Abstract
Lectin conjugated, fluorescent silica nanoparticles (fNP) have been developed for carbohydrate based histopathology evaluations of epithelial tissue biopsies. The fNP platform was selected for its enhanced emissive brightness compared to direct dye labeling. Carbohydrate microarray studies were performed to compare the carbohydrate selectivity of the mannose recognizing lectin Concanavalin A (ConA) before and after conjugation to fluorescent silica nanoparticles (ConA-fNP). These studies revealed surprisingly low emission intensities upon staining with ConA-fNP compared to biotin-ConA / Cy3-strepavidin staining. A series of photophysical and biophysical characterizations of the fNP and ConA-fNP conjugates were performed to probe the low sensitivity from fNP in the microarray assays. Up to 1200 fluorescein (FL) and 80 tetramethylrhodamine (TR) dye molecules were incorporated into 46 nm diameter fNP, yielding emissive brightness’ 400 and 35 times larger than the individual dye molecules, respectively. ConA lectin conjugated to carboxylic acid surface modified nanoparticles covers 15–30% of the fNP surface. The CD spectra and mannose substrate selectivity of ConA conjugated to the fNP differed slightly compared to soluble ConA. Although, the high emissive brightness of fNP enhances detection sensitivity for samples with low analyte densities, large fNP diameters limit fNP recruitment and binding to samples with high analyte densities. The high analyte density and nearly two-dimensional target format of carbohydrate microarrays make probe size a critical parameter. In this application, fNP labels afford minimal sensitivity advantage compared to direct dye labeling.
Introduction
Silica nanoparticles containing multiple copies of fluorescent inorganic1 or organic2 dyes offer high emissive brightness3a–d, flexible surface modification3e–g and preparative simplicity. Biomolecule - fluorescent silica nanoparticle conjugates have been used in numerous imaging4 and diagnostic5 applications, in which they are reported to provide high detection sensitivity. Human cells are coated with carbohydrates in the form of glycoproteins, proteoglycans and glycolipids.6 These carbohydrates can be interrogated using lectins, a class of carbohydrate binding proteins.7,8 As many cancers alter glycoconjugate composition and density during disease progression, lectin staining provides useful characterization of biopsied tissue and of carcinoma cell lines.9 Strong correlations have been found between specific lectin’s affinity for cells and disease phenotypes such as metastatic potential.9,10 Lectin conjugated, emissive nanoparticles have been used to detect glycosylation differences between non-cancerous and cancerous cells.11 Encouraged by these reports, we developed lectin-conjugated fluorescent silica nanoparticles (fNP) for quantitative histopathology evaluations of epithelial tissue biopsies.
Lectin histopathology is usually carried out using a qualitative scoring scheme based on staining intensity. To perform quantitative biopsy studies, emission intensities from lectin-modified fluorescent silica nanoparticles (lec-fNP) and from other fluorescent cell markers must be evaluated to determine glycoconjugate levels as a function of, and as an indicator of, disease state. In order to ascertain how lectin conjugation to fNP alters carbohydrate selectivity, carbohydrate microarray12 studies using the lectin Concanavalin A conjugated to fNP (ConA-fNP) and to biotin (biotin-ConA) were performed. Given prior reports of fNP emissive brightness, we were surprised by the consistently weaker emission intensities obtained from microarrays treated with ConA-fNP relative to those treated with biotin-ConA followed by Cy3-streptavidin. A series of photophysical and biophysical characterizations of the fNP and ConA-fNP conjugates were performed to understand the origin of the unexpectedly low sensitivity of lec-fNP conjugate in these assays. The results and analyses identify previously unreported constraints on the sensitivity advantage conferred by fluorescent silica nanoparticles.
Experimental Procedures
Materials
All commercially available reagents were used without further purification. Fluorescein isothiocyanate (FITC), tetramethylrhodamine isothiocyanate (TRITC), 3-aminopropyl-triethoxysilane (APTS), cyclohexane, polyoxyethylene(5)nonylphenylether (Igepal CO 520), lectin from Canavalia ensiformis (ConA) and N-hydroxysuccinimide (NHS) were purchased from Sigma-Aldrich. Biotinylated ConA was purchased from Vector Labs (Burlingame, CA). Ammonium hydroxide (NH4OH) (28.0–30.0%) was purchased from VWR. N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) was purchased from Fluka. Tetraethoxysilane (TEOS) and N-(trimethoxysilyl-propyl) ethylene-diamine triacetic acid trisodium salt (45% in water) (EDATAS) were purchased from Gelest.
Instruments
TEM images were recorded on a Philips EM420 Transmission Electron Microscope at 120keV. Fluorescent spectra were recorded on a Spex Fluorolog III spectrofluorometer. UV-Vis spectra were recorded on an Agilent 8453 UV-visible diode-array spectrometer. Light scattering measurements were performed on a Zetasizer Nano ZS particle analyzer. Carbohydrate microarray staining images were recorded on a GenePix®4000B microarray scanner with 532nm and 635nm dual lasers. CD spectra were recorded on a Jasco J-815 spectrometer using 0.2mm quartz cuvettes (50 nm/min, 1 nm bandwidth).
Preparation of fluorescent silica nanoparticles
Fluorescent silica nanoparticles were prepared using the inverse micelle procedure described by Yi et. al..13 The covalent adduct of 3-aminopropyltriethoxysilane (APTS) with fluoresceinisothiocyanate or tetramethylrhodamineisothiocyanate was used to incorporate dye into the silica matrix. Solutions of dye-silane conjugate were prepared by reacting 5mg FITC or 2.5 mg TRITC with 16 μL APTS (> 5:1 molar ratio) in 1mL solvent (ethanol or tetrahydrofuran (THF)) for twelve hours in a microwave vial sealed with a Teflon lined cap.3f Fluorescent nanoparticles were prepared by adding 40μL of the dye-silane conjugate solution and 120 μL TEOS to a water-in-oil micelle system formed from 480μL of Igepal CO520 surfactant and 240μL ammonium hydroxide in 9mL cyclohexane. Nanoparticle assembly proceeded at room temperature for 24 hours in a 20mL glass vial, tightly sealed with a Teflon lined cap. Next, the nanoparticle surface was capped by adding 40μL of TEOS to the micelle solution. After 2.5hr, 96μL of EDATAS solution was added to the micelle solution and vial was stirred at room temperature for 24 hours. Incorporation of each EDATAS molecule at the NP surface introduces three carboxylic acid groups. The f(dye)NP particles were isolated by addition of 10ml ethanol and centrifugation. Surfactant and unincorporated dye-silane conjugate were removed by suspending the nanoparticles in organic solvent followed by centrifugation and removal of the supernatant (4 washes with cyclohexane, 2 washes with acetone). The washed f(dye)NP samples were air dried. Particle optical properties were determined as a function of the concentration, time and solvent in which dye-silane conjugate was added to the growing nanoparticle solution. For the dye concentration studies, 40 μL of one of the following dye-conjugate solutions was added immediately after adding TEOS (0 hour): 5mg dye in either 1mL solvent (1x), 0.33 mL solvent (3x), 0.2 mL solvent (5x) or 0.13 mL solvent (8x). For the time delay experiments, 1x dye-silane conjugate was added 0, 2.5, 5 or 8 hours after initiation of NP growth (TEOS addition). Ethanol and THF were tested as solvents for the preparation and addition of the dye-silane conjugate.
Nanoparticle Optical Studies
1mg/mL f(dye)NP solutions in PBS (pH 7.3) were prepared by dissolving 10mg f(dye)NP in 10mL PBS solution. Absorbance spectra of these f(dye)NP solutions and of identically prepared, dye-free silica nanoparticle solutions dye were recorded. The dye absorbance contribution to the f(dye)NP UV-Vis spectrum was obtained by subtracting the light scattering spectrum of the corresponding dye-free silica nanoparticle solution. The absorbance and scattering contributions at 493nm are reported in Table 2 for the fluorescein nanoparticles, f(FL)NP. Fluorescent spectra and intensities of matched absorbance_(scattering corrected) f(FL)NP and FL-APTS solutions were recorded with excitation at 493 nm (0.5 mm excitation and emission slits; 2 nm resolution). Ratios of the f(FL)NP and FL-APTS fluorescence intensities were determined as the ratios of the integrated fluorescence bands from the absorbance matched solutions. Identical experiments were performed on absorbance matched TRITC nanoparticle, f(TR)NP, and TR-APTS solutions that were excited at 544 nm.
