Abstract
Protein radicals are implicated in oxidative stress and are associated with a wide range of diseases and disorders. In the present work, we describe the specific application of a newly synthesized nitrone spin trap, Bio-SS-DMPO, for the detection of these highly reactive species by mass spectrometry (MS). Bio-SS-DMPO is a biotinylated analog of the spin-trap DMPO that allows for specific capture of the protein(s)/peptide(s) labeled by the spin-trap on a (strept)avidin-bound solid matrix. The disulfide bond in the linker arm joining biotin to DMPO can be cleaved to release captured spin-adduct peptide from the solid matrix. This (strept)avidin-based affinity purification reduces the complexity of the samples prior to MS analyses, thereby facilitating the location of the sites of spin trap addition. In addition, the biotin moiety on the spin-trap can efficiently be probed with (strept)avidin-conjugated reporter. This offers an effective means to visualize the presence of DMPO-adducted proteins in intact cells.
Keywords: DMPO, biotin, avidin, streptavidin, protein radical, amino-acid radical, mass spectrometry, myoglobin
INTRODUCTION
A fundamental challenge for protein-centered radicals characterization by combined use of spin-trapping and mass spectrometry (MS) is the low abundance of the spin-trapped protein radical adducts in complex protein mixtures. Here we report a novel biotinylated spin trap derivative that allows for the effective enrichment of trapped protein radicals prior to MS analyses.
Protein radicals have traditionally been studied either through direct electron paramagnetic resonance (EPR) or by spin trapping with EPR detection.1,2 A major limitation associated with these methods is the rapid conversion of paramagnetic protein radical species into EPR-silent diamagnetic products. To overcome this limitation MS-based strategies that focus on the identification of the diamagnetic products have been developed. One highly sensitive technique that has been used by a number of groups3–10 combines spin trapping, digestion of the protein adducts with proteolytic agents, and coupled chromatographic/tandem MS procedures (LC-MS/MS) to localize the actual protein residues labeled by the spin trap.
Standard LC-MS/MS approaches are generally successful in identifying sites of spin-trapping when proteins contain only a few sites to which the spin trap is attached, and/or when the spin-trapping efficiency is high. However, even when other analytical approaches, such as EPR, confirm that the spin trap is attached to a protein, LC-MS/MS analysis frequently fails to provide information about the sites of spin trap addition. One inherent reason is the low trapping efficiencies of the spin-traps for protein radicals. Consequently, the peptide adducts are generally highly underrepresented in the generally complex peptide mixtures. Thus, to improve the detection of low-abundance peptide adducts from proteolytic digests, selective enrichment methods must be employed prior to analysis.
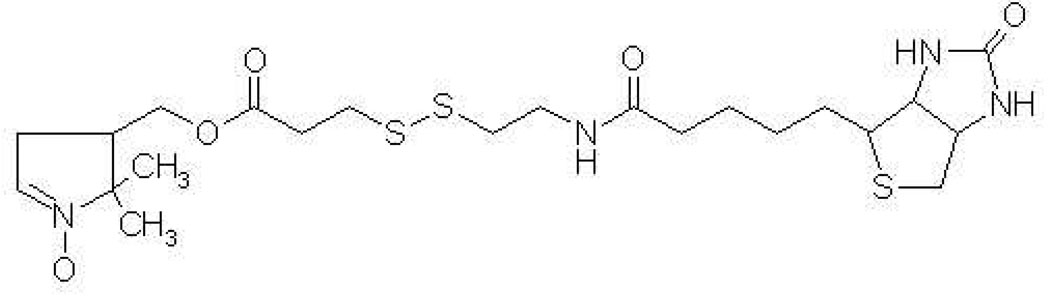
Herein, we have explored the use of a disulfide-bonded biotinylated analog of the nitrone spin-trap 5,5-dimethyl-1-pyrroline-N-oxide (DMPO) that allows for specific capture of the peptide(s) labeled by the spin-trap on a solid matrix. This method reduces the complexity of the samples prior to the MS analyses, thereby facilitating the location of DMPO adducts. The chemical structure of the biotinylated-DMPO analog (Bio-SS-DMPO) is presented in Figure 1. Synthesis of biotinylated derivatives of nitrone spin-traps have been reported by others.11,12 However, a unique feature of Bio-SS-DMPO is the disulfide bond in the linker arm joining biotin to DMPO that can be cleaved to release captured spin-adduct peptides or proteins from the solid matrix. The strategy is demonstrated for horse heart myoglobin. This globin protein is known to form a DMPO-trappable protein radical species on treatment of the met form with H2O2.13,14
Figure 1.
Chemical structure of Bio-SS-DMPO, a biotinylated analog of the spin-trap DMPO containing a chemically cleavable disulfide bond in the linker arm joining biotin to DMPO.
EXPERIMENTAL SECTION
Materials
Horse heart metmyoglobin (Mb) (USB, Cleveland, Ohio) was purified before use by passage through a Sephadex G-25 (PD10) gel filtration column (GE Healthcare Bio-Sciences, Piscataway, NJ) and elution with 50 mM potassium phosphate buffer, pH 7.4. Catalase from beef liver (suspension, 64000 units per mg) and trypsin (from bovine pancreas, modified, sequencing grade) were obtained from Roche Molecular Biochemicals (Indianapolis, IN). All aqueous solutions were prepared using Picopure 2UV Plus system (Hydro Services and Supplies, Inc, RTP, NC) equipped with a 0.2 µm pore size filter. Diluted H2O2 solutions, prepared from a 30% solution (Fisher Scientific Co., Fairlawn, NJ), were used within 1 h of preparation. The spin trap 3-[[2-(Biotinamido)ethyl] dithio] propionic Acid 4’-(Hydroxymethyl) DMPO Ester (Bio-SS-DMPO) was synthesized by Toronto Research Chemicals Inc. (Toronto, ON, Canada). All other chemicals were of analytical grade and were purchased from Sigma (St Louis, MO) or Roche Molecular Biochemicals (Indianapolis, IN).
Spin-trapping
The trapping experiments were conducted in 50 mM potassium phosphate buffer, pH 7.4, using 500 µM protein in the presence of 100 mM Bio-SS-DMPO. The reactions were initiated by the addition of 1 eq of H2O2 (e.g., a final concentration of 500 µM). After a 30-s incubation, the excess H2O2 was removed by adding catalase (26 units).
Protein Digestion
The resulting solutions were cooled to 4 °C and acidified to pH 2.5 by addition of concentrated HCl. The heme was extracted with 1 volume of cold 2-butanone. The organic layers containing the extracted heme were discarded. The aqueous solutions containing the apoprotein were passed over a Sephadex G-25 (PD10) gel filtration column previously equilibrated and eluted with 100 mM Tris-HCl, pH 8.5. Samples were diluted to a final concentration of ~ 50 µM with 100 mM Tris-HCl, pH 8.5, and were digested using a 20:1 substrate-to-protease ratio for 16 h at 37 °C.
Pull Down Assay
The digests were added to streptavidin-coated magnetic beads (Dynabeads M-270; Invitrogen, San Diego, CA). The beads were washed two times with phosphate buffer saline, pH 7.4 (100 mM potassium phosphate, 150 mM NaCl). The disulfide bond in the spacer arm was cleaved with 5 mM DTT, alkylated with 15 mM iodoacetamide and the beads were separated using a permanent magnet. Supernatants were analyzed by LC/ESI/MS.
Electrospray Mass Spectrometry
