Abstract
One of the major hurdles in the development of safe and effective drugs targeting G-protein coupled receptors (GPCRs) is finding ligands that are highly selective for a specific receptor subtype. Structural understanding of subtype-specific binding pocket variations and ligand-receptor interactions may greatly facilitate design of selective ligands. To gain insights into the structural basis of ligand subtype selectivity within the family of adenosine receptors (AR: A1, A2A, A2B, and A3) we generated 3D models of all four subtypes using the recently determined crystal structure of the AA2AR as a template, and employing the methodology of ligand-guided receptor optimization for refinement. This approach produced 3D conformational models of AR subtypes that effectively explain binding modes and subtype selectivity for a diverse set of known AR antagonists. Analysis of the subtype-specific ligand-receptor interactions allowed identification of the major determinants of ligand selectivity, which may facilitate discovery of more efficient drug candidates.
Introduction
Major targets for drug discovery and development, proteins of the G-protein coupled receptor (GPCR) family are involved in recognition of a great variety of extracellular signals including ions, small molecules, peptides and globular proteins1–2. Despite the diversity of natural GPCR ligands, there exist several receptor subfamilies in which all proteins respond to a single endogenous agonist: for example, all GPCRs in the adrenergic subfamily are activated by epinephrine while all muscarinic receptors naturally bind acetylcholine and its derivatives. GPCR subtypes within a subfamily usually have distinct amino acid sequences, tissue distributions, effector coupling, and/or functional and pharmacological profiles; however, their ligand binding pockets are highly conserved within the subfamily. The similarity of the orthosteric binding pockets poses a challenge for design of subtype selective ligands which remains one of the main hurdles in development of safe and effective medications targeting GPCRs.
The Adenosine Receptor (AR) subfamily is comprised of four member subtypes (A1, A2A, A2B, A3) that present a prominent example of closely related GPCRs activated by a single endogenous agonist, adenosine3. All four AR subtypes have been considered as potential therapies for neurodegenerative4–5, cardiac6–7, immune, and inflammatory disorders8–9 and cancer10. Functional importance of different AR subtypes in various body functions and tissues imposes very high requirements on subtype selectivity of AR antagonists and agonists as candidate drugs11–12 and leads to significant challenges in clinical development of the candidate drugs. Despite early setbacks, 2008 has been marked by successful FDA approval of the new generation A2AAR selective agonist regadenoson as a coronary vasodilator for use in myocardial perfusion imaging13. This breakthrough, along with other advances in pre-clinical and clinical studies3 boosts interest to development of a new generation of bioavailable and safe agonists and antagonists for adenosine receptors.
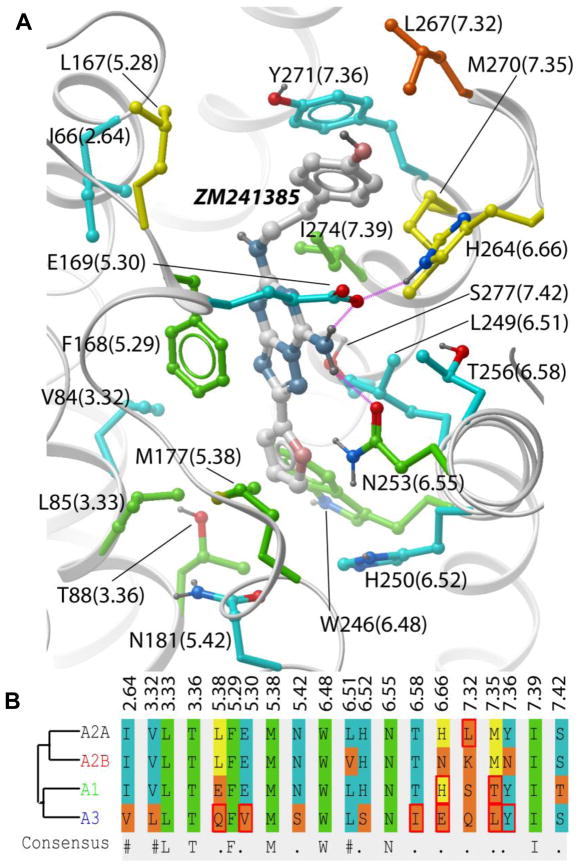
The recent crystal structure of AA2AR in complex with an antagonist (PDB code: 3EML)14 elucidates the details of ligand-receptor interactions at atomic resolution (Figure 1), and provides an excellent template for virtual ligand screening and structure-based drug discovery. This was recently demonstrated by exceptionally high hit rates (~40%) in prospective virtual ligand screening (VLS) for novel chemotypes of AA2AR antagonists15–16. The core ligand-receptor interactions are defined by aromatic stacking of the ligand aromatic ring system with the side chain of Phe168(5.29), hydrophobic interactions with Leu249(6.51), Ile274(7.39) and M177(5.38), and the hydrogen bonding to Asn253(6.55). (Residue numbering is for SWISSPROT human AA2AR sequence P29274, while Ballesteros-Weinstein numbering17 in parenthesis represents position of the residue relative to the most conserved position in each TM helix, extended to loop regions as in ref.14). As shown by mutations and computational analysis18, these residues make a major contribution to the ligand binding affinity. Note that the above mentioned “core” pocket side chains are fully conserved in all four AR subtypes, with the exception of Leu replaced by a similar Val side chain in position 6.51 of A2B. Moreover, there are only a few side chains in the binding pocket of crystallographic antagonist ZM241385, which are conserved in less than 3 subtypes (positions 5.28, 6.66 7.32 and 7.35), all of them located in the extracellular loop (ECL) region.
Figure 1.
Residue variations in the ligand binding pocket between four human adenosine receptor subtypes. (A) In the 3D structure of the AA2AR binding pocket, residue numbers are shown for AA2 subtype (number in brackets as in ref14, based on Ballesteros-Weinstein GPCR numbering)17. Side chain carbons are colored according to their conservation: green – fully identical in all 4 subtypes, cyan – in 3 subtypes, yellow – in 2 subtypes, orange - in only one subtype. Binding pocket residue alignment (B) uses the same color coding. Residues that vary between clinically relevant species (human, rat, mouse, dog) in the same subtype are marked with a red box.
The subfamily-wide conservation of the core pocket residues and the peripheral location of nonconserved amino-acids apparently pose serious challenges for the discovery of subtype selective ligands. Previous SAR studies for major AR-binding chemotypes, including xanthine and adenine derivatives and other polyheterocyclic compounds (for example reviewed in ref.19), suggest that the core chemical scaffolds themselves do not provide significant subtype selectivity. Extensive SAR analysis of multipoint substitutions in the scaffolds, in some cases guided by pharmacophore models, was required to achieve high affinity and reliable selectivity for each of the subtypes. Our virtual ligand screening efforts against the AA2AR crystal structure15 also suggest that ligand subtype selectivity, especially between A2A, A2B and A1 subtypes, does not come automatically with the high affinity binding to one of the subtypes. Indeed, while most of novel A2A binders found in the above VLS study among commercially available compounds had reduced affinity to A3 subtype, only one of them showed ~10-fold selectivity vs. A1 subtype.
In the past 20 years, intensive drug discovery efforts have resulted in hundreds of SAR profiles and yielded a number of AR ligands with respectable (~100 fold) selectivity for each of the subtypes 12. However, there is an ongoing interest in new chemical scaffolds and new highly selective compounds with improved pharmacological properties. As there is still no clear understanding of structural basis of ligand selectivity in ARs, structural models of AR subtypes that may explain and predict ligand selectivity are of paramount importance for rational drug discovery.
In this study we set out to generate accurate 3D models of all four AR subtypes that successfully predict the subtype selectivity of known antagonists. We used the crystal structure of AA2AR as a 3D template, and refined the models with a set of known subtype-selective ligands (Figure SM2) using the recently developed Ligand Guided Receptor Optimization approach20–21. While the starting raw homology models showed poor discrimination between ligands and decoys in our VLS benchmarks, the performance of the optimized models improved dramatically through iterations of the procedure. For A1 and A3 subtypes, the screening performance approached that of the AA2AR crystal structure. We then built the “virtual selectivity panel” that demonstrated the excellent discrimination between subtype-selective ligands. We also suggested the structural features of AR subtypes that play the key roles in selective ligand recognition. The developed selectivity panel can be applied to rational design of novel subtype-selective AR antagonists. With the rapid progress of GPCR crystallography, the approach will become applicable to other GPCR families with at least one representative crystal structure and known high affinity ligands.
Methods
Subtype selective ligand sets
High affinity adenosine receptor antagonists with at least 50-fold selectivity for each of the AR subtypes were collected from literature (see Supplementary Materials). The ligand set includes preclinical and clinical candidates listed in a recent review12, a diverse set of A2AAR selective compounds from ref.22, as well as several series of AR binding compounds for A1AR23–24, A3AR25, and A2BAR26–27, for which full AR selectivity profiles were available. The ligand set included 22 selective compounds for each of the four subtypes, 88 compounds total.
A set of 1000 decoy compounds was randomly selected from a Chemdiv Inc. database of drug-like compounds so that distributions of chemical properties (molecular weight and predicted LogP and LogS values) of ligand and decoy sets were similar.
Homology modeling of A1, A2B and A3 adenosine receptor subtypes
The initial homology models of A1AR, A2BAR and A3AR were generated using ICM homology modeling tool with extensive side chain sampling and refinement28–29. For stretches of residues that aligned against gaps in the AA2AR template, the algorithm used an automated search for initial loop placement. The portion of the loop EL2 between positions Leu141(4.62) and Cys166(5.27) was omitted. For short loop EL3, extensive conformational modeling was performed using ICM global optimization algorithm, which included sampling of backbone conformations30.
Ligand guided optimization of AR subtype models
Initial models were optimized using Ligand-guided Receptor Optimization algorithm (LiBERO) described previously15. Briefly, the protocol consists of repeated steps of model conformational sampling and selection based on their VLS screening performance evaluated on a small compound benchmark. The version of the LiBERO algorithm employed in this paper did not use large scale sampling of the protein backbone. At each step of the optimization procedure, a set of 100 conformational models was generated for each adenosine receptor subtype by co-optimization of representative subtype-selective antagonists in the model binding pocket. The optimization was performed using BPMC method31, which allowed extensive sampling of the flexible ligand and flexible receptor side chain conformations, while limiting protein backbone movements using harmonic restraints. For each of the resulting conformers, a small-scale VLS benchmarking was performed and the model screening performance was quantified using the NSQ_AUC measure described below.
ICM Grid Docking
ICM molecular modeling software30, 32 was used for ligand docking and scoring. ICM ligand docking is based on biased probability Monte Carlo (BPMC) optimization of the ligand internal coordinates in the set of grid potential maps of the receptor33. Compounds in two-dimensional representation were converted to 3D and optimized using MMFF-94 force field34. The generated conformers were then placed into the binding pocket in four principal orientations and used as starting points for Monte Carlo optimization. The optimized energy function included the ligand internal strain and a weighted sum of the grid map values in ligand atom centers. To ensure convergence of the Monte Carlo optimization, three independent runs of the docking procedure were performed, and the best scoring pose per compound was kept. No distance restraints or any other experimentally derived information was used in the ligand docking procedure. The docking procedure took about 30 seconds of Intel Xeon 2.8 Ghz CPU time per compound, and was performed using a 100 processor Linux cluster.