Table 2.
Optical Properties in PBS (7.3 pH) solution
| Samplea | Absorbance (493 nm)b | Fluorescence | ||||
|---|---|---|---|---|---|---|
| Dye amount-solvent-add time | FWHM (nm) | εABS(λMAX) | εSCAT(λMAX) | Dyes/f(FL)NP | Integrated Emission fNP/FITC | Brightness |
| 1x-Et-0 hr | 36 | 0.12 | 0.05 | 103 ± 33 | 0.87 | 89 ± 33 |
| 1x-Et-2.5 hr | 34 | 0.10 | 0.09 | 88 ± 31 | 0.82 | 72 ± 29 |
| 1x-Et-5 hr | 34 | 0.05 | 0.11 | 69 ± 24 | 0.79 | 55 ± 22 |
| 1x-Et-8 hr | 35 | 0.10 | 0.09 | 95 ± 40 | 0.81 | 77 ± 36 |
| 1x-T-0hr | 36 | 0.14 | 0.01 | 120 ± 36 | 0.77 | 92 ± 33 |
| 1x-T-2.5 hr | 35 | 0.14 | 0.02 | 113 ± 23 | 0.83 | 94 ± 24 |
| 1x-T-5 hr | 35 | 0.11 | 0.01 | 100 ± 17 | 0.85 | 85 ± 18 |
| 1x-T-8-hr | 35 | 0.09 | 0.10 | 111 ± 41 | 0.85 | 94 ± 39 |
| 8x-Et-0 hr | 51 | 1.50 | 0.08 | 1150 ± 170 | 0.33 | 390 ± 61 |
| 8x-Et-2.5 hr | 49 | 1.30 | 0.06 | 1070 | 0.39 | 420 |
| 8x-Et-5 hr | 50 | 1.09 | 0.11 | 880 | 0.39 | 350 |
| 8x-Et-8 hr | 50 | 1.08 | 0.07 | 830 | 0.41 | 340 |
| FL-APTS | 33 | - | - | - | - | 1 |
1x = 0.2 mg FL/40μL solvent; 8x = 1.6mg FL/40μL solvent; Et = ethanol, T = THF
molar extinction coefficient (absorption or scattering, M−1 cm−1) divided by fNP MW (~ 6×107 g/mol)
ConA conjugation
A solution of 0.5mg/mL f(dye)NP in MES buffer (pH 5.2) was stirred for 12 hours to achieve complete nanoparticle dissolution prior to surface activation. 5mg EDC and 1.2mg NHS were added to activate the EDATAS carboxylic acids and the f(dye)NP solution was gently rocked at room temperature for 20min. The fNPs were isolated by centrifugation, redissolved in 0.3ml PBS (pH 6.6, 0.1mM CaCl2, 0.1mM MnCl2) and then added dropwise to 1mL of a 0.5mg/mL solution of ConA in PBS (pH 6.6, 0.1mM Ca2+, 0.1mM Mn2+) solution. The mixture was rocked at room temperature for 1h. ConA-f(dye)NP were isolated by centrifugation and then redissolved in 1ml PBS (pH 6.6, 0.1mM CaCl2, 0.1mM MnCl2).
Carbohydrate Microarray Profiling
Arrays were fabricated as reported previously.12e–g The array contained 108 total array components, including 79 neoglycoproteins, 24 glycoproteins, and 5 controls. A full list of array components and their abbreviations is provided in the Supporting Information. Biotinylated lectins were assayed as described previously, with minor modifications.12g Briefly, slides were blocked with 3% BSA/PBS for 2 hours, incubated for 2 hours with biotinylated ConA in PBS containing 3% BSA, 0.1 mM CaCl2, and 0.1 mM MgCl2, washed, and then incubated for 1 hour in the dark with a 2 μg/mL solution of Cy3-labeled streptavidin (Zymed Laboratories of Invitrogen; Carlsbad, CA; 2–4 Cy3′s per streptavidin) in PBS containing 3% BSA, 0.1 mM CaCl2, and 0.1 mM MgCl2. Slides were washed 5 times with PBS containing 0.1 mM CaCl2 and 0.1 mM MgCl2, dried, and then scanned using a Genepix 4000B microarray scanner at 10 μm resolution (Molecular Devices Corporation, Union City, CA). Image analysis was carried out with Genepix Pro 6.0 analysis software (Molecular Devices Corporation, Union City, CA). Spots were defined as circular features with maximum diameter of 100 μm and a minimum diameter of 70 μm. The background-corrected median feature intensity, F532median-B532, was used for each spot. Spots corresponding to a given array component were averaged to give a single RFU value using Microsoft Excel. All spots with signal intensities below 50 were set to 50. Binding of ConA-f(dye)NP was evaluated using a simplified procedure. Briefly, slides were blocked with 3% BSA/PBS for 2 hours, incubated for 2 hours in the dark at room temperature with nanoparticles diluted in PBS containing 3% BSA, 0.1 mM CaCl2, and 0.1 mM MgCl2, washed 5 times with PBS containing 0.1 mM CaCl2 and 0.1 mM MgCl2, dried, and then scanned and processed as above.
Results and Discussion
Carbohydrate Microarray Studies
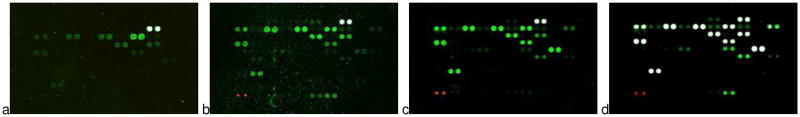
Figure 1 displays Genepix scans of carbohydrate microarrays (103 targets, in duplicate) incubated with ConA-f(TR)NP at 0.1 and 0.5 mg/mL (1×1012 and 5×1012 f(TR)NP/mL) and biotin-ConA at 0.5 μg/mL and 5 μg/mL 6×1012 and 60×1012 biotin-ConA/mL).14 Biotinylated ConA was detected with Cy3-labeled streptavidin (SA). Roughly 20 carbohydrate targets are labeled by the ConA conjugates, albeit with different relative intensities (Table 1). Biotin-ConA labels Man-T, Man7D1 and Man8D1D3 most intensely, Man9 and Man7D3 slightly less and Man5, Man3 and Man6 significantly less. ConA-f(TR)NP labels Man7D1 and Man8D1D3 most intensely. Man5, Man7D3 and Man6 acquire significantly less fluorescence intensity. The rather weak labeling of Man-T and Man9 by ConA-f(TR)NP contrasts with these carbohydrates’ strong labeling by biotin-ConA. ConA conjugated to nanoparticles or to biotin retains its known preference for mannose, albeit with slightly different mannose derivative selectivities from the different conjugates.
Figure 1.
Carbohydrate microarray scans: a) (left) 0.1 mg ConA-f(TR)NP/mL; b) 0.5 mg ConA-f(TR)-NP/mL; c) 0.5 μg biotin-ConA/mL; d) 5 μg biotin-ConA/mL. The rightmost pair of white spots in the upper row of each scan are Cy3-BSA controls. The pair of red spots in b) – d) are Cy5-BSA controls.
Table 1.
Normalized Fluorescence Intensities from Carbohydrate Microarrays
| Targeta | Typeb | Biotin-ConA 500 ng/mL | ConA-f(TR)NP 0.5 mg/mL |
|---|---|---|---|
| ManT | carb | 100c | 37 |
| Man7D1 | carb | 98 | 100c |
| Man8D1D3 | carb | 95 | 96 |
| Man9 | carb | 89 | 23 |
| Man7D3 | carb | 87 | 49 |
| Man5 | carb | 66 | 67 |
| Man3 | carb | 48 | 28 |
| Tgl | GP | 39 | 9 |
| CEA | GP | 31 | 29 |
| Man6 | carb | 16 | 48 |
See Supporting Information for these targets’ full names.
carb = carbohydrate, GP = glycoprotein
Counts scaled to 100. All other data for the same ConA-conjugate scaled by the same factor.