Flow injection electrospray ionization (ESI/MS) analyses were performed with a Micromass Q-TOF Micro (Waters Micromass Corporation, Manchester, UK) mass spectrometer. Samples of reaction mixtures, prepared as described in the spin-trapping section, were diluted 1:1000 just prior to analysis with a solution of water/acetonitrile 50:50 (v/v) with 0.1% formic acid and infused into the mass spectrometer at ~300 nL/min using a pressure injection vessel.15 The instrumental parameters were as follow: capillary voltage, 3.3 kV; cone voltage, 40 V; collision energy, 10 eV; source temperature, 80 °C. A Waters Q-TOF Premier mass spectrometer equipped with a nanoAcquity UPLC system and NanoLockspray source was used for the acquisition of the LC-ESI/MS and LC-ESI/MS/MS data. Separations were performed using a 3 µm nanoAcquity Atlantis dC18 (100 µm × 100 mm) column (Waters) at a flow rate of 300 nL/min. A 5 µm nanoAcquity Symmetry C18 (100 µm × 20 mm) trapping column (Waters) was positioned in-line with the analytical column. The digested samples were diluted to 0.5 pmol/µL just prior to analysis with their respective buffers as described above, and a 5 µL aliquot was injected. Peptides were eluted using a linear gradient from 98% solvent A [water/0.1%formic acid (v/v)] and 2% solvent B [acetonitrile/0.1% formic acid (v/v)] to 95% solvent B over 120 min. Mass spectrometer settings for MS analyses were a capillary voltage of 3.5 kV, a cone voltage of 30 V, a source temperature of 80 °C, and a collision energy of 8 eV. MS/MS data were acquired in the data-dependent mode, using collision energies based on mass and charge state of the candidate ions. For calibration, an external lock mass was used with a separate reference spray (LockSpray) using a solution of glu-fibrinopeptide B (300 fmol/µL) in water/acetonitrile 80:20 (v/v) with 0.1% formic acid and a mass of 785.8496 (2+). Data analysis was accomplished with a MassLynx data system, MaxEnt deconvolution software, and ProteinLynx software supplied by the manufacturer
Confocal Fluorescence Imaging
The mouse macrophage cell line Raw 264.7 was grown and maintained in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum, penicillin and streptomycin. Upon reaching 80% confluence, cells were counted, seeded at 1×106 cells/ml into 35 mm2 glass bottomed dishes and treated with 25 mM Bio-SS-DMPO for 6 hrs at 37°C. Cells were washed, fixed with 4% paraformaldehyde for 15 min at room temperature, and then permeabilized with methanol for 5 min at room temperature. Cells were incubated for 1h at room temperature with a streptavidin fluorescent conjugate (Alexa 488) diluted in PBS to a final concentration of 5 µg/ml. After washing (7×), cells were visualized with a laser scanning confocal fluorescence microscope.
RESULTS AND DISCUSSION
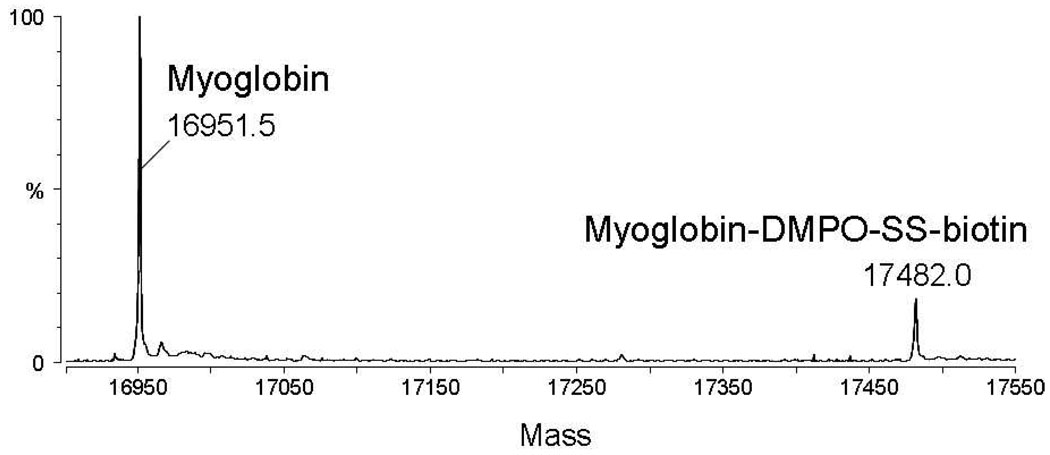
The occurrence of protein Bio-SS-DMPO adducts and the trapping yields were ascertained by flow-injection electrospray ionization (ESI) measurements. The deconvoluted ESI mass spectrum of the reaction mixture of horse heart myoglobin/H2O2/Bio-SS-DMPO revealed that Bio-SS-DMPO adducts (Δm = +530 Da) are observed at 15% relative abundance to the unmodified protein (Figure 2). The formation of the adducts was dependent on the presence of both the hydrogen peroxide and the Bio-SS-DMPO spin trap in the reaction mixture (data not shown).
Figure 2.
Deconvoluted electrospray mass spectrum obtained from the reaction of horse metmyoglobin with hydrogen peroxide in the presence of Bio-SS-DMPO.