ICM Full-Atom Scoring
The top-scoring ligand poses were merged with their receptors to obtain full-atom models of the complexes. The models were evaluated with all-atom ICM ligand binding score35–36 that has been previously derived from a multi-receptor screening benchmark as a compromise between approximated Gibbs free energy of binding and numerical errors. The score was calculated as:
| (4) |
where Evw, Eel, Ehb, Ehp, and Esf are Van der Waals, electrostatic, hydrogen bonding, non-polar and polar atom solvation energy differences between bound and unbound states, Eint is the ligand internal strain, ΔSTor is its conformational entropy loss upon binding, T = 300 K, and αi are ligand- and receptor-independent constants.
Evaluation of the model screening performance using normalized square-root AUC
Benchmark compounds were docked into each model and ranked by their full-atom score. To build the compound recognition curve, for each compound rank i, the fraction of true positive among i top-scoring compounds in the list was plotted against Sqrt(i)/n, where n is the number of false positive compounds in the list. The area under curve was calculated and normalized to the range of 0 to 100 to obtain the Normalized Square Root Area Under Curve, or NSQ_AUC. Calculated as described, NSQ_AUC mildly emphasizes the initial enrichment in small molecule screening which makes it a suitable objective function for pocket optimization37.
Results and Discussion
Docking and virtual ligand screening for A2A subtype identifies core ligand-pocket interactions
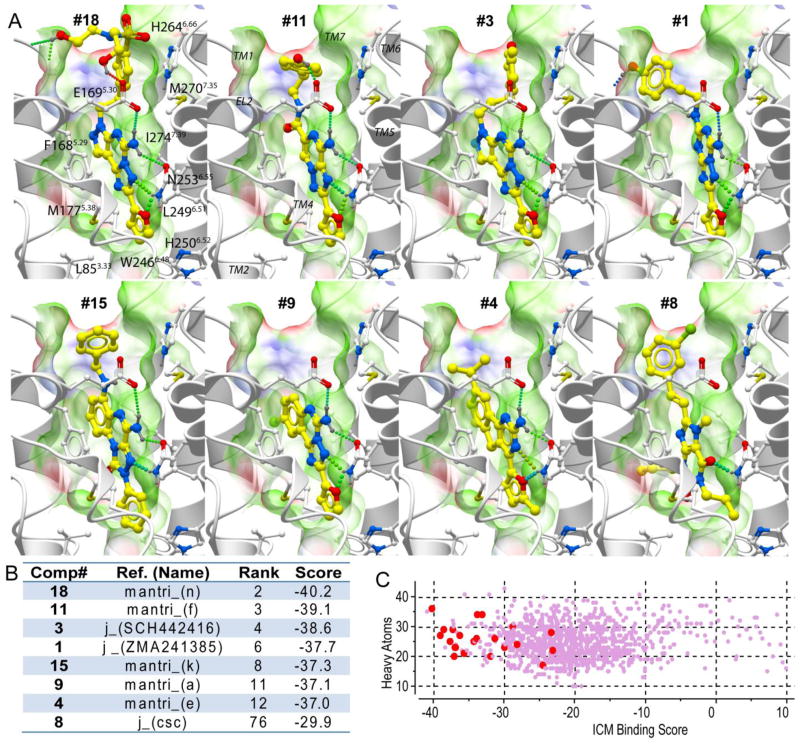
We first evaluated the ability of the A2AAR crystal structure-based models14 to accommodate known A2AAR specific antagonists and recognize them in a set of random drug-like decoys. A benchmark compound set included 22 selective A2AAR antagonists and 1000 decoys (see Methods section). In Figure 2A, the predicted binding poses are shown for seven A2A antagonists that were found among 12 highest-scoring compounds, as well as for compound #8 (CSC12), which is the highest scoring xanthine-like compound.
Figure 2.
Virtual screening with a crystal structure of adenosine A2A receptor. A) Examples of the binding modes for the top seven ranked ligands (in order of their ranking), as well as for the top ranked xanthine-based compound 8. Ligands are shown with yellow carbon atoms, while receptor side chains carbons are white. Ligand-receptor hydrogen bonds are indicated by green spheres. The A2AAR binding pocket is illustrated by molecular skin colored by properties (green – hydrophobic, red and blue – hydrogen bond acceptor and donor respectively). (B) Binding Scores and ranking for these compounds. C) Scatter plot of ICM binding scores for the whole benchmark set, with the experimentally validated ligands shown by larger red dots and decoys with small purple dots. The Y axis shows distribution of compound size as a number of heavy atoms.
The predicted docking poses for known A2AAR-selective antagonists suggest that these chemically diverse compounds bind similarly to the ZM241385 antagonist in the published crystal structure14. The common core interaction for all ligands involves aromatic stacking with the conserved Phe168(5.29) of the receptor and additional hydrophobic interactions with the conserved Ile274(7.39) and Leu249(6.51) side chains. Strong polar interactions are formed with the side-chain of the conserved Asn253(6.55), where the role of the hydrogen bond donor in most high-affinity ligands is played by the exocyclic amine group, with a notable exception of methylxanthine analogues (e.g. compound #8) that lack this group. The exocyclic amine group also forms a hydrogen bond to Glu169(5.30) side chain in the EL2, which is present in subtypes A2A, A2B and A1, but replaced by Val in the A3AR. Most of the A2A ligands also have an acceptor for H-bonding with Asn253(6.55) amide donor. This interaction pattern is consistent with the previous mutation data summarized in ref.38 and19 showing loss of affinity for Asn253(6.55) and Ile274(7.39) mutants, as well as with recent mutagenesis data18 showing the critical role of Phe168(5.29) and Leu249(6.51) for both agonist and antagonist binding.