A tenfold increase of the biotin-ConA incubation concentration (0.5→5.0 μg/mL: Fig. 1c→1d) produced large intensity increases for all active carbohydrate targets. By contrast, a fivefold increase of the ConA-f(TR)NP incubation concentration (0.1 → 0.5 mg/mL: Fig. 1a→1b) produced moderate intensity increases for the “low intensity” carbohydrate targets in 1a and small intensity increases for the “high intensity” carbohydrate targets in 1a. The latter, small intensity increases suggest that these carbohydrate targets’ fluorescence is “saturating” for incubation with ConA-f(TR)NP. The “saturated” carbohydrate targets’ fluorescence intensities for incubation with 0.5 mg ConA-f(TR)NP/mL (Fig. 1b) are comparable to those produced by incubation with 0.5 μg biotin-ConA/mL (Fig. 1c) and much weaker than the intensities produced by incubation with 5 μg biotin-ConA/mL (Fig. 1d). These observations indicate that fluorescence conferred by incubation with ConA-f(TR)NP saturates at a lower intensity than fluorescence conferred by incubation with biotin-ConA.
Langmuir models have been used to investigate biomolecule binding to microarrays.15 The saturation characteristics of biotin-ConA and ConA-f(TR)NP were analyzed using a simple Langmuir model (eqn 1), which relates a carbohydrate target’s fluorescence intensity to the effective binding constant for the carbohydrate target/lectin pair, KC, and the lectin concentration, where IntC([Lectin]) is the observed target fluorescence
| (eqn 1) |
intensity for a specific lectin concentration and IntMAX,C is the carbohydrate target’s maximum possible fluorescence intensity for that lectin-dye conjugate. This simple Langmuir model predicts a non-linear dependence on KC of the relative fluorescence intensities for carbohydrate targets at two different lectin concentrations (eqn 2); where IntC([Lectin]) is the target fluorescence intensity at a low lectin concentration and
| (eqn 2) |
IntC(N × [Lectin]) is the same target’s fluorescence intensity at an N-fold higher lectin concentration. If KC × [Lectin] ≪ N−1 at the low lectin concentration, the target’s fluorescence intensity will be N-fold greater at the N-fold higher lectin concentration. If KC × [Lectin] ≫ 1 at the low lectin concentration, the target’s fluorescence intensity will be the same at the N-fold higher lectin concentration.
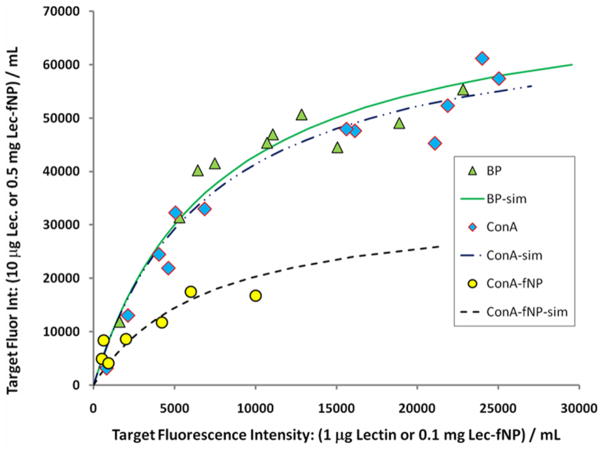
The numerous carbohydrate structures in a microarray provide a wide range of effective binding constants for the incubating lectin. An xy plot of the set of carbohydrate targets’ intensities at low (x) and high (y) lectin concentrations should vary with KC according to eqn. 2. Figure 2 presents these plots for the 7 – 12 most fluorescent carbohydrate targets upon incubation with the fluorophore-lectin conjugates biotin-ConA, ConA-f(TR)NP and biotin-BP.16 The Langmuir model was used to estimate17 the maximum fluorescence intensity (IntMAX,C) for each of the carbohydrate targets, based on its intensities at the two lectin concentrations. The average of these targets’ IntMAX,C values for each fluorophore-lectin conjugate, <IntMAX,C>, was used in the simple Langmuir model to calculate the lines in Figure 2. The data and the calculated curves indicate that incubation of the carbohydrate microarray with ConA-f(TR)NP generates a maximum target fluorescence intensity, 27400 counts, that is less than half those generated by incubation with either biotin-lectin conjugate (67800 and 63600).
Figure 2.
Fluorescence intensities from incubation of carbohydrate microarrays with lectin conjugates. X-axis data: intensity from incubation at lower lectin concentration (1μg/mL biotin-lectin or 0.1 mg/mL lectin-fNP) ( = biotin-BP [67800],
= biotin-BP [67800],  = biotin-ConA [63600],
= biotin-ConA [63600],  = ConA-f(TR)NP [27400].) Y-axis data: intensity from incubation at higher lectin concentration (10 μg/mL biotin-lectin or 0.5 mg/mL lectin-fNP). The mean value (σ ~ ± 30%) of the maximum fluorescence counts that each lectin conjugate can impart to a carbohydrate microarray target is indicated in [brackets] (see text).
= ConA-f(TR)NP [27400].) Y-axis data: intensity from incubation at higher lectin concentration (10 μg/mL biotin-lectin or 0.5 mg/mL lectin-fNP). The mean value (σ ~ ± 30%) of the maximum fluorescence counts that each lectin conjugate can impart to a carbohydrate microarray target is indicated in [brackets] (see text).
Given literature reports3a–d of fluorescent silica nanoparticles’ high brightness relative to single dye molecules, the weaker fluorescence produced by microarray staining with ConA-f(TR)NP was surprising.18 Microarray studies characterize the selectivity and dosing behavior of the lectin-f(TR)NP but provide no characterization of the attached lectin or of the f(TR)NP in the conjugate. f(dye)NP brightness and the amount, activity and stability of lectin in the conjugate must be known in order to understand the saturation behavior observed in Figure 2 and to perform quantitative histology applications. The following studies were performed to (i) quantify and maximize fNP brightness, (ii) characterize the lectin component conjugated to fNPs and (iii) elucidate the origin of the low fluorescence signal imparted to carbohydrate microarray targets by lectin-f(dye)NP conjugates..
Nanoparticle Optical Properties
Absorbance and scattering spectra, absorption and scattering extinction coefficients, fluorescence spectra, relative emission quantum yields and fNP brightness relative to single dye molecules were determined as a function of dye amount added to the inverse micelles, the solvent used for dye addition and the time delay of dye addition. FITC and its silane adduct (FL-APTS) were more soluble in ethanol and tetrahydrofuran (THF) than TRITC and its adduct (TR-APTS), allowing greater variation of dye concentration in the effort to maximize f(dye)NP brightness.
Characterization of dye properties within the nanoparticles, such as band width (FWHM), number of dyes per NP and NP brightness, required corrections for light scattering by the nanoparticles. Light scattering “line shapes” (supporting information) were recorded from NP samples prepared identically to the f(dye)NP, except for the absence of dye in the APTS/ethanol or THF solution added to the inverse micelle preparation. Scattering line shapes were subtracted from raw f(FL)NP spectra to obtain the FL absorbance line shape. Addition of 1 equivalent (1x) of FL-APTS in ethanol (0.2 mg FL/40 μL) during the first five hours of NP synthesis (Table 2: 1x-Et-0 hr – 1x-Et-5 hr) generated 5 – 10 times more extinction via light scattering at 493 nm (the FL absorbance maximum) than addition in THF (Table 2: 1x-T-0 hr – 1x-T-5 hr). The f(FL)NP diameter determined by TEM, 46±3 nm (σ=6%), was the same whether FL-APTS was added in ethanol or THF. However, the mean f(FL)NP diameter determined by dynamic light scattering (DLS) in DI water was larger when FL-APTS was added in ethanol (94 nm) compared to when it was added in THF (46 nm).19 Filtration assays using fixed pore diameter filters also revealed larger f(FL)NP when FL-APTS was added in ethanol.20 The TEM diameter, DLS diameter and filtration assay data can be reconciled if the f(FL)NP prepared with FL-APTS in ethanol aggregate more strongly than f(FL)NP prepared using THF. Closely packed nanoparticles were observed in all TEM images (supporting information). The strength of fNP aggregation was not discernable from the TEM data. Moreover, close packing did not alter TEM determined diameters. By contrast, f(FL)NP aggregation would produce larger DLS diameter values. More extensive aggregation of f(FL)NP prepared using ethanol similarly explains their larger scattering extinction coefficients. The undesirable f(FL)NP extinction from scattering worsened with increased time between initiation of NP growth and FL-APTS addition. The least aggregated f(FL)NP are prepared by adding FL-APTS in THF at the onset of NP growth.
f(FL)NP solutions in water or PBS exhibited absorption maxima within 1 nm of 493 nm, the maximum of the FL–APTS adduct in PBS. After correcting for NP light scattering, the absorption FWHM of f(FL)NP prepared using 1x FL-APTS in THF or ethanol was 1 - 3 nm wider than the FWHM of FL-APTS in PBS. f(FL)NP prepared using the same volume (40 μL) of an 8-fold more concentrated FL-APTS solution in ethanol (Table 2: 8x-Et-0 hr – 8x-Et-8 hr) yielded scattering corrected absorption FWHM that were 50% wider than for FL-APTS in PBS. The additional broadening in the 8x samples was equally distributed toward the red and blue edges of the absorption band.
The average number of FL molecules incorporated per f(FL)NP was determined from scattering corrected absorbance spectra of 10 mg f(FL)NP/10 mL PBS solutions (Table 2). The analyses assumed that all f(FL)NP have 46 nm diameters and a density of 1.9 g/mL.21 The extinction coefficient of FL within f(FL)NP and FL-APTS are the same.22 The number of FL per NP was determined as a function of (i) the solvent used to add 1x FL-APTS, (ii) the time delay between initiation of NP growth and FL-APTS addition23 and (iii) the FL-APTS concentration in the addition solvent. Averaged over all 1x studies, addition of FL-APTS in THF yielded NP with an average of 110 fluorescein molecules, whereas FL-APTS addition in EtOH yielded NP with 90 fluoresceins.24a Addition of 1x of FL-APTS 0, 2.5, 5 or 8 hours after initiating NP growth had minimal impact on FL incorporation, whether dye was added using THF or EtOH. Use of 8-fold more concentrated FL-APTS aliquots (8x in EtOH) greatly increased the number of FL incorporated per nanoparticle (1150 FL per NP at 0 hr). Delayed addition of the 8-fold more concentrated FL-APTS aliquots markedly reduced FL incorporation into the NP (830 Fl NP at 8 hr).25 Both the number of FL per NP and the percentage of added FL-APTS incorporated into NP increased with increasing concentration of the FL-APTS solution.25b Addition of 1x FL-APTS in THF produced slightly higher FL incorporation and fNP that were less prone to aggregate. However, the higher solubility of FL-APTS in EtOH enabled preparation of f(FL)NP with a much larger number of fluorophores. Adding more than 40 μL EtOH produced larger and more highly aggregated fNP. Thus, 0 hr addition of 8x FL-APTS in EtOH yielded 46 nm diameter fNP with the highest fluorescein concentration (~ 40 mM).
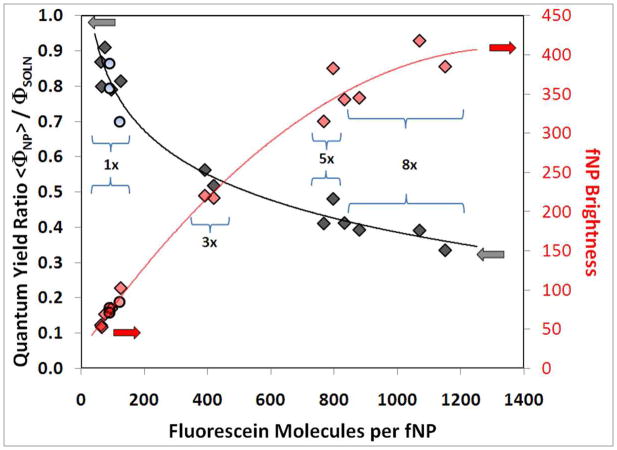
f(FL)NP brightness may be defined as the number of FL molecules per NP times the average emission quantum yield of FL molecules in the NP, <ΦNP>, divided by the emission quantum yield of FL molecules in solution, ΦSOLN. With this definition, a dye molecule in solution has a brightness of 1. The ratio of emission quantum yields, <ΦNP>/ΦSOLN, was determined as the ratio of integrated emission intensities from matched absorption (scattering corrected) samples of f(FL)NP and FL-APTS in PBS (Table 2, Figure 3-left axis). f(FL)NP containing 100 FL exhibited emission quantum yields that were nearly as large (80%) as from FL-APTS in PBS. By contrast, the emission quantum yields from f(FL)NP containing 1000 or more FL were only 35% as large as for FL-APTS in PBS. Decreasing emission efficiency with increasing FL concentration26 produced a sub-linear increase of f(FL)NP brightness. 46 nm diameter f(FL)NP with more than 800 FL are, at most, 400 times brighter than a single FL molecule.
Figure 3.
Emission quantum yield and brightness of 46 nm diameter f(FL)NP relative to FL-APTS in PBS buffer. Left axis: relative emission quantum yield,  = FL-APTS added in EtOH, ○ = FL-APTS added in THF. Right axis: f(FL)NP brightness relative to FL-APTS,
= FL-APTS added in EtOH, ○ = FL-APTS added in THF. Right axis: f(FL)NP brightness relative to FL-APTS,  = FL-APTS added in EtOH,
= FL-APTS added in EtOH,  = FL-APTS added in THF. The solid lines are provided as guides to the eye.
= FL-APTS added in THF. The solid lines are provided as guides to the eye.
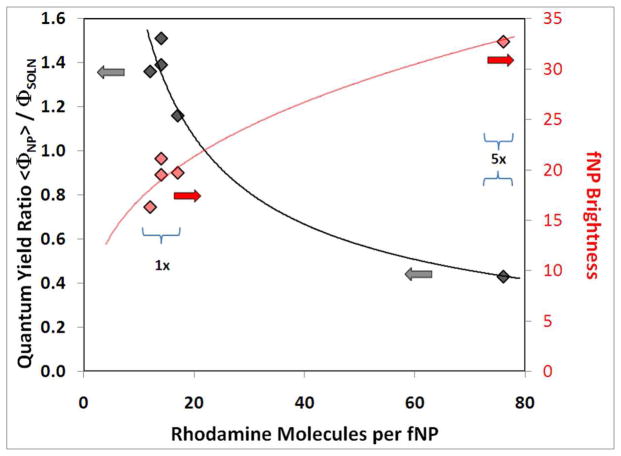
Analogous studies of tetramethylrhodamine incorporation, relative emission quantum yield and brightness were performed for f(TR)NP prepared from 40 μL of 1x (2.5 mg TR/mL EtOH) and 5x (12.5 mg TR/mL EtOH) solutions. Analyses of scattering corrected absorption spectra (TR absorption maximum at 544 nm) indicated fewer than 20 TR molecules per NP in 1x samples and fewer than 80 TR molecules per NP in 5x samples (Figure 4). The TR emission quantum yields from f(TR)NP with fewer than 20 dye molecules were 20 – 50% larger than from TR-APTS in PBS. The TR emission quantum yields from f(TR)NP containing ~ 80 dye molecules were ~ 60% smaller than from TR-APTS in PBS buffer. A previous study reported a 2 – 3 fold increase of TR emission quantum yield from 32±2 nm diameter silica nanoparticles containing fewer than 9 TR molecules.23b Thus, two studies find increased emission efficiency from low density TR within a silica NP matrix compared to TR in aqueous buffer (albeit with different enhancement factors).
Figure 4.
Emission quantum yield and brightness of 46 nm diameter f(TR)NP relative to TR-APTS in PBS. Left axis: relative emission quantum yield,  = TR-APTS added in EtOH. Right axis: f(TR)NP brightness relative to TR-APTS,
= TR-APTS added in EtOH. Right axis: f(TR)NP brightness relative to TR-APTS,  = TR-APTS added in EtOH. The solid lines are provided as guides to the eye.
= TR-APTS added in EtOH. The solid lines are provided as guides to the eye.