To determine which amino acid(s) was modified by the spin-trap, the reaction mixture of horse heart myoglobin/H2O2/Bio-SS-DMPO was subjected to tryptic digestion and analysis by LC-ESI/MS. The ions of m/z 604.5 (+4) and 805.7 (+3) were observed which correspond in mass to tryptic peptide T16 plus the Bio-SS-DMPO. These two ions were subjected to MS/MS analysis. The MS/MS spectrum of the +4 charge state showed a series of carboxy-terminal y ions (y1-y10) and amino-terminal b ions (b2-b4) that provides the necessary data to identify Tyr-103 as the site of modification by Bio-SS-DMPO in the spin trap experiment (data not shown). This observation is consistent with previous mass spectrometric studies performed with non-biotinylated DMPO molecule.13,14
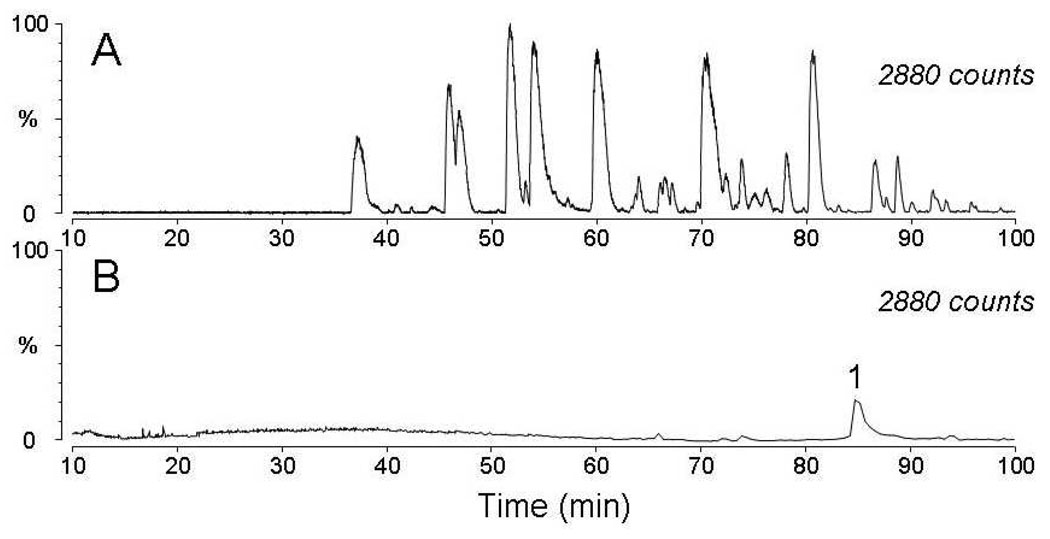
We next investigated whether the biotinylated DMPO peptide adducts in the tryptic digest could be bound to streptavidin-coated magnetic beads and if the adducts could be recovered from the beads after reduction of the disulfide bound with dithiothreitol, alkylation of the thiol moieties with iodoacetamide, and removal of the beads using a permanent magnet. Figure 3 shows the base peak intensity (BPI) chromatogram of the LC/MS analysis of the tryptic digest obtained before and after streptavidin purification. One peak (peak 1) can be readily discerned in the chromatogram after streptavidin purification. The ESI mass spectrum of this chromatographic peak (Figure S-1) exhibited two abundant ions of m/z 543.5 (+4) and 724.4 (+3) which correspond in mass to tryptic peptide T16 modified by the spin trap.
Figure 3.
BPI chromatograms acquired during the LC-ESI/MS analysis of the horse heart myoglobin/H2O2/Bio-SS-DMPO tryptic digest (A) prior to purification with streptavidin beads, and (B) after capture and release of DMPO-adducted peptides. Two abundant ions detected in Peak 1 correspond in mass to tryptic peptide T16 modified by the spin trap. The low abundance peaks (<5% relative abundance) at 65–75 min correspond to minimal nonspecific binding of myoglobin tryptic peptides to the streptavidin beads.