The results of benchmark screening with the A2AAR crystal structure model, illustrated in Figure 2B,C shows very favorable binding scores for most A2AAR antagonists and high efficiency of the model in separating those antagonists from decoy compounds. As mentioned above, seven out of twelve top- ranking compounds are A2AAR antagonists. Only 6 out of 22 A2AAR ligands in the set were not predicted as A2AAR ligands based on their ICM binding score >-32.0 kJ/mol, including two xanthine analogues and two other compounds lacking the exocyclic amine.
Initial modeling of A1, A2B, and A3 adenosine receptor subtypes
The initial models of AR subtypes A1, A2B, and A3 were generated with standard homology modeling tools using sequence alignments shown in Figure SM1. The alignment demonstrates the high variability of the EL2 part between residues Leu141(4.62) and Cys166(5.27). The length and the disulfide bonding patterns in this region differ between the AR subtypes; moreover, this region is partially disordered in A2AAR crystal structure. Because this ambiguous portion of the EL2 is located far from the binding pocket and is unlikely to make a significant contribution to orthosteric ligand binding, it was excluded from the models. In contrast, the short extracellular loop EL3, ranging from 9 amino acid length in A2BAR to just 5 amino acids in A3AR, can be involved in subtype-specific binding properties, at least indirectly via a hydrogen bonding network with the side-chain of E169(5.30), as seen in the crystal structure14. To account for possible effects of EL3, the conformation of EL3 backbone and side chains was predicted using ICM global optimization algorithm. The standard homology building procedure also included the initial refinement of the modified side chains through their global energy optimization39.
The initial models were tested for their ability to discriminate between known subtype-selective compounds and decoys in the ligand benchmark (see Materials and Methods). The initial A2A model derived from the high-resolution crystal structure, demonstrated high compound recognition with AUC of 90% (NSQ_AUC=75). In stark contrast to A2A, the initial models of other subtypes had very poor performance. In particular, the model of the most distant subtype, A3, had a pronounced negative selectivity (NSQ_AUC = −11), suggesting some steric hindrance for ligand binding. Our analysis showed that this hindrance was associated with a specific rotamer state of the Val(5.30) side chain in the initial model.
Ligand-Guided Optimization of adenosine receptor subtype models
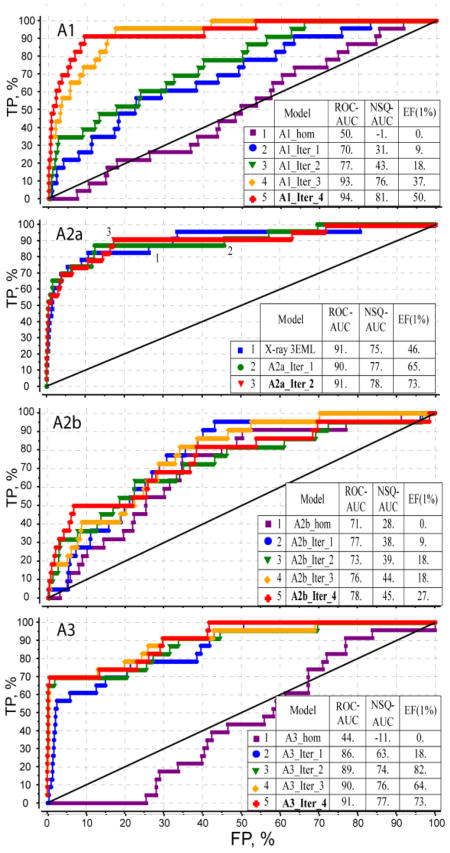
The initial models of the AR subtypes were subsequently optimized using an automated LiBERO algorithm as described previously for A2AAR15, see Methods. In each iteration step of the procedure, at least 100 distinct conformations of the binding pocket were generated with a stochastic sampling procedure (see Methods). The performance of each conformational model in separating the subtype selective ligands from decoy compounds was assessed by calculating ROC curves and corresponding values of linear AUC (ROC_AUC), normalized square root AUC (NSQ_AUC) and initial enrichment factors (EF1%).
Figure 3 presents the results of the optimization procedure for A1, A3 and A2B adenosine receptor subtypes. For the A2A optimization, which started with the high-performing crystal structure model, the procedure did not affect the ROC_AUC value and only modestly improved NSQ_AUC and initial enrichment EF(1%) after two optimization steps. In contrast, for A3, a change of the Val5.30 rotamer in the first iteration models led to a dramatic rise in the screening performance. The iterative improvement of VLS performance for A1 and A2B subtypes was more gradual, suggesting more complicated adjustments in the binding pocket involving multiple side chain movements and minor backbone adjustments. After four iterations sampling the total of 400 binding pocket conformations the screening performance apparently reached a plateau for all AR subtypes. The best optimized models represented by red ROC curves in Figure 3 show reasonable performance for A1 and A3 subtype screening models, which is on par with the X-ray-based A2A model. The lower performance of the A2B model may be explained by the fact that the A2B selective ligand set was comprised of xanthine derivatives that generally have less favorable scores in screening benchmarks (compare compound 8 in Figure 2).