The key result from Figure 4 is that 5x f(TR)NP are about 33 times brighter than a single TR molecule. Following conjugation to ConA, these 5x f(TR)NP were used in the carbohydrate microarray studies. The densest, 2D-hexagonal packing of these 5x ConA-f(TR)NP on a flat surface yields a dye surface density of 0.041 TR/nm2 and a “2D-brightness” of 0.018 TR equivalents/nm2.27 For comparison, the ConA tetramer is tetrahedral, with less than 9nm separation between corners.28 Assuming each ConA tetramer binds a Streptavidin with three Cy3 dyes, the densest, 2D-hexagonal packing of ConA tetramers29 on a flat surface yields a dye surface density of 0.083 Cy3/nm2. This over-simplified model indicates that the packing-limited, surface-dye density from directly labeled ConA is two times higher than from the 5x ConA-f(TR)NP. Including the 2.6 – 3.7 fold more favorable spectral properties for TR relative to Cy3 in the Genepix measurement18 and the 2.5-fold lower brightness of TR in f(TR)NP compared to solution, the dense packing model predicts the maximum ConA-f(TR)NP signal is 0.32 to 0.46 times as large as the maximum biotin-ConA signal. This is in good agreement with the value of 0.40 for the ConA-f(TR)NP: biotin-ConA ratio of maximum target intensities determined from the microarray data (27400: 67800).
In all likelihood, neither the carbohydrates within the microarray targets nor the bound ConA-f(TR)NP or biotin-ConA assemble densely packed surfaces. In addition, carbohydrate placements within the targets are unlikely to define a flat surface. Although a “roughened” target surface affords higher total surface area, the accessible surface area decreases with increasing probe size. Carbohydrates lying in surface “nooks and crannies” will be less accessible to the 46 nm diameter ConA-f(TR)NP than to the < 10 nm diameter biotin-ConA. Thus, surface roughness at the 10’s of nm length scales would reduce further the signal from ConA-f(TR)NP compared to biotin-ConA. Binding of small ConA-f(TR)NP aggregates or small biotin-ConA aggregates30 to the carbohydrate targets would alter the relative emission intensities from the two types of ConA conjugates. If the Genepix instrument measured fluorescein emission, a similar line of reasoning predicts that incubation of microarrays with 8x ConA-f(FL)NP would produce a maximum target fluorescence intensity that is two to three times larger than from biotin-ConA incubated with Streptavidin bearing three fluorescein dyes.31
Nanoparticle Lectin characterization
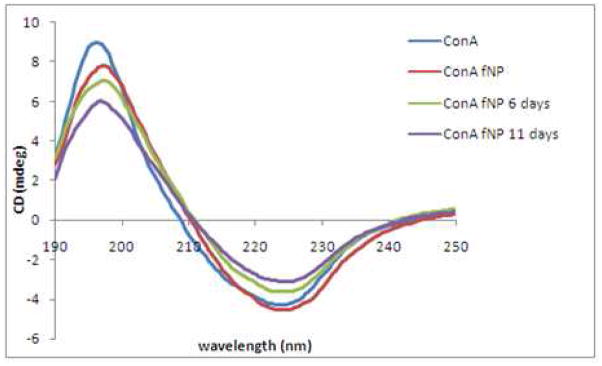
Circular dichroism spectroscopy (CD) was used to characterize ConA loading and structure after conjugation to fNP. Significant absorbance by NaCl in PBS made the CD spectra unusable in the 190–200nm region. Thus, native ConA and ConA-fNP were dissolved in 0.01mM phosphate buffered solution (pH = 6). The secondary structure content of native ConA and ConA-fNP were analyzed using the CDSSTR32 algorithm. Both the ConA33 and ConA-fNP exhibit atypical β-sheet band shapes (Figure 5). The CD of ConA exhibited negative and positive extrema at 223nm and 196nm, respectively, and a positive to negative crossover at 208.2nm.33 The negative and positive extrema in the CD of ConA-fNP appeared at 224 and 197 nm, respectively, with the crossover at 210 nm. These values for ConA-fNP are red shifted 1 – 2 nm from the native ConA values. CDSSTR analyses of the CD spectral shapes (Table 4) revealed similar secondary structure compositions of ConA and freshly prepared ConA-fNP, with 6–7% α-helix and 35–40% β-sheet. Storage of the ConA-fNP in PBS at 4 °C for a period of eleven days resulted in slight changes to the CD spectra, which CDSSTR analyses ascribed to decreasing amounts of α-helix. The majority of amino acid residues that contribute to carbohydrate binding are located in α-helix and turn regions.34 Changes in these regions may contribute to the time dependent loss of ConA-fNP activity.35
Figure 5.
CD spectra of ConA (43 μg/mL) and ConA-fNP (0.85 mg/mL) in 0.01 M phosphate buffer (pH 6.3) without NaCl. The CD spectra of ConA-fNP in this buffer evolve over 12 days. Low NaCl concentrations hasten the loss of ConA activity.36b
Table 4.
CDSSTR Secondary Structure Analyses of 43μg/ml ConA and 0.85mg/ml ConA-fNP
| α-helix (%) | β-sheet (%) | Turns(%) | Unordered (%) | |
|---|---|---|---|---|
| ConA | 6.8 | 35.4 | 23.6 | 33.6 |
| ConA fNP | 6.4 | 40.4 | 23.4 | 29.8 |
| ConA fNP (6 days) | 4.6 | 39.2 | 23.2 | 32 |
| ConA fNP (11 days) | 3.2 | 41.4 | 22.6 | 32 |
In the absence of protein-protein interactions, the intensity of a CD spectrum varies linearly with concentration. The values of the ConA and ConA-fNP extrema were used to estimate the relative concentrations of the ConA and ConA-fNP samples. The absolute concentrations of the soluble ConA samples were determined from the protein OD at 280 nm.36 With the assumption that ConA and ConA-fNP have identical positive and negative per residue molar absorption (Δε: mdeg·M−1·cm−1) at their respective extrema, the CD spectra indicate 50±10μg of ConA was conjugated per mg of ConA-fNP.37 This corresponds to 28 ConA tetramers bound per fNP. An upper limit of 100μg ConA per mg of fNP (< 56 ConA tetramers per fNP) was determined from the loss of 280nm optical density in the ConA solutions used to conjugate lectin and fNP.
The surface of a 46nm diameter NP can fit a maximum of 190 densely packed ConA tetramers29a or 250μg ConA tetramer per mg of ConA-fNP.38 Both the CD and UV analyses indicate the fNP surface is not dense-packed with ConA tetramers. In addition to the unlikely assembly of a dense-packed tetramer layer during conjugation, ConA exhibits tetramer - dimer equilibrium, with lower pH (< 6.3) and temperatures (< 20° C) favoring the dimer.39 Acylation of surface lysines also shifts the equilibrium toward ConA dimers.39,40 ConA dimer units not covalently linked to the NP surface during the conjugation step may dissociate during ConA-fNP purification, resulting in reduced ConA loading. Subunit dissociation during cold temperature storage may contribute to the time dependent CD spectrum.
Limits on fNP sensitivity enhancement
The CD and dye studies reveal that the ConA-f(TR)NP used for the carbohydrate microarray studies contained 76 TR per 28 ConA tetramer. The reduced dye emission yield in 5x f(TR)NP provided an effective brightness of 1.2 TR per ConA. These ConA-f(TR)NP afford lower emissive brightness per lectin than the biotin-ConA/Strepavidin with two to four Cy3 molecules. However, TR brightness per lectin is not the most relevant measure of fNP sensitivity in detection applications. When carbohydrate substrates for ConA are present at very low density, sensitivity is determined by the number of emission equivalents linked per binding event. Under these conditions, ConA-f(TR)NP will provide greater sensitivity than biotin-labeled ConA due to the larger number of emission equivalents (here, 33 for ConA-f(TR)NP, 2–4 for Cy3-Streptavidin/biotin-ConA) per rare binding event and because the large number of ConA per fNP should enhance binding to analyte. The sensitivity advantage afforded by fNP at low analyte densities could be even greater if their spatial distribution, such as in lipid-rafts, allows polyvalent binding to lec-fNP.41
At high analyte densities, detection sensitivity is determined by the numbers of emission equivalents per unit area. The larger number of dyes provided per bound fNP is countered by the larger surface area blocked by each fNP. Moderate size biomolecules labeled with only a few dye molecules, such as the dye-Streptavidin/biotin-lectin conjugates employed here, benefit from a much smaller binding foot print. Dye loadings, dye emission quantum yields and the cross sectional areas of dye-biomolecule or biomolecule-fNP conjugates collectively determine which biomolecule conjugate provides higher sensitivity. As observed in these microarray studies, 33 TR emission equivalents per 46nm fNP affords less fluorescence signal than the Cy3-streptavidin/biotin-lectin conjugates, despite the lower emission quantum yield of Cy3. Using the brightest nanoparticle prepared in this study, the 46nm 8x f(FL)NP offering 400 Fl emission equivalents, would provide only a 2- to 3-fold sensitivity advantage over FL-lectin conjugates.31 These results indicate that the optimal scenario for use of biomolecule-fNP conjugates occurs in cases where the targeted analyte’s density is low. In systems where the targeted analyte’s density is high, the large fNP footprint can reduce detection sensitivity compared to smaller, dye labeled probes. However, size disadvantages of biomolecule-fNP conjugates at high analyte densities may be ameliorated by more favorable conjugate stability and accessibility/delivery to the target, particularly for applications in cells and in vivo.
Conclusions
The brightness of fluorescein (FL) and tetramethyrhodamine (TR) fNP were optimized by varying the concentration, the solvent and the time when dye solution was introduced during NP growth. Up to 1200 FL molecules were captured per fNP through addition of increasingly concentrated FL solutions, while retaining a constant f(FL)NP diameter of 46 nm. The f(FL)NP brightness increased 7-fold between 100 and 800 trapped FL molecules. f(FL)NP brightness starts to plateau with more dye, growing only 15% for an increase from 800 to 1200 (40 mM bulk concentration) FL molecules. The brightest f(FL)NP were ~ 400 times brighter than a single fluorescein molecule in PBS buffer. The TR derivative used to prepare f(TR)NP was much less soluble in ethanol and other solvents than its FL analog. This low solubility limited the number of dyes that incorporated per f(TR)NP. Larger volumes of dye solution were not used as more ethanol increased both fNP diameter and aggregation. The brightest f(TR)NP contained fewer than 80 TR molecules (2–3 mM) and were ~ 35 times brighter than a single TR molecule in PBS. The TR emission quantum yield from f(TR)NP containing 15 – 20 dye molecules was higher than for TR molecule in PBS. Larger enhancements of TR fluorescence quantum yield in dilute f(TR)NP were reported previously.23b For the preparation of f(TR)NP or f(FL)NP, delaying dye addition up to eight hours after initiating NP growth had minimal impact on dye incorporation or fNP brightness.
fNP surface modification using N-(trimethoxysilyl-propyl) ethylene-diamine triacetic acid trisodium salt (EDATAS)/tetraethoxysilane mixtures enhanced fNP water solubility and enabled fNP-protein conjugation following EDC/NHS activation. Between 50 and 100 μg ConA was conjugated per mg of fNP (and per mg of ConA). This amount of ConA (equivalent to 28 – 56 ConA tetramers per fNP) covers 15 – 30% of the fNP surface area. Carbohydrate microarray analyses confirmed typical lectin activity of the ConA-fNP, with a slightly modified mannose substrate selectivity compared to soluble ConA. The CD spectra of ConA-fNP exhibit maxima (196 nm) and minima (223n m) red-shifted by 1 – 2 nm relative to soluble ConA. CDSSTR spectral analyses of ConA-fNP CD spectra indicates a loss of α-helix upon storage in phosphate buffer. Changes to the lectin structure upon storage may be the origin of reduced ConA-fNP activity with time.
Emission intensities from silica nanoparticles containing organic fluorophores can be 101 – 103 times brighter than from a single fluorophore. The higher brightness of fNP greatly increases detection sensitivity for low density analytes in fluorescence assays. In systems with high analyte densities, the large footprint of an fNP can erode its brightness advantage compared to smaller, dye labeled probes. The nearly two-dimensional target format and high analyte density within a carbohydrate microarray makes this a “worst-case” application for fNP. This study found nearly identical emission intensities (within a factor of two) from carbohydrate microarrays stained using ConA-f(dye)NP and dye-Streptavidin/biotin-ConA. Fluorescence and bio-imaging analyses of thicker, more three dimensional, samples with lower analyte densities should benefit from the enhanced brightness of lectin-fNP. The larger diameter and multiple lectins per fNP will yield flatter intensity variation with analyte concentration, making lectin-fNP useful complements to soluble lectin probes at all analyte densities. Despite the abatement of the brightness advantage at high carbohydrate densities, lectin-fNP are a useful probe for glycobiology studies.
Supplementary Material
Acknowledgments
This research was supported, in part, by the Intramural Research Program of the NIH, NCI (JCG), by a Brown University Seed Fund Award (AB), by a Chemistry Department Seed Fund Award (AB, MZ), by a Brown University Wernig Fellowship (YW) and by the National Science Foundation (CHE0616474).
Footnotes
Supporting Information Available: TEM of f(FL)NP. Absorption and emission plots of f(FL)NP and f(TR)NP. Scattering extinction versus wavelength plots for fNP prepared in ethanol and tetrahydrofuran. Full names of ConA primary targets in the microarrays. A list of all carbohydrate microarray components. Estimates of ConA density, particle (fNP or lectin) number density and dye number density for the four solutions used to incubate carbohydrate microarrays. Raw data from the microarray studies. This material is available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Yaqi Wang, Department of Chemistry, Brown University, Providence, RI 02912.
Jeffrey C. Gildersleeve, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702
Amit Basu, Department of Chemistry, Brown University, Providence, RI 02912.