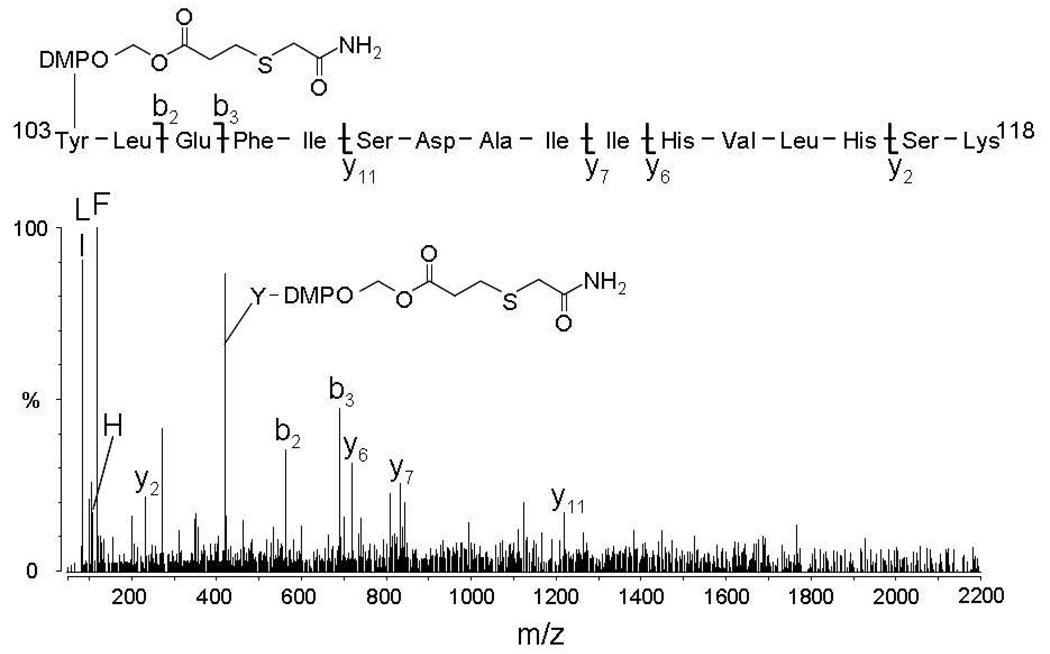
The two ions were subjected to MS/MS analysis. The MS/MS spectrum for the +4 charge state ion is presented in Figure 4. The observation of abundant b2 and b3 ions and of one abundant ion of m/z 422.2 corresponding in mass to the immonium ion of tyrosine plus a S-carboxyamidomethyl-modified DMPO tag, provide the necessary data to unequivocally assign the site of DMPO attachment to Tyr-103.
Figure 4.
MS/MS of the peptide released from the streptavidin beads allow identification of the site of DMPO adduction. The spectrum was acquired from the ion of m/z 543.5 (+4) which corresponds in mass to tryptic peptide T16 plus a S-carboxyamidomethyl-modified DMPO tag.
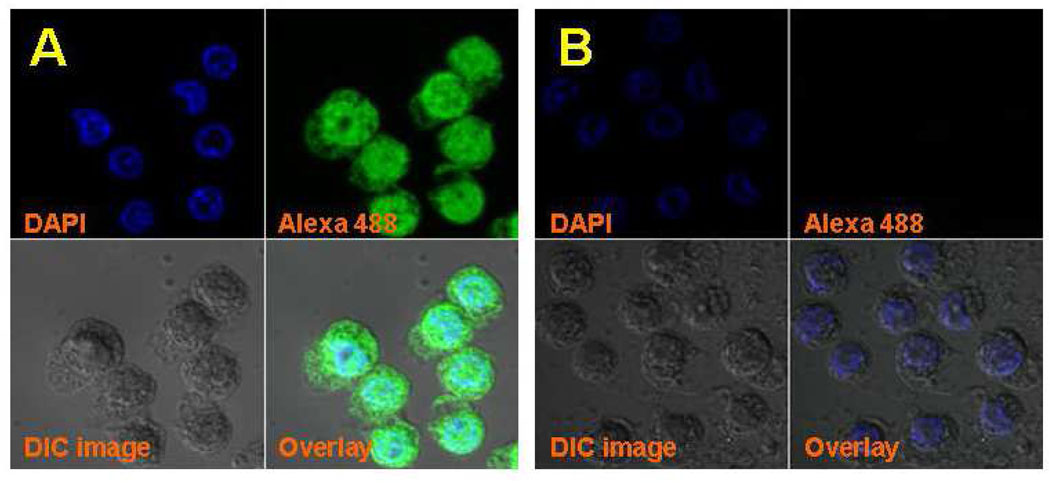
To ascertain whether Bio-SS-DMPO can be used as a probe to visualize the formation of protein-centered radicals in intact cells, we next performed confocal fluorescence imaging studies in mouse macrophage Raw-264.7 cells. Staining with a streptavidin fluorescent conjugate (Alexa 488) revealed that Bio-SS-DMPO could be taken up by intact metabolizing cells and the probe appears to accumulate both in the cytoplasm and in the nucleus (Figure 5A). No green fluorescence could be detected in negative controls (Figure 5B).
Figure 5.