Figure 3.
Ligand guided optimization of AR subtype models. Progression of VLS selectivity from an initial homology model through four iterations of the procedure is shown for each of the AR subtypes (only 2 iterations for A2AAR model). Tables show three different metrics of VLS performance of the models: linear ROC_AUC, normalized square root NSQ_AUC and enrichment factor at 1% dataset cutoff, EF(1%).
Note, that such distinct behavior of the optimization procedure for different AR subtypes reflects not only differences in the receptor structures, but also composition of the corresponding ligand sets, some of which are less diverse than others. For example, most of the A3 selective ligands in our set are based on the pyrazolo-triazolo-pyrimidine scaffold {Baraldi, 2003 #1728}, and therefore applicability of the optimized A3 model may be limited to this and some similar chemotypes. On the other hand, such single-scaffold optimized models can be especially useful in structure based SAR analysis and lead optimization for this specific scaffold, as they may reflect scaffold-specific induced fit in the binding pocket. One may also find that some of the compounds most dissimilar to the major chemotypes (e.g. mantri_l, mantri_r, veld_3 in our sets), which probably represent alternative binding modes or allosteric sites, and their presence in the training set does not improve the overall model quality. The construction of ligand sets therefore should be guided on practice not only by availability and quality of the ligand binding data, but also by the intended application of the optimized models in a specific drug discovery project.
Subtype Selectivity Profiles for the optimized AR models
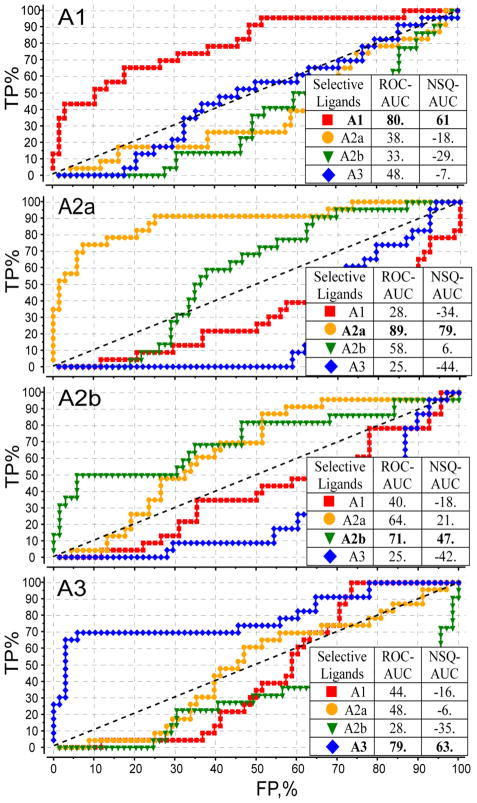
Ligand selectivity profiles were generated by docking a set of all 88 AR ligands from Figure SM1 into the optimized models A1, A2A, A2B and A3 adenosine receptor subtypes. Figure 4 shows individual selectivity ROC curves for these benchmarks. The ROC curves were calculated for each subtype model and for each subset of selective ligands. For example, the red curve in A1 model panel of Figure 4 uses the subset of A1 selective ligands as “positives” and the other 66 ligands as “negatives”. High ROC_AUC and NSQ_AUC values for this curve, shown in bold type, suggest that the optimized A1AR model is highly efficient in discriminating selective A1 antagonists from other adenosine receptor binders.
Figure 4.
Selectivity profiles for all four AR subtypes. Each ROC curve represents performance of an optimized conformation model of an AR subtype in discriminating subtype-selective antagonists from all other AR antagonists in the set. Tables show linear ROC_AUC and normalized square root NSQ_AUC for each of the curves, with results for each matching model-ligand subset shown in bold font.
Analysis of individual curves suggests a specific pattern of selectivity between the individual subtypes. For example, in both A2A and A2B panels, the separation between A2A and A2B ligand sets (yellow and green curves respectively), though still significant, is the smallest as compared to other subtypes. This is likely to reflect the fact that A2A and A2B receptor subtypes have the highest level of sequence conservation, especially in the binding pocket. This “convergence” of selectivity for A2A and A2B binding models was observed even despite the fact that ligand set for A2A and A2B (Figure SM1) are quite dissimilar chemically.
3D models suggest insight into structural basis of ligand subtype selectivity
The optimized models of four human AR subtypes with representative subtype-selective ligands are illustrated by Figure 5. Similar to the A2AAR models, all subtypes share core interactions with residues conserved across ARs, including aromatic stacking with Phe(5.29), hydrophobic interactions with conserved Ile(7.39) and Leu(6.51) side chains and strong hydrogen bonding with Asn(6.55). At the same time, model comparison suggests some subtype specific interactions that may serve as key selectivity determinants for individual AR subtypes.
Figure 5.
Predicted binding modes of subtype-specific antagonists in the corresponding optimized models of the AR subtypes. Residues critical for subtype selectivity are labeled in bold font. Side chain carbons are colored as in Figure 1. PDB files for all optimized models in complex with antagonists can be found in Supplementary Materials.
The A3 subtype is the most divergent from other ARs, with 10 out of 20 side chains in the ligand binding pocket unique for this subtype. The most important difference is a valine in position 5.30: in all other subtypes this position is occupied by a glutamate that plays an important role in high affinity ligand binding by forming a hydrogen bond with the unsubstituted exocyclic amine. With Val in this position, the A3AR loses this interaction and therefore allows bulky amine substitutions protruding towards the extracellular opening of the pocket. As mentioned before, some Val rotamer conformations can also partially block this opening, so conformational optimization is required to adequately represent specific ligand binding in A3. Importantly, there are two other mutations deeper in the pocket, H(6.52)S and N(5.42)S, in both cases to the smaller Ser side chain. These substitutions create an additional subpocket in A3 that can be exploited in the design of selective inhibitors. However, none of the A3 selective ligands in our set seem to take advantage of this sub-pocket, as they have a furan group similar to many other AR ligands.