Matthew B. Zimmt, Department of Chemistry, Brown University, Providence, RI 02912
References
- 1.a) Santra S, Zhang P, Wang K, Tan WH. Anal Chem. 2001;73:4988–93. doi: 10.1021/ac010406+. [DOI] [PubMed] [Google Scholar]; b) Zhao XJ, Tapec-Dytioco R, Tan WH. J Am Chem Soc. 2003;125:11474–5. doi: 10.1021/ja0358854. [DOI] [PubMed] [Google Scholar]
- 2.a) Zhao XJ, Bagwe RP, Tan WH. Adv Mater. 2004;16:173–6. [Google Scholar]; b) Zhou XC, Zhou JZ. Anal Chem. 2004;76:5302–12. doi: 10.1021/ac049472c. [DOI] [PubMed] [Google Scholar]
- 3.a) Sokolov I, Naik S. Small. 2008;4:934–39. doi: 10.1002/smll.200700236. [DOI] [PubMed] [Google Scholar]; b) Burns A, Ow H, Wiesner U. Chem Soc Rev. 2006;35:1028–42. doi: 10.1039/b600562b. [DOI] [PubMed] [Google Scholar]; c) Zhao XJ, Bagwe RP, Tan WH. Adv Mater. 2004;16:173–76. [Google Scholar]; d) Zhou XC, Zhou JZ. Anal Chem. 2004;76:5302–12. doi: 10.1021/ac049472c. [DOI] [PubMed] [Google Scholar]; e) Zhao XJ, Tapec-Dytioco R, Tan WH. J Am Chem Soc. 2003;125:11474–75. doi: 10.1021/ja0358854. [DOI] [PubMed] [Google Scholar]; f) Ow H, Larson DR, Srivastava M, Baird BA, Webb WW, Wiesner U. Nano Lett. 2005;5:113–17. doi: 10.1021/nl0482478. [DOI] [PubMed] [Google Scholar]; g) Wang L, Wang KM, Santra S, Zhao XJ, Hilliard LR, Smith JE, Wu JR, Tan WH. Anal Chem. 2006;78:646–54. [Google Scholar]; h) Santra S, Liesenfeld B, Bertolino C, Dutta D, Cao Z, Tan WH, Moudgil MB, Mericle AR. J Lumin. 2006;117:75–82. [Google Scholar]
- 4.a) Fuller EJ, Zugates TG, Ferreira SL, Ow SH, Nguyen NN, Wiesner BU, Langer RS. Biomaterials. 2008;29:1526–32. doi: 10.1016/j.biomaterials.2007.11.025. [DOI] [PubMed] [Google Scholar]; b) Roy I, Ohulchanskyy TY, Bharali DJ, Pudavar HE, Mistretta RA, Kaur N, Prasad PN. Proc Natl Acad Sci USA. 2005;102:279–84. doi: 10.1073/pnas.0408039101. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Yoon TJ, Kim SJ, Kim GB, Yu NK, Cho MH, Lee JK. Angew Chem Int Ed. 2005;44:106871. doi: 10.1002/anie.200461910. [DOI] [PubMed] [Google Scholar]
- 5.a) He X, Duan J, Wang K, Tan K, Lin X, He C. J Nanosci Nanotechnol. 2004;4:585–9. doi: 10.1166/jnn.2004.011. [DOI] [PubMed] [Google Scholar]; b) Zhao X, Hilliard LR, Mechery SJ, Wang Y, Bagwe RP, Jin S, Tan W. Proc Natl Acad Sci USA. 2004;101:15027–32. doi: 10.1073/pnas.0404806101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabius HJ, Nagel GA. Lectins and Glycoconjugates in Oncology. Springer-Verlag; New York: 1988. pp. 5–24. [Google Scholar]
- 7.a) Wu A, Lisowska E, Duk M, Yang Z. Glycoconj J. 2009;26:899–913. doi: 10.1007/s10719-008-9119-7. [DOI] [PubMed] [Google Scholar]; b) Gemeiner P, Mislovičová D, Tkáč J, Švitel J, Pätoprstý V, Hrabárová E, Kogan G, Kožár T. Biotechnology Advances. 2009;27:1–15. doi: 10.1016/j.biotechadv.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 8.a) Chen S, Zheng T, Shortreed MR, Alexander C, Smith LM, Smith Anal Chem. 2007;79:5698–702. doi: 10.1021/ac070423k. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Pilobello KT, Slawek DE, Mahal LK. Proc Natl Acad Sci USA. 2007;104:11534. doi: 10.1073/pnas.0704954104. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hsu KL, Mahal LK. Nature Protocols. 2006;1:543. doi: 10.1038/nprot.2006.76. [DOI] [PubMed] [Google Scholar]; c) Matsuda A, Kuno A, Ishida H, Kawamoto T, Shoda JI, Hirabayashi J. Biochem Biophys Res Comm. 2008;370:259. doi: 10.1016/j.bbrc.2008.03.090. [DOI] [PubMed] [Google Scholar]; d) Leipold M, Herrera I, Ornatsky O, Baranov V, Nitz M. J Proteome Res. 2009;8:443–49. doi: 10.1021/pr800645r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Gorelik E, Galili U, Raz A. Cancer Metastasis Rev. 2001;20:245–77. doi: 10.1023/a:1015535427597. [DOI] [PubMed] [Google Scholar]; b) Babál P, Janega P, Cerná A, Kholavá I, Brabencová E. Acta Histochemica. 2006;108:133. doi: 10.1016/j.acthis.2006.03.003. [DOI] [PubMed] [Google Scholar]; c) Brooks SA, Hall DMS. Methods Mol Med. 2001;57:49–65. doi: 10.1385/1-59259-136-1:49. [DOI] [PubMed] [Google Scholar]
- 10.Nemoto-Sasaki Y, Mitsuki M, Morimoto-Tomita M, Maeda A, Tsuji M, Irimura T. Glycoconj J. 2001;18:895–906. doi: 10.1023/a:1022252509765. [DOI] [PubMed] [Google Scholar]
- 11.a) Santos BS, De Farias PMA, de Menezes FD, de C Ferreira R, Júnior SA, Figueiredo RCBQ, de Carvalho Junior LB, Beltrão EIC. Phys Stat Solid C. 2006;3:4017–22. [Google Scholar]; b) Zhelev Z, Ohba H, Bakalova R, Jose R, Fukuoka S, Nagase T, Ishikawa M, Baba Y. Chem Comm. 2005:1980–82. doi: 10.1039/b419305a. [DOI] [PubMed] [Google Scholar]
- 12.a) Wu CY, Liang PH, Wong CH. Org Biomol Chem. 2009;7:2247–54. doi: 10.1039/b902510n. [DOI] [PubMed] [Google Scholar]; b) Song EH, Pohl NLB. Curr Opin Chem Biol. 2009;13:626–32. doi: 10.1016/j.cbpa.2009.09.021. [DOI] [PubMed] [Google Scholar]; c) Oyelaran O, Gildersleeve JC. Curr Opin Chem Biol. 2009;13:406–13. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Liu Y, Palma AS, Feizi T. Biol Chem. 2009;390:647–56. doi: 10.1515/BC.2009.071. [DOI] [PubMed] [Google Scholar]; e) Oyelaran OO, Li Q, Farnsworth DF, Gildersleeve JC. J Proteome Res. 2009;8:3529–38. doi: 10.1021/pr9002245. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Oyelaran O, McShane LM, Dodd L, Gildersleeve JC. J Proteome Res. 2009;8:4301–10. doi: 10.1021/pr900515y. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Manimala JC, Roach TA, Li ZT, Gildersleeve JC. Angew Chem Int Ed. 2006;45:3607–10. doi: 10.1002/anie.200600591. [DOI] [PubMed] [Google Scholar]
- 13.Yi DK, Selvan ST, Lee SS, Papaefthymiou GC, Kundaliya D, Ying JY. J Am Chem Soc. 2005;127:4990–91. doi: 10.1021/ja0428863. [DOI] [PubMed] [Google Scholar]
- 14.To facilitate comparisons of these ConA-conjugate solutions, Table S3 in the supporting information lists the ConA content, particle number density and dye molecule density for each of the four solutions used to incubate carbohydrate microarrays.
- 15.a) Held GA, Grinstein G, Tu Y. Proc Natl Acad Sci USA. 2003;100:7575–80. doi: 10.1073/pnas.0832500100. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hekstra D, Taussig AR, Magnasco M, Naef F. Nucleic Acids Res. 2003;31:1962–68. doi: 10.1093/nar/gkg283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Bauhinia purpurea lectin, BP, is a Gal-β-1,3-GalNAc binder with a broad specificity. (www.eylabs.com). The data from BP lectin demonstrates that the fluorescence saturation observed from ConA at mannose targets is produced by another lectin at its preferred carbohydrate targets.
-
17.Using the fluorescence intensity data from two different lectin concentrations, [Lectin] and [Lectin × N], the maximum signal intensity can be calculated as
- 18.a) Cy3 and TR have different optical properties. For the Genepix scanner configuration used in the microarray studies, (532 nm excitation, 557–592 nm detection), TR will generate 2.6–3.7 times more emission than Cy3 from identical, low concentration samples. These numbers were determined from (i) emission quantum yields: (TR) 0.2018b–0.2818c, (Cy3) 0.0418d; (ii) extinction coefficients @ 532 nm: ε(TR)/ε(Cy3) ~ 0.66 = 103000 M−1cm−1 (552 nm)18b/150000 M−1cm−1 (554 nm)18d × [0.52/0.54]. Each number in square brackets is the respective compound’s relative extinction coefficient at 532 nm and at the absorption maximum (ε(532nm)/ε(λMAX).18e The detector bandwidth captures 46% of the TR spectrum and 55% of Cy3 spectrum,18e yielding an emission detection ratio of 0.8. The product of the ratios of extinction coefficient, quantum yield and detection yield limits of 2.6–3.7. b) http://www.promega.com/geneticidproc/ussymp8proc/21.html. Selvin PR, Hearst JR. Proc Natl Acad Sci USA. 1994;91:10024–28. doi: 10.1073/pnas.91.21.10024.Mujumdar RB, Ernst LA, Mujumdar SR, Lewis CJ, Waggoner AS. Bioconj Chem. 1993;4:105–11. doi: 10.1021/bc00020a001.e) http://www.invitrogen.com/site/us/en/home/support/Research-Tools/Fluorescence-SpectraViewer.html
- 19.In PBS (pH 7.3), DLS values of the f(FL)NP diameters were 124 nm (FL-APTS added in ethanol) and 80 nm (FL-APTS added in THF).
- 20.The percentage of fNP that passed through 0.10 μm Anatop filters was 95~100% for FL-APTS addition in THF and 70~80 % for FL-APTS addition in ethanol.
- 21.a) Rajagopalan R, Merritt A, Tseytlin A, Foley HC. Carbon. 2004;44:2051–58. [Google Scholar]; b) Green DL, Lin JS, Lam YF, Hu MZC, Schaefer DW, Harris MT. J Colloid Interface Sci. 2003;266:346–58. doi: 10.1016/s0021-9797(03)00610-6. [DOI] [PubMed] [Google Scholar]
- 22.a) Equal absorbance (scattering corrected) solutions of FL-APTS and f(FL)NP (2mL) were added to 2 mL of 0.4 M NaOH. After four hours, the fNP dissolved (verified by the disappearance of the scattering background)22b,c. The absorbances of the resulting solutions differed by less than 5%. Viger ML, Live SL, Therrien DO, Boudreau D. Plasmonics. 2008;3:33–40.Ritter R, Bruhwiler D. J Phys Chem C. 2009;113:10667–74.
- 23.a) A prior study23b reported that the time interval between dye addition and the initiation of NP growth influences the amount of dye incorporated and the fluorescence efficiency. Larson RD, Ow H, Vishwasrao DH, Heikal AA, Wiesner U, Webb WW. Chem Mater. 2008;20:2677–84.
- 24.a) For 1x additions, the FL concentration inside an NP is 3–4 mM. 15–20% of the FL-APTS added to the inverse micelle solution is cross linked into the NP silica matrix. b) For 3x additions in EtOH, the FL concentration inside an NP, [FL]-NP = 12–15 mM (24–28% incorporation of FL-APTS). For 5x additions in EtOH, [FL]-NP = 25–28 mM (28–31% incorporation). For 8x additions, [FL]-NP = 38–40 mM (27–28% incorporation).
- 25.The difference of FL incorporation for 8x FL-APTS addition at 0-hr and 8-hr (28%) is twice as large as the standard deviation of FL incorporation in 8x 0-hr samples (15%).
- 26.a) Concentration dependent FL emission yields are frequently observed.26b,cHill CS, Pinnick GR, Niles S, Fell FN, Jr, Pan YL, Bottiger J, Bronk VB, Holler S, Chang KR. Appl Opt. 2001;40:3005–13. doi: 10.1364/ao.40.003005.Isak JS, Eyring ME. J Phys Chem. 1992;96:1738–42.
- 27.The 2D density of hexagonally packed circles is 0.907. Assuming an average of 76 dyes (35 dye brightness equivalents) per 46 nm diameter circle yields 0.041 TR/nm2 and 0.018 TR-brightness equivalents per nm2 of surface.
- 28.Moothoo ND, Naismith HJ. J Biol Chem. 1996;271:972–76. doi: 10.1074/jbc.271.2.972. [DOI] [PubMed] [Google Scholar]
- 29.a) JMOL29b measurements of the ConA tetramer crystal structure29c yield corner separations of 7.2nm, 7.3nm, 7.6nm and 8.7nm. Assuming a 9 nm edge length, dense packing of equilateral triangles produces three Cy3 dyes per 35.1 nm2 or 0.083 Cy3/nm2. The streptavidin tetramer has a smaller footprint (5.4 nm × 5.8 nm × 4.8 nm)29d than the ConA tetramer. Thus, the ConA tetramer was used for this intensity calculation. b) Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/. Deacon A, Gleichmann T, Kalb (Gilboa) AJ, Price H, Raftery J, Bradbrook G, Yariv J, Helliwell JR. J Chem Soc Faraday Trans. 1997;93:4305–12.Hendrickson WA, Pähler A, Smith JL, Satow Y, Merritt EA, Phizackerley RP. Proc Natl Acad Sci USA. 1989;86:2190–94. doi: 10.1073/pnas.86.7.2190.
- 30.a) Maeda H, Hattori H, Kanoh H. Int J Biol Macromol. 1989;11:290–96. doi: 10.1016/0141-8130(89)90022-6. [DOI] [PubMed] [Google Scholar]; b) Vetri V, Canale C, Relini A, Librizzi F, Militello V, Gliozzi A, Leone M. Biophys Chem. 2007;125:184–90. doi: 10.1016/j.bpc.2006.07.012. [DOI] [PubMed] [Google Scholar]; (c) Fatima S, Ahmad B, Khan HR. Archive Biochem Biophysics. 2006;454:170–80. doi: 10.1016/j.abb.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 31.Assuming an average of 1150 dyes (400 dye brightness equivalents) per 46 nm diameter circle and dense packed fNP yields 0.62 FL/nm2 and 0.22 FL-brightness equivalents/nm2, compared to 0.083 FL (brightness equivalents)/nm2 for ConA tagged with three FL.29a
- 32.Sreerama N, Woody RW. Anal Biochem. 2000;287:252–60. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 33.Monteiro ACO, Muniz-Filho WE, Horta ACG, Beltramini ML, Moreira AR. R Bras Fisiol Veg. 1998;10:167–72. [Google Scholar]
- 34.a) Sanders DA, Moothoo DN, Helliwell JR, Naismith JH. J Mol Biol. 2001;310:875–84. doi: 10.1006/jmbi.2001.4806. [DOI] [PubMed] [Google Scholar]; b) Derewenda Z, Yariv J, Helliwell JR, Kalb AJ, Dodson EJ, Papiz MZ, Wan T, Campbell J. EMBO J. 1989;8:2189–93. doi: 10.1002/j.1460-2075.1989.tb08341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carbohydrate inhibition experiments were performed to assess ConA-fNP activity. TentaGel beads (0.3mmol/g NH2) reacted with α-D-mannopyranosylphenyl isothiocyanate (M-beads) were incubated with ConA-f(TR)-NP solution for 30min. The beads were washed three times with PBS containing 0.05% Tween20. Fluorescence intensity conferred by ConA-fNP binding to the M-beads was quantified using a Zeiss Axiovert 200M fluorescence microscope. The binding between ConA-fNP and M-beads could be inhibited by pre-incubating ConA-fNP with 100mM Methyl-α-D-mannopyranoside for 30min before incubating with M-beads. This preincubation of freshly prepared ConA-fNP reduced M-bead fluorescence signal by 35%. After storing ConA-fNP 12 days, preincubation with 100mM free mannnoside reduced M-bead fluorescence by only 3%.
- 36.a) The 280 nm extinction coefficient37b of the ConA tetramer, at pH 6.8, PBS 0.2 M NaCl, corresponds to a molar extinction coefficient of 1.44×105 cm−1 M−1 (MW (tetramer) = 1.06×105 g/mole). b) Concanavalin A Product Information Sheet - Sigma Prod. No. C2010.
-
37.CD Values used to estimate ConA loading per mg fNP.
CD/mdeg 196nm maximum 223nm minimum Native ConA 8.98 −4.28 43 μg/mL ConA fNP 7.52 −4.59 0.85 mg/mL → 42 μg ConA / mg fNP → 54 μg ConA / mg fNP - 38.A 46nm diameter fNP has a surface area of 6650 nm2. Treating the ConA as an equilateral triangle with 9nm edges yields a 35.1nm2 footprint. 189 triangles (ConA tetrahedra) would densely cover the fNP surface. The mass of 190 ConA is 33 attograms. The mass of a 46 nm diameter fNP (1.9 g/cm3)21 is 97attograms. This loading corresponds to 250μg ConA/mg of ConA-fNP.
- 39.Huet M. Eur J Biochem. 1975;59:627–32. doi: 10.1111/j.1432-1033.1975.tb02491.x. [DOI] [PubMed] [Google Scholar]
- 40.Gunther GR, Wang JL, Yahara I, Cunningham BA, Edelman GM. Proc Natl Acad Sci U S A. 1973;70:1012–16. doi: 10.1073/pnas.70.4.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prinetti A, Loberto N, Chigorno V, Sonnino S. Biochim Biophys Acta. 2009;1788:184–93. doi: 10.1016/j.bbamem.2008.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.