could be identified in 2 patients (WC: heterozygous; TE: homozygous). The β3 subunit contains more polymorphisms. Patients DZ and YF show the Leu33Pro polymorphism in the homozygous and patient SB in the heterozygous form. Furthermore, the polymorphisms Val381Val (TE: heterozygous; PJ: homozygous) and Arg489Gln (TE: heterozygous; PJ: homozygous) could be ascertained. The possibility of a founder effect for the mutations V585G, 2232Gins, V1021A, G286A + 1129Ains, 1550delG, and 2068delGT seems unlikely but cannot be excluded because the patients carrying polymorphisms all seem to suffer from GT type I, except for one. However, a coherency is not established.
Platelet Function Studies
The results of flow cytometric and platelet aggregation studies are summarized in table 6. As expected, platelets from patients with type I GT (0–5% of αIIbβ3 on platelets: TA, TG, GE, TE, LN, HM, OK, MM, WC, PJ) were unable to aggregate in response to ADP (5 μmol/l) and collagen (1 μg/ml). Platelets of these patients failed to aggregate even with higher concentrations of ADP (10 μmol/l) and collagen (2 μg/ml) (data not shown). Furthermore, analysis of αIIbβ3 activation-dependent epitope using flow cytometry showed that mab PAC-1 was unable to bind after activation with TRAP or ADP (table 6). However, a larger variability was seen between the different tests in patients with >20% of αIIbβ3 on platelets. For example, the heterozygous parents of TE and TG had a normal amount of αIIbβ3 and slightly inhibited platelet aggregation but showed distinct inhibition of PAC-1 binding. The parents of patient WC also showed normal αIIbβ3 expression on platelets. PAC-1 binding was markedly decreased with a more pronounced effect for the mother's platelets. Surprisingly, the father's platelets were unable to aggregate in response to ADP and collagen. In the mother, we observed a markedly decreased platelet aggregation response to ADP and collagen. Patient SB had a residual αIIbβ3 surface expression of 23%. Functional analysis of αIIbβ3 revealed low PAC-1 binding capacity of about 10% after activation with ADP or TRAP. These results indicate that both a quantitative and qualitative abnormality in the αIIbβ3 receptor exist in this patient.
Table 6.
Flow cytometric analysis and agonist-induced platelet aggregation in GT patients
| Patient | FACS analysis |
Platelet aggregation |
||||
|---|---|---|---|---|---|---|
| CD41 anti-αIIb % | PAC-1, % |
ADP (5 urnol/1) | collagen (1 μg/ml) | ristocetin (1.3 mg/ml) | ||
| 10 μmol/l ADP | 10 μmol/1 TRAP | |||||
| TA (M) | <1 | <1 | <1 | Ø | Ø | N |
| TG (M) | <1 | <1 | <1 | Ø | Ø | N |
| TN (F) | 90 | 37 | 55 | (▾) | (▾) | N |
| TF (M) | 78 | 38 | 25 | (▾) | (▾) | N |
| GE (F) | <1 | <1 | <1 | Ø | Ø | N |
| CM (F) | 58 | 44 | 51 | (▾) | ▾ | N |
| SB (F) | 23 | 13 | 12 | ▾ | (▾) | N |
| YF (M) | 3 | n.a. | n.a. | n.a. | n.a. | n.a. |
| TE (M) | <1 | <1 | <1 | Ø | Ø | N |
| DZ (F) | 0 | n.a. | n.a. | n.a. | n.a. | n.a. |
| LN (F) | <1 | <1 | n.a. | Ø | Ø | N |
| HM (M) | 0 | <1 | <1 | n.a. | n.a. | n.a. |
| OK (F) | 4 | <1 | <1 | Ø | Ø | N |
| MM (M) | <1 | <1 | <1 | ▾ | ▾ | (▾) |
| MU (F) | 82 | 57 | 42 | n | N | N |
| WC (M) | 0 | <1 | <1 | Ø | Ø | N |
| WV (F) | 68 | 35 | 25 | ▾ | ▾ | N |
| WA (M) | 78 | 66 | 22 | Ø | Ø | (▾) |
| PJ (M) | <1 | n.a. | n.a. | Ø | Ø | n |
N = Normal; Ø = absent; ▾ = inhibited or reduced; (▾) = slightly inhibited; n.a. = not analyzed.
Discussion
GT is a rare congenital bleeding syndrome which results in deficient platelet aggregation due to the absence or dysfunction of the fibrinogen receptor αIIbβ3. Due to the dimension and diversity of the occurring mutations in αIIbβ3, it is rather complex to identify mutations that are responsible for GT. In the present study, 10 novel and 2 previously described gene alterations that are known to cause GT were identified in 12 unrelated patients (table 3) and their first-degree relatives. To ascertain the mutations, we sequenced the coding regions of the αIIb and β3 subunits including the exon/intron boundaries. Our study revealed 4 point mutations which are all non-conservative substitutions in strictly conserved residues among vertebrates in both genes. Two of the point mutations in αIIb, Thr 207 to Ile and Leu 214 to Pro, are located in the FG-GAP 3 of the β propeller. Leu 214 to Pro has already been described by Grimaldi et al. [31]. It has been shown that the FG-GAP is important for ligand binding, so the Leu 214 to Pro mutation disrupts the structural conformation and impairs the ligandbinding properties of the heterodimeric complex. Additionally, the mutation seems to confer susceptibility to proteolysis [31, 32]. Concerning the Thr 207 to Ile mutation, it has been described to impair fibrinogen binding and affect the expression of αIIbβ3 on the platelet surface [28]. Further, we found a point mutation at position 684 leading to a Leu to Arg substitution. This mutation is located in the calf-1 domain of exon 20, also in the αIIb subunit. The calf-1 domain creates minor interfaces with the β3 subunit [33]. Therefore, conformational changes due to this amino acid substitution could impede contact between αIIb and β3 resulting in a failure of complex-formation. If the subunits do not form a complex, the protein subunits are not protected from intracellular proteolysis, and therefore they cannot be anchored in the cell membrane.
Gly 286 to Arg is located in the VWFA domain, also called βA domain, of the β3 subunit. The βA domain is a ligand-binding head region of the β subunit. Conformational changes in VWFA have an impact on the ligand-binding activity, and ligand recognition also causes conformational changes transduced from this domain [34].
In 5 unrelated individuals and their relatives we detected splicing site defects. Three of those splicing site mutations are located in the splicing donor site in exon 29, and 2 of them (3062T→C; SB, LN) are even identical although no apparent consanguinity was reported. However, both patients were from the same region in Germany. Both of those splicing site mutations lead to a complete loss of exon 29, which was proven by cDNA sequencing. The mutations are located in the cytoplasmatic domain proximal to the transmembrane region in the N-terminal area of the GFFKR motif. Regions proximal to the transmembrane domains of the α and β subunits contain highly conserved sequences, such as the GFFKR motif referring to the αIIb subunit, and are found to be important for maintaining the inactive low-affinity state of the integrin. It is confirmed that the cytoplasmatic domains are essential for the formation of the heterodimer, and the GFFKR motif appears to be required for association of the α and β subunits [35, 36]. The 3062T→C substitution at position V1021 is located at only 1 amino acid prior to the start of the GFFKR motif and leads to an alternative splicing. Therefore, it could be assumed that due to the alternative splicing, N-terminal to the motif, the conformation of the protein is changed in such a way that it could be impossible for the above-mentioned motif to associate with the corresponding motif in the β3 subunit.
The other two splicing site defects were detected in exons 17 and 18 of the αIIb subunit. The 1754T→C (DZ) substitution situated in the splicing donor site of exon 17 is located in the thigh domain of the αIIb subunit. The thigh domain has substantial contact with the β-propeller. Losonczy et al. [37] previously suggested that this contact might be important for the proper orientation of the β-propeller to interact properly with the β3 subunit. An alteration in this domain could therefore change the orientation of the β-propeller and inhibit complex formation with the β3 subunit [37]. The homozygous splicing donor site mutation 1878G→C (HM) affecting exon 18 is situated in the calf-1 domain of the GPIIb subunit. The calf-1 and calf-2 domains form minor interfaces with GPIIIa [36]. Rosenberg et al. [33] formerly published that significant changes in the calf-1 and calf-2 domains do not avert complex formation of αIIbβ3 but affect its transport to the cell surface, probably by its retention and subsequent degradation in the endoplasmic reticulum [33]. We assume that due to the found splicing site defects, the affected exons are non-existent and the concerned domains are thereby sustainably influenced in their conformations.
In 4 GT patients, we found frameshift mutations caused by insertions or deletions. Three out of 4 are situated in the β3 subunit. Insertions or deletions lead to a shift in the reading frame and often cause a premature stop codon resulting in premature termination of the protein or an abnormal protein product with an incorrect amino acid sequence.
In 1 patient, we could identify a heterozygous insertion of G at nucleotide position 2232. This frameshift does not result in a premature stop codon but the amino acid sequence is severely altered, probably resulting in an abnormal protein. Since this mutation is also situated in the calf-1 domain of the αIIb, we assume once more that complex formation is impaired resulting in a premature intracellular proteolysis.
A compound heterozygous patient and his mother showed a heterozygous insertion of A in exon 9 at nucleotide position 1129. This insertion results in a stop codon at amino acid position 380, probably leading to premature termination. Here, the hybrid domain is affected.
Concerning frameshift mutations we could additionally identify 2 patients with deletions within the β3 subunit. A deletion of 1 nucleotide (1550delG) was detected in exon 10 of β3. This deletion is situated in the EGF-2 domain and does not lead to a premature stop codon, nevertheless we assume that the nearby located disulphide bond can not be formed due to a modified amino acid sequence. The EGF domains are cysteine-rich modules, containing multiple disulphide bonds. In particular, the EGF-2 region is wedged in the bent between integrin stalk and headpiece domains, poised to communicate structural rearrangements between the cytoplasmic domains and the ligand-binding headpiece [38]. Also located in β3, a 2-nucleotide deletion (2082delGT) was identified in exon 13, affecting the β-tail domain (β-TD). This mutation leads to premature termination of the translation at amino acid position 696 and therefore to a truncated protein.
Several of these patients were carrier of diverse polymorphisms (table 6). We cannot exclude founder effects of polymorphisms regarding the newly revealed mutations in αIIb and β3 but it seems improbable. Solely in 3 heterozygous patients, a second mutation forming the compound heterozygous status could not yet be detected. A genotype/phenotype correlation is only vaguely viable. Therefore, a profound genotype/phenotype coherence could not be implemented. However, we came to know that the patients with splicing site defects suffer from mild to moderate bleeding, whereas point mutations and insertions/deletions tendencially result in moderate to severe bleeding. In conclusion, our investigation of several patients, and in some cases including their first-degree relatives, revealed a number of different mutations within the αIIb and β3 subunits associated with causing GT. This vast number of different mutations indicates the genetic heterogeneity in the studied group as well as in GT in general.
Disclosure
The authors declared no conflict of interest.
References
- 1.Caen JP. Glanzmann's thrombasthenia. Clin Hae-matol. 1989;2:609–625. doi: 10.1016/s0950-3536(89)80036-8. [DOI] [PubMed] [Google Scholar]
- 2.Bellucci S, Caen J. Molecular basis of Glanzmann's thrombasthenia and current strategies in treatment. Blood Rev. 2002;16:193–202. doi: 10.1016/s0268-960x(02)00030-9. [DOI] [PubMed] [Google Scholar]
- 3.Nurden AT. Glanzmann thrombasthenia. Orphan-et J Rare Dis. 2006;1:10. doi: 10.1186/1750-1172-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shattil SJ, Kashiwagi H, Pampori N. Integrin signaling: the platelet paradigm. Blood. 1998;91:2645–2657. [PubMed] [Google Scholar]
- 5.D'Souza SE, Ginsberg MH, Burke TA, Lam CT, Plow EF. Localization of an Arg-GlyAsp recognition site within an integrin adhesion receptor. Science. 1988;242:91–93. doi: 10.1126/science.3262922. [DOI] [PubMed] [Google Scholar]
- 6.Farrell DH, Thiagarajan P, Chung DW, Davie EW. Role of fibrinogen alpha an gamma chain sites in platelet aggregation. Proc Nati Acad Sei USA. 1992;89:10729–10732. doi: 10.1073/pnas.89.22.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Souza SE, Ginsberg MH, Burke TA, Lam CT, Plow EF. Localization of an Arg-Gly-Asp recognition site within an integrin adhesion receptor. Science. 1988;242:91–93. doi: 10.1126/science.3262922. [DOI] [PubMed] [Google Scholar]
- 8.Loftus JC, Smith JW, Ginsberg MH. Integrin-me-diated cell adhesion: the extracellular face. J Biol Chem. 1994;269:25235–25238. [PubMed] [Google Scholar]
- 9.Loftus JC, O'Toole TE, Plow EF, Glass A, Frelin-ger AL, 3rd, Ginsberg MH. A beta 3 integrin mutation abolishes ligand binding and alters divalent cation-dependent conformation. Science. 1990;249:915–918. doi: 10.1126/science.2392682. [DOI] [PubMed] [Google Scholar]
- 10.D'Souza SE, Ginsberg MH, Burke TA, Plow EF. The ligand binding site of the platelet integrin receptor GPIIb-IIIa is proximal to the second calcium binding domain of its alpha subunit. J Biol Chem. 1990;265:3440–3446. [PubMed] [Google Scholar]
- 11.Kamata T, Irie A, Tokuhira M, Takada Y. Critical residues of integrin alphallb subunit for binding of alphallbbeta3 (glycoprotein Ilb-IIIa) to fibrinogen and ligand-mimetic antibodies (PAC-1, OP-G2, and LJ-CP3) J Biol Chem. 1996;271:18610–18615. doi: 10.1074/jbc.271.31.18610. [DOI] [PubMed] [Google Scholar]
- 12.Calvete JJ. Platelet integrin GPIIb/IIIa: structure-function correlations. An update and lessons from other integrins. Proc Soc Exp Biol Med. 1999;222:9–38. doi: 10.1111/j.1525-1373.1999.09993.x. [DOI] [PubMed] [Google Scholar]
- 13.George JN, Caen JP, Nurden AT. Glanzmann's thrombasthenia: the spectrum of clinical disease. Blood. 1990;75:1383–1395. [PubMed] [Google Scholar]
- 14.Bellucci S, Caen J. Molecular basis of Glanzmann's thrombasthenia and current strategies in treatment. Blood Rev. 2002;16:193–202. doi: 10.1016/s0268-960x(02)00030-9. [DOI] [PubMed] [Google Scholar]
- 15.Phillips DR, Charo IF, Parise LV, Fitzgerald LA. The platelet membrane glycoprotein Ilb-IIIa complex. Blood. 1988;71:831–843. [PubMed] [Google Scholar]
- 16.www.sinaicentral.mssm.edu/intranet/research/glanz-mann
- 17.www.uwcm.ac.uk/uwcm/mg/hgmdO.html
- 18.French DL, Coller BS. Hematologically important mutations: Glanzmann thrombasthenia. Blood Cells Mol Dis. 1996;23:39–51. doi: 10.1006/bcmd.1997.0117. [DOI] [PubMed] [Google Scholar]
- 19.French DL. The molecular genetics of Glanz-mann's thrombasthenia. Platelets. 1998;9:5–20. doi: 10.1080/09537109876951. [DOI] [PubMed] [Google Scholar]
- 20.Jayo A, Pabón D, Lastres P, Jiménez-Yuste V, González-Manchón C. Type II Glanzmann thrombasthenia in a compound hétérozygote for the alpha lib gene. A novel missense mutation in exon 27. Haematologica. 2006;91:1352–1359. [PubMed] [Google Scholar]
- 21.Tao J, Arias-Salgado EG, González-Manchón C, Díaz-Cremades J, Ayuso MS, Parrilla R. A novel (288delC) mutation in exon 2 of GPIIb associated with type I Glanzmann's thrombasthenia. Br J Haematol. 2000;111:96–103. doi: 10.1046/j.1365-2141.2000.02336.x. [DOI] [PubMed] [Google Scholar]
- 22.Arias-Salgado EG, Tao J, González-Manchón C, Butta N, Vicente, Ayuso MS, et al. Nonsense mutation in exon 19 of GPIIb associated with throm-basthenic phenotype. Failure of GPIIb (D597-1008) to form stable complexes with GPIIIa. Thromb Haemost. 2002;87:684–691. [PubMed] [Google Scholar]
- 23.González-Manchón C, Arias-Salgado EG, Butta N, Martin G, Rodríguez RB, Elalamy I, et al. A novel homozygous splice junction mutation in GPIIb associated with alternative splicing, nonsense-mediated decay of GPIIb mRNA, and type II Glanzmann's thrombasthenia. J Thromb Haemost. 2003;1:1071–1078. doi: 10.1046/j.1538-7836.2003.00204.x. [DOI] [PubMed] [Google Scholar]
- 24.Ambo H, Kamata T, Handa M, Taki M, Kuwajima M, Kawai Y, Oda A, Murata M, Takada Y, Watan-abe K, Ikeda Y. Three novel integrin beta3 subunit missense mutations (H280P, C560F, and G579S) in thrombasthenia, including one (H280P) prevalent in Japanese patients. Biochem Biophys Res Commun. 1998;251:763–768. doi: 10.1006/bbrc.1998.9526. [DOI] [PubMed] [Google Scholar]
- 25.Coller BS, Cheresh DA, Asch E, Seligsohn U. Platelet vitronectin receptor expression differentiates Iraqi-Jewish from Arab patients with Glanzmann thrombasthenia in Israel. Blood. 1991;77:75–83. [PubMed] [Google Scholar]
- 26.Peretz H, Seligsohn U, Zwang E, Coller BS, Newman PJ. Detection of the Glanzmann's thrombasthenia mutations in Arab and Iraqi-Jewish patients by polymerase chain reaction and restriction analysis of blood or urine samples. Thromb Haemost. 1991;66:500–504. [PubMed] [Google Scholar]
- 27.Tadokoro S, Tomiyama Y, Honda S, Arai M, Yamamoto N, Shiraga M, Kosugj S, Kanakura Y, Kurata Y, Matsuzawa Y. A Gln747→Pro substitution in the lib subunit is responsible for a moderate IIbbeta3 deficiency in Glanzmann thrombasthenia. Blood. 1998;92:2750–2758. [PubMed] [Google Scholar]
- 28.Westrup D, Santoso S, Follert-Hagendorff K, Bas-sus S, Just M, Jablonka B, Kirchmaier CM. Glanz-mann thrombasthenia Frankfurt I is associated with a point mutation Thrl76Ile in the N-terminal region of alpha lib subunit integrin. Thromb Haemost. 2004;92:1040–1051. doi: 10.1160/TH04-03-0170. [DOI] [PubMed] [Google Scholar]
- 29.Xiong J-P, Sthele T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnout MA. Crystal structure of the extracellular segment of integrin alpha Vbeta3. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calvete JJ, Henschen A, Gonzalez-Rodriguez J. Assignment of the disulphide binds in human platelet GPIIIa: a disulphide pattern for the b subunits of the integrin family. Biochem J. 1991;274:63–71. doi: 10.1042/bj2740063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimaldi CM, Chen F, Wu C, Weiss HJ, Coller BS, French DL. Glycoprotein lib Leu214Pro Mutation produces Glanzmann thrombasthenia with both quantitative and qualitative abnormalities in GPIIb/IIIa. Blood. 1998;91:1562–1571. [PubMed] [Google Scholar]
- 32.Loftus JC, Smith JW, Ginsberg MH. Integrin-me-diated cell adhesion: the extracellular face. J Biol Chem. 1994;269:25235–25238. [PubMed] [Google Scholar]
- 33.Rosenberg N, Yatuv R, Sobolev V, Peretz H, Zive-lin A, Seligsohn U. Major mutations in calf-1 and calf-2 domains of glycoprotein lib in patients with Glanzmann thrombasthenia enable GPIIb/IIIa complex formation, but impair its transport from the en-doplasmic reticulum to the Golgi apparatus. Blood. 2003;101:4808–4815. doi: 10.1182/blood-2002-08-2452. [DOI] [PubMed] [Google Scholar]
- 34.Mould AP, Barton SJ, Askari JA, McEwan PA, Buckley PA, Craig SE, Humphries MJ. Conformational changes in the integrin beta A domain provide a mechanism for signal transduction via hybrid domain movement. J Biol Chem. 2003;278:17028–17035. [Google Scholar]
- 35.De Melker A, Kramer D, Kuikman I, Sonnenberg A. The two phenylalanins in the GFFKR motif of the integrin alpha6A subunit are essential for het-erodimerization. Biochem J. 1997;328:529–537. doi: 10.1042/bj3280529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bennett JS. Structure and function of the platelet integrin alphallbbeta3. J Clin Invest. 2005;115:3363–3369. doi: 10.1172/JCI26989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Losonczy G, Rosenberg N, Boda Z, Vereb G, Kap-pelmayer J, Hauschner H, Bereczky, Muszbek L. Three novel mutations in the glycoprotein lib gene in a patient with type II Glanzmann thrombasthenia. Heamatologica. 2007;92:698–701. doi: 10.3324/haematol.10847. [DOI] [PubMed] [Google Scholar]
- 38.Beglova N, Blacklow SC, Takagj J, Springer TA. Cysteine-rich module structure reveals a fulcrum for integrin rearrangement upon activation. Nature structural biology. 2002;9:282–287. doi: 10.1038/nsb779. [DOI] [PubMed] [Google Scholar]