The A1 subtype has a much closer homology to A2A and A2B subtypes, with only 4 side chain substitutions on the periphery of the binding pocket. The change in one of these positions, from Met(7.35) to a smaller Thr, apparently creates an additional sub-pocket in the loop region. This mutation, combined with slightly shifted conformation of E(5.30) in the optimized A1AR model makes possible accommodation of relatively small aliphatic and aromatic substitutions at exocyclic amine, which are characteristic of most A1-selective ligands. Interestingly, the shift in E(5.30) positions is absolutely required for binding of N-substituted compounds, but is not directly guided by amino acid differences between A1 and A2A pockets. One possible indirect explanation is in the modified EL3 loop, which is one amino acid shorter in A1 than in A2A; this difference may impose modified preferences for the position of His(6.66) side chain which plays a key role in stabilizing E(5.30). Other variations in the extracellular loops of the A1 sequence, M(5.28)E and L(7.32)S, are not predicted to be involved in binding of known A1 selective ligands. The engagement of these groups would require ligands that have additional hydrophilic extensions. This limited number of “reachable” mutations may explain why A1 selectivity is usually much harder to achieve than A3 selectivity.
The A2B subtype has only two deviations from A2A in the proximity of the ligand binding pocket. One of them is H(6.66)N in loop EL3, which leads to less stable conformation of E(5.30) in EL2 and thus reduced importance of an unsubstituted exocyclic amine in the ligands. Indeed, most ligands with A2B vs. A2A selectivity are based on chemical scaffolds that lack this amino group. The second residue substitution is L(7.32)K at the extracellular opening of the pocket. In the optimized model of A2B the basic Lys side chain is posed to interact with the acidic groups of the ligands. Some examples of such groups in known A2B-selective ligands are oxygen in the carbamoyl-methoxy-phenyl moiety of the antagonists40 or in sulphonyl moiety41. Interestingly, the same substituted carbamoyl-methoxy-phenyl motif was also found to confer A2B selectivity in agonists based on a different adenine-like scaffold42, which suggests general importance of this interaction for A2B ligand selectivity.
Conclusions
While high resolution 3D structure of adenosine A2A receptor represents an excellent template for virtual ligand screening for A2A antagonists, its utility in discovery and optimization of subtype selective adenosine receptor antagonists has yet to be shown. Our results suggest that AR subtype models directly built by homology with A2AAR crystal structure do not automatically guarantee recognition of subtype specific ligands and may have poor VLS performance. However, optimization of the models with the ligand-guided receptor optimization (LiBERO) methodology improves their VLS performance to the level comparable to that of the crystal structure. The analysis of the optimized models points to some of the non-conserved residues in the extracellular loop region which serve as determinants of ligand selectivity exploited by previously discovered subtype-selective ligands. The resulting models may serve as 3D selectivity panels to predict subtype selectivity of novel AR antagonists, or as structural templates for direct rational design of novel chemotypes of subtype selective AR ligands. The method can be applied to other families of closely related receptor subtypes, for which new 3D information is emerging from the structural genomics initiatives. The only intrinsic limitation of the technique is availability of quality ligand sets, which ideally have to comprise high affinity and high selectivity compounds, and be sufficiently diverse to avoid model overtraining. Fortunately, such ligand sets can be readily constructed for many clinically relevant GPCR targets, making the methodology applicable to rational structure-based design of subtype selective tools compounds and drug candidates.
Supplementary Material
Acknowledgments
We gratefully acknowledge support from the National Institutes of Health, R01-GM071872 and 1R43MH084374-01A1.
Abbreviations
- GPCR
G protein-coupled receptor
- AR
adenosine receptor
- A2AAR
subtype A2A
- TM
transmembrane
- EL2
extracellular loop 2
- VLS
virtual ligands screening
- PDB
protein databank
- RMSD
root mean square deviation
- ROC
receiver operating characteristic
- AUC
area under ROC curve
- NSQ_AUC
normalized square root AUC
- BPMC
biased probability Monte Carlo method
Footnotes
Supporting Information Available: Chemical structures of 88 subtype selective AR compounds and their docking poses in the optimized AR models of are presented in supplementary materials. This material is available free of charge via the Internet the journal website.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Tyndall JD, Sandilya R. GPCR agonists and antagonists in the clinic. Med Chem. 2005;1:405–421. doi: 10.2174/1573406054368675. [DOI] [PubMed] [Google Scholar]
- 2.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 3.Jacobson KA. Introduction to adenosine receptors as therapeutic targets. Handb Exp Pharmacol. 2009:1–24. doi: 10.1007/978-3-540-89615-9_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morelli M, Carta AR, Jenner P. Adenosine A(2A) Receptors and Parkinson’s Disease. Handb Exp Pharmacol. 2009:589–615. doi: 10.1007/978-3-540-89615-9_18. [DOI] [PubMed] [Google Scholar]
- 5.Sebastiao AM, Ribeiro JA. Adenosine receptors and the central nervous system. Handb Exp Pharmacol. 2009:471–534. doi: 10.1007/978-3-540-89615-9_16. [DOI] [PubMed] [Google Scholar]
- 6.Mustafa SJ, Morrison RR, Teng B, Pelleg A. Adenosine receptors and the heart: role in regulation of coronary blood flow and cardiac electrophysiology. Handb Exp Pharmacol. 2009:161–188. doi: 10.1007/978-3-540-89615-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Headrick JP, Lasley RD. Adenosine receptors and reperfusion injury of the heart. Handb Exp Pharmacol. 2009:189–214. doi: 10.1007/978-3-540-89615-9_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson CN, Nadeem A, Spina D, Brown R, Page CP, Mustafa SJ. Adenosine receptors and asthma. Handb Exp Pharmacol. 2009:329–362. doi: 10.1007/978-3-540-89615-9_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackburn MR, Vance CO, Morschl E, Wilson CN. Adenosine receptors and inflammation. Handb Exp Pharmacol. 2009:215–269. doi: 10.1007/978-3-540-89615-9_8. [DOI] [PubMed] [Google Scholar]
- 10.Fishman P, Bar-Yehuda S, Synowitz M, Powell JD, Klotz KN, Gessi S, Borea PA. Adenosine receptors and cancer. Handb Exp Pharmacol. 2009:399–441. doi: 10.1007/978-3-540-89615-9_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan L, Burbiel JC, Maass A, Muller CE. Adenosine receptor agonists: from basic medicinal chemistry to clinical development. Expert Opin Emerg Drugs. 2003;8:537–576. doi: 10.1517/14728214.8.2.537. [DOI] [PubMed] [Google Scholar]
- 12.Jacobson KA, Gao ZG. Adenosine receptors as therapeutic targets. Nat Rev Drug Discov. 2006;5:247–264. doi: 10.1038/nrd1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas GS, Thompson RC, Miyamoto MI, Ip TK, Rice DL, Milikien D, Lieu HD, Mathur VS. The RegEx trial: a randomized, double-blind, placebo- and active-controlled pilot study combining regadenoson, a selective A(2A) adenosine agonist, with low-level exercise, in patients undergoing myocardial perfusion imaging. J Nucl Cardiol. 2009;16:63–72. doi: 10.1007/s12350-008-9001-9. [DOI] [PubMed] [Google Scholar]
- 14.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katritch V, Jaakola VP, Lane JR, Lin J, Ijzerman AP, Yeager M, Kufareva I, Stevens RC, Abagyan R. Structure-Based Discovery of Novel Chemotypes for Adenosine A(2A) Receptor Antagonists. J Med Chem. 2010 doi: 10.1021/jm901647p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlsson J, Yoo L, Gao ZG, Irwin JJ, Shoichet BK, Jacobson KA. Structure-Based Discovery of A(2A) Adenosine Receptor Ligands. J Med Chem. 2010 doi: 10.1021/jm100240h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballesteros JA, Weinstein H. Integrated methods for the construction of three dimensional models and computational probing of structure–function relations in G-protein coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 18.Jaakola VP, Lane RJ, Lin JY, Katritch V, IJzerman AP, Stevens RC. Identification and Characterization of Amino Acid Residues Essential for Human A2A Adenosine Receptor:ZM241385 Binding and Subtype Selectivity. J Biol Chem. 2010 doi: 10.1074/jbc.M109.096974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cristalli G, Lambertucci C, Marucci G, Volpini R, Dal Ben D. A2A adenosine receptor and its modulators: overview on a druggable GPCR and on structure-activity relationship analysis and binding requirements of agonists and antagonists. Curr Pharm Des. 2008;14:1525–1552. doi: 10.2174/138161208784480081. [DOI] [PubMed] [Google Scholar]
- 20.Cavasotto CN, Orry AJ, Murgolo NJ, Czarniecki MF, Kocsi SA, Hawes BE, O’Neill KA, Hine H, Burton MS, Voigt JH, Abagyan RA, Bayne ML, Monsma FJ., Jr Discovery of novel chemotypes to a G-protein-coupled receptor through ligand-steered homology modeling and structure-based virtual screening. J Med Chem. 2008;51:581–588. doi: 10.1021/jm070759m. [DOI] [PubMed] [Google Scholar]
- 21.Reynolds KA, Katritch V, Abagyan R. Identifying conformational changes of the beta(2) adrenoceptor that enable accurate prediction of ligand/receptor interactions and screening for GPCR modulators. J Comput Aided Mol Des. 2009;23:273–288. doi: 10.1007/s10822-008-9257-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mantri M, de Graaf O, van Veldhoven J, Goblyos A, von Frijtag Drabbe Kunzel JK, Mulder-Krieger T, Link R, de Vries H, Beukers MW, Brussee J, Ijzerman AP. 2-Amino-6-furan-2-yl-4-substituted nicotinonitriles as A2A adenosine receptor antagonists. J Med Chem. 2008;51:4449–4455. doi: 10.1021/jm701594y. [DOI] [PubMed] [Google Scholar]
- 23.van Veldhoven JP, Chang LC, von Frijtag Drabbe Kunzel JK, Mulder-Krieger T, Struensee-Link R, Beukers MW, Brussee J, APIJ A new generation of adenosine receptor antagonists: from di- to trisubstituted aminopyrimidines. Bioorg Med Chem. 2008;16:2741–2752. doi: 10.1016/j.bmc.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 24.Colotta V, Catarzi D, Varano F, Cecchi L, Filacchioni G, Martini C, Trincavelli L, Lucacchini A. 1,2,4-Triazolo[4,3-a]quinoxalin-1-one: a versatile tool for the synthesis of potent and selective adenosine receptor antagonists. J Med Chem. 2000;43:1158–1164. doi: 10.1021/jm991096e. [DOI] [PubMed] [Google Scholar]
- 25.Baraldi PG, Tabrizi MA, Bovero A, Avitabile B, Preti D, Fruttarolo F, Romagnoli R, Varani K, Borea PA. Recent developments in the field of A2A and A3 adenosine receptor antagonists. Eur J Med Chem. 2003;38:367–382. doi: 10.1016/s0223-5234(03)00042-4. [DOI] [PubMed] [Google Scholar]
- 26.Stefanachi A, Brea JM, Cadavid MI, Centeno NB, Esteve C, Loza MI, Martinez A, Nieto R, Ravina E, Sanz F, Segarra V, Sotelo E, Vidal B, Carotti A. 1-, 3- and 8-substituted-9-deazaxanthines as potent and selective antagonists at the human A2B adenosine receptor. Bioorg Med Chem. 2008;16:2852–2869. doi: 10.1016/j.bmc.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Kalla RV, Zablocki J. Progress in the discovery of selective, high affinity A(2B) adenosine receptor antagonists as clinical candidates. Purinergic Signal. 2009;5:21–29. doi: 10.1007/s11302-008-9119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abagyan R, Batalov S, Cardozo T, Totrov M, Webber J, Zhou Y. Homology modeling with internal coordinate mechanics: deformation zone mapping and improvements of models via conformational search. Proteins. 1997;(Suppl 1):29–37. doi: 10.1002/(sici)1097-0134(1997)1+<29::aid-prot5>3.3.co;2-4. [DOI] [PubMed] [Google Scholar]
- 29.Cardozo T, Batalov S, Abagyan R. Estimating local backbone structural deviation in homology models. Comput Chem. 2000;24:13–31. doi: 10.1016/s0097-8485(99)00044-3. [DOI] [PubMed] [Google Scholar]
- 30.Abagyan RA, Totrov MM, Kuznetsov DA. Icm: A New Method For Protein Modeling and Design: Applications To Docking and Structure Prediction From The Distorted Native Conformation. J Comp Chem. 1994;15:488–506. [Google Scholar]
- 31.Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 32.Abagyan RA, Orry A, Raush E, Budagyan L, Totrov M. ICM Manual, 3.0. MolSoft LLC; La Jolla, CA: 2009. [Google Scholar]
- 33.Totrov M, Abagyan R. Flexible protein-ligand docking by global energy optimization in internal coordinates. Proteins. 1997;(Suppl 1):215–220. doi: 10.1002/(sici)1097-0134(1997)1+<215::aid-prot29>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
- 34.Halgren T. Merck molecular force field I-V. J Comp Chem. 1995;17:490–641. [Google Scholar]
- 35.Bursulaya BD, Totrov M, Abagyan R, Brooks CL., 3rd Comparative study of several algorithms for flexible ligand docking. J Comput Aided Mol Des. 2003;17:755–763. doi: 10.1023/b:jcam.0000017496.76572.6f. [DOI] [PubMed] [Google Scholar]
- 36.Schapira M, Totrov M, Abagyan R. Prediction of the binding energy for small molecules, peptides and proteins. J Mol Recognit. 1999;12:177–190. doi: 10.1002/(SICI)1099-1352(199905/06)12:3<177::AID-JMR451>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 37.Katritch V, Rueda M, Lam PC, Yeager M, Abagyan R. GPCR 3D homology models for ligand screening: lessons learned from blind predictions of adenosine A2A receptor complex. Proteins. 2010;78:197–211. doi: 10.1002/prot.22507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim SK, Gao ZG, Van Rompaey P, Gross AS, Chen A, Van Calenbergh S, Jacobson KA. Modeling the adenosine receptors: comparison of the binding domains of A2A agonists and antagonists. J Med Chem. 2003;46:4847–4859. doi: 10.1021/jm0300431. [DOI] [PubMed] [Google Scholar]
- 39.Cardozo T, Totrov M, Abagyan R. Homology modeling by the ICM method. Proteins. 1995;23:403–414. doi: 10.1002/prot.340230314. [DOI] [PubMed] [Google Scholar]
- 40.Stefanachi A, Nicolotti O, Leonetti F, Cellamare S, Campagna F, Loza MI, Brea JM, Mazza F, Gavuzzo E, Carotti A. 1,3-Dialkyl-8-(hetero)aryl-9-OH-9-deazaxanthines as potent A2B adenosine receptor antagonists: design, synthesis, structure-affinity and structure-selectivity relationships. Bioorg Med Chem. 2008;16:9780–9789. doi: 10.1016/j.bmc.2008.09.067. [DOI] [PubMed] [Google Scholar]
- 41.Borrmann T, Hinz S, Bertarelli DC, Li W, Florin NC, Scheiff AB, Muller CE. 1-alkyl-8-(piperazine-1-sulfonyl)phenylxanthines: development and characterization of adenosine A2B receptor antagonists and a new radioligand with subnanomolar affinity and subtype specificity. J Med Chem. 2009;52:3994–4006. doi: 10.1021/jm900413e. [DOI] [PubMed] [Google Scholar]
- 42.Baraldi PG, Preti D, Tabrizi MA, Fruttarolo F, Romagnoli R, Carrion MD, Cara LC, Moorman AR, Varani K, Borea PA. Synthesis and biological evaluation of novel 1-deoxy-1-[6-[((hetero)arylcarbonyl)hydrazino]-9H-purin-9-yl]-N-ethyl-beta-D-ribofuranuronamide derivatives as useful templates for the development of A2B adenosine receptor agonists. J Med Chem. 2007;50:374–380. doi: 10.1021/jm061170a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.