Confocal fluorescence imaging studies of the distribution of Bio-SS-DMPO in Raw-264.7 cells. (A) Cells were treated with 25 mM Bio-SS-DMPO for 6 hrs at 37°C; (B) same as A, but in the absence of Bio-SS-DMPO. Alexa 488 (green) was used to visualize the accumulation of Bio-SS-DMPO in the cells. 4’,6-diamino-2-phenylindole (DAPI, in blue) was used to visualize nuclei. Differential interference contrast microscopy (DIC, in gray) was used to enhance contrast between phases by polarized light.
CONCLUSIONS
Methods using mass spectrometry in combination with spin-trapping are among the most powerful approaches for the identification of protein-based radicals. Herein, we present a variant to this approach, which combines spin-trapping with Bio-SS-DMPO, a biotinylated analog of the spin-trap DMPO, and a pull-down assay with streptavidin-coated magnetic beads. This method adds a dimension of selectivity prior to the MS analyses, thereby facilitating the location of DMPO nitrone adducts for the unambiguous structural determination of modified sites in the polypeptide chains. In addition, we show that Bio-SS-DMPO is accumulated inside intact metabolizing cells. The use of biotinylated spin trap analogs should allow for the identification of proteins as well as the site(s) of protein radical spin trapping from complex mixtures and/or proteomic-like studies.
Supplementary Material
ACKNOWLEDGMENT
This work has been supported by the Intramural Research Program of the National Institutes of Health/National Institute of Environmental Health Sciences (projects ES050171 and ES050139). The authors would like to thank the NIEHS Protein Microcharacterization Core Facility and the NIEHS Fluorescence Microscopy and Imaging Center for helpful discussions and assistance.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Davies MJ, Hawkins CL. Free Radic Biol Med. 2004;36:1072–1086. doi: 10.1016/j.freeradbiomed.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 2.Augusto O, Muntz Vaz S. Amino Acids. 2007;32:535–542. doi: 10.1007/s00726-006-0429-4. [DOI] [PubMed] [Google Scholar]
- 3.Deterding LJ, Ramirez DC, Dubin JR, Mason RP, Tomer KB. J. Biol. Chem. 2004;279:11600–11607. doi: 10.1074/jbc.M310704200. [DOI] [PubMed] [Google Scholar]
- 4.Lardinois OM, Tomer KB, Mason RP, Deterding LJ. Biochemistry. 2008;47:10440–10448. doi: 10.1021/bi800771k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deterding LJ, Bhattacharjee S, Ramirez DC, Mason RP, Tomer KB. Anal. Chem. 2007;79:6236–6248. doi: 10.1021/ac070935z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen YR, Chen CL, Zhang L, Green-Church KB, Zweier JL. J. Biol. Chem. 2005;280:37339–37348. doi: 10.1074/jbc.M503936200. [DOI] [PubMed] [Google Scholar]
- 7.Tsaprailis G, English AM. J. Biol. Inorg. Chem. 2003;8:248–255. doi: 10.1007/s00775-002-0407-6. [DOI] [PubMed] [Google Scholar]
- 8.Harris MN, Burchiel SW, Winyard PG, Engen JR, Mobarak CD, Timmins GS. Chem. Res. Toxicol. 2002;15:1589–1594. doi: 10.1021/tx025594t. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, He S, Mauk AG. Biochemistry. 2002;41:13507–13513. doi: 10.1021/bi026122g. [DOI] [PubMed] [Google Scholar]
- 10.Fenwick CW, English AM. J. Am. Chem. Soc. 1996;118:12236–12237. [Google Scholar]
- 11.Arya P. Heterocycles. 1996;43:397–406. [Google Scholar]
- 12.Chalier F, Hardy M, Ouari O, Rockenbauer A, Tordo P. J. Org. Chem. 2007;72:7886–7892. doi: 10.1021/jo071070s. [DOI] [PubMed] [Google Scholar]
- 13.Lardinois OM, Detweiler CD, Tomer KB, Mason RP, Deterding LJ. Free Radic. Biol. Med. 2008;44:893–906. doi: 10.1016/j.freeradbiomed.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Detweiler CD, Lardinois OM, Deterding LJ, Ortiz de Montellano PR, Tomer KB, Mason RP. Free Radic. Biol. Med. 2005;38:969–976. doi: 10.1016/j.freeradbiomed.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 15.Deterding LJ, Moseley MA, Tomer KB, Jorgenson JW. Anal. Chem. 1989;61:2504–2511. doi: 10.1021/ac00197a011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.