Abstract
The mitochondrial retrograde response has been extensively described in Saccharomyces cerevisiae, where it has been found to extend life span during times of mitochondrial dysfunction, damage, or low nutrient levels. In yeast, the retrograde response genes (RTG) convey these stress responses to the nucleus to change the gene expression adaptively. Similarly, most classes of higher organisms have been shown to have some version of a central stress-mediating transcription factor, NF-κB. There have been several modifications along the phylogenetic tree as NF-κB has taken a larger role in managing cellular stresses. Here we review similarities and differences of mechanisms and pathways between RTG genes in yeast and NF-κB as seen in more complex organisms. We perform a structural homology search and reveal similarities of Rtg proteins with eukaryotic transcription factors involved in development and metabolism. NF-κB shows more sophisticated functions as compared to RTG genes including participation in immune responses and induction of apoptosis under high levels of ROS-induced mitochondrial and nuclear DNA damage. Involvement of NF-κB in chromosomal stability, co-regulation of mitochondrial respiration and crosstalk with the TOR pathway points to a conserved mechanism also found in yeast.
Introduction: Evolutionary Basis
Several stress-sensitive pathways, both oxidative stress-related and others, have been conserved in humans. While humans and other mammals have developed specialized stress responses and methods to regulate them, such as the transcription factor Nuclear Factor – kappaB (NF-κB), the basis is still present in S. cerevisiae and other simple eukaryotes such as C. elegans (Pujol et al. 2001). One stress response mechanism in S. cerevisiae is the retrograde response, which is controlled by the stress-sensitive retrograde response (RTG) pathway that elicits transcriptional changes. The retrograde response links stress response to nutritional changes, mitochondrial dysfunction and aging, with adaptation of metabolism, chromatin-dependent gene regulation and genome stability (Jazwinski 2005). Hints that both pathways, NF-κB in humans, and RTG in yeast, may be related, stems from the fact that NF-κB is activated in response to the introduction of mitochondrial dysfunction (Biswas et al. 1999; Amuthan et al. 2002). This warrants discussion of potential similarities between RTG genes and NF-κB.
Currently, there is a clear gap between the understanding of the NF-κB pathway that has been established in C. elegans, D. melanogaster, and C. rotundicauda (horseshoe crab) and higher organisms and the more primitive RTG pathway. Both pathways have in common that they are tuned to mitochondrial dysfunction and levels of oxidative stress (Butow & Avadhani 2004). For instance, oxygen consumption and free radical production as the byproduct of mitochondrial respiration, as initially outlined in the free radical theory of aging (Gerschman et al. 1954; Harman 1956), contribute to the accumulation of oxidized proteins and dysfunction that activates either pathway and extends life span. However, there are several clear differences between mammals and simple eukaryotes to consider that affect the conservation of these responses. The first is the development of organ systems and the fact that selection occurs at the organismal level – thus a response that is beneficial to a particular cell may not be so for the organism as a whole. The other is the development of an immune system and changes in stresses experienced, based on nutritional and environmental dependencies. The development of these other systems and stresses has led to the allocation of additional roles for these stress mediating molecules. In this review, we take an evolutionary perspective to bridge the gap between these two well-characterized pathways: the yeast retrograde response and the mammalian/human NF-κB mediated stress response.
RTG pathway and aging
The retrograde response was discovered through the observation of a curious accumulation of nuclear transcripts in yeast cells lacking mitochondrial DNA (Parikh et al. 1987). Subsequent studies identified some of these transcripts, and a genome-wide analysis demonstrated that the retrograde response activates genes involved in metabolism and stress responses that encode proteins destined for the mitochondrion, the cytoplasm, and peroxisomes (Epstein et al. 2001). Concomitantly, several genes were implicated in the retrograde signaling pathway, including RTG1-3, discussed in detail here (Liao & Butow 1993; Jia et al. 1997a). Further efforts demonstrated the translocation of the Rtg1–Rtg3 transcription factor from the cytoplasm to the nucleus and the involvement of RTG2 in this process (Sekito et al. 2000). Rtg2 is a phosphatase with an ATP binding domain similar to that of the Hsp70/actin/sugar kinase superfamily (Ferreira Junior et al. 2005). When bound to Rtg2, the heterodimeric transcription factor complex Rtg1/Rtg3 can shuttle to the nucleus. Rtg3 contains several sites in its N-terminal region that are phosphorylated upon activation, and it translocates to the nucleus when dimerized with Rtg1. Otherwise, the Rtg3 nuclear localization sequence (NLS) is blocked, the protein becomes dephosphorylated at these sites in the N-terminal region, and returns to the cytoplasm (Ferreira Junior et al. 2005).
A key element in the regulation of this pathway is the interaction of Rtg2 with Mks1 (Sekito et al. 2002; Liu et al. 2003; Ferreira Junior et al. 2005). Mks1 is a negative regulator of the retrograde response; it was originally identified as a negative regulator of the Ras2-cAMP pathway (Matsuura & Anraku 1993). In hyperphosphorylated form, Mks1 is bound to Bmh1 and Bmh2, yeast homologues of mammalian 14-3-3 proteins (Dilova et al. 2004). This complex maintains Rtg3 in a hyperphosphorylated, cytoplasmic form. The efficient switch between the hypo- and hyperphosphorylated forms of Mks1 is facilitated by ubiquitination and degradation of partially phosphorylated Mks1. Strong activation of Mks1p therefore suppresses the retrograde response (Liu et al. 2003; Liu et al. 2005).
The discovery of the involvement of Lst8, a component of the TOR complex, in retrograde signaling was the first clue of the interaction between TOR signaling and the retrograde response (Liu et al. 2001). It too is a negative regulator of the retrograde response like Mks1. However, TOR regulation of retrograde target gene expression is distinct from the activation of the retrograde response by mitochondrial dysfunction (Giannattasio et al. 2005). In addition to its role in the translocation of Rtg1–Rtg3 to the nucleus, Rtg2 is a component of the transactivation complex SLIK (SAGA-like), and it is found at the promoters of retrograde target genes (Pray-Grant et al. 2002). It is also essential for the suppression of trinucleotide repeat expansion (Bhattacharyya et al. 2002). It has recently been shown that there is some overlap in the target genes of the retrograde response and caloric restriction in growing yeasts (Wang et al. 2010). This overlap primarily involves the activation of the glyoxylate cycle, which facilitates the production of biosynthetic intermediates for cell growth.
Activation of the retrograde response increases yeast replicative life span (Kirchman et al. 1999). This increase in yeast longevity requires RTG2 and RTG3 (Kirchman et al. 1999; Borghouts et al. 2004). It also requires RAS2 (Kirchman et al. 1999). Activation of the retrograde response in these studies was accomplished either by elimination of mitochondrial DNA to generate mitochondrial petite yeast or by the deletion of the COX4 gene, both of which interrupt the electron transport chain to generate dysfunctional mitochondria. In some yeast strains, these manipulations do not activate the retrograde response and do not enhance replicative life span. However, switching to growth on raffinose rather than glucose, which relieves glucose repression allows activation and increased longevity (Kirchman et al. 1999). Loss of mitochondrial DNA or lack of respiration ability is not essential for activation of the retrograde response. During normal yeast aging, there is a gradual loss of mitochondrial membrane potential which is not the result of interruption of the electron transport chain, because the cells can still grow on non-fermentable carbon source (Muller 1971; Lai et al. 2002; Borghouts et al. 2004). There is also a corresponding gradual activation of the retrograde response (Borghouts et al. 2004). This activation compensates for the increasing mitochondrial dysfunction to enhance yeast longevity because it has been shown that the higher the induction of the retrograde response the greater is the extension of yeast life span (Jazwinski 2000). Consistent with the known negative effect of TOR signaling on the retrograde response, disruption of TOR signaling increases yeast replicative life span (Kaeberlein et al. 2005). RTG2 is also involved in chronological life span in yeast, as deletion of the gene curtails it (Barros et al. 2004). Survival in stationary phase involves mitophagy in yeast, which is regulated by a conserved protein phosphatase, Aup1 (Journo et al. 2009). The activation of retrograde signaling is defective in mutants missing this enzyme, and the deletion of RTG3 results in defective autophagy in stationary phase (Journo et al. 2009). This constitutes additional evidence that the retrograde response may be involved in chronological aging.
As mentioned above, RTG2 is a component of the SLIK complex. SLIK is a transcriptional co-activator that shares many of its subunits with SAGA (Pray-Grant et al. 2002). One of these is the histone acetyltransferase Gcn5. Deletion of GCN5 attenuates the retrograde response in similar fashion to the deletion of RTG2 (Kim et al. 2004). SAGA overlaps with another transcriptional co-activator, TFIID, in its choice of promoters (Huisinga & Pugh 2004). However, SAGA is specific for stress response genes, while TFIID activates housekeeping genes. These stress response genes are suppressed by co-repressor complexes containing Rpd3 or Hda1 (Huisinga & Pugh 2004). It has been suggested, that the SLIK version of SAGA may extend the activity of this co-activator to the metabolic genes that are the targets of retrograde signaling (Jazwinski 2005). RTG2 links metabolism to stress responses, because it is proximal to the signal generated by dysfunctional mitochondria in the retrograde response and it is a component of SLIK which is involved in the activation of certain metabolic and stress response genes. Deletion of RTG2 abrogates the retrograde response and prevents extension of replicative life span. Importantly, deletion of RPD3 or HDA1 has the expected opposite effect of enhancing yeast longevity (Kim et al. 1999). The involvement of SLIK in the retrograde response links metabolism with chromatin-dependent gene regulation through RTG2.
The retrograde response also links metabolism with genome stability. RTG2 is responsible for suppressing the generation of extrachromosomal ribosomal DNA circles or ERC (Borghouts et al. 2004),whose accumulation can cause yeast death (Sinclair & Guarente 1997). However, Rtg2 can only suppress ERC production when it is not engaged in retrograde signaling. Nevertheless, cells in which the retrograde response is induced are more resistant to the deleterious effects of ERC despite the unavailability of Rtg2 (Borghouts et al. 2004). Disruption of the SLIK complex by deletion of GCN5 prevents retrograde signaling but it reduces ERC production in mitochondrial petites, presumably by making Rtg2 available to suppress their formation (Kim et al. 2004).
NF-κB pathway and aging
NF-κB has accumulated various adaptive functions through its evolutionary history, and it has acquired “multichannel processor” capabilities canalizing a spectrum of exogenous and endogenous stressors, immune responses, organelle dysfunction and aging (Salminen et al. 2008a; Kriete & Mayo 2009). The pathway participates in the signal transduction of the immune system, representing a bow-tie architecture with many upstream inputs, a number of central key proteins, and downstream fan outs to regulate a broad number of target genes (Kitano & Oda 2006). The NF-κB family consists of several proteins, which can be activated in part or jointly. For instance, TNF alpha induces five mammalian NF-kappaB/Rel proteins, c-Rel, NF-kappaB1 (p50/p105), NF-kappaB2 (p52/p100), RelA/p65 and RelB. These complexes associate with the cytosolic inhibitor IκB for retention in the cytoplasm. Phosphorylation and subsequent degradation of IκB by the IKK complex (IKKα, IKKβ and IKKγ /NEMO) unmasks a nuclear translocation signal, which shuttles NF-κB to the nucleus where it transcribes anti-apoptotic genes, cytokines, immunoreceptors and adhesion molecules (Pahl 1999; Hayden & Ghosh 2004).
Activation of NF-κB by pro-inflammatory cytokines like TNFalpha is the canonical pathway, while activation in B-cells by a specific IKK signalosome complex is termed an alternative pathway. Other stress factors including oxidative stress are termed atypical mechanisms (Pahl 1999). They include oxidative (Li & Karin 1999), genotoxic (Janssens & Tschopp 2006), organelle stress including the endoplasmic overdose response (EOR) (Pahl & Baeuerle 1995), and change in calcium homeostasis involving calcineurin and IκBβ (Biswas et al. 2008). Dynamic regulation of NF-κB requires modulation of interorganelle trafficking by phosporylation, masking and unmasking of the nuclear localization signal (NLS). More recently, 14-3-3 phosphoserine and phosphothreonine-binding proteins, which play a role in many signaling pathways (Dougherty & Morrison 2004), have been found to be involved in regulating location and binding of both IKBα and p65, both of which contain 14-3-3 binding domains (Aguilera et al. 2006). Disrupting 14-3-3 activity by transfection with a dominant-negative 14-3-3 leads to the accumulation of nuclear p65-IκBα complexes and the constitutive association of p65 with the chromatin. It is unknown, whether 14-3-3 proteins interact with other NF-κB heterodimers or whether 14-3-3 homologs, Bmh1p and Bmh2p, may play a similar role in RTG responses in yeast.
While most widely studied and characterized in mice and humans, the earliest traces of NF-κB can be found in the NF-κB p100 homologue in C. rotundicauda Relish (CrRelish), an early antibacterial defense mechanism in the horseshoe crab (Fan et al. 2008). Orthologous proteins and domains have been found in even simpler organisms, such as the nematode C. elegans, though in much more primitive form and function (Fan et al. 2008). The three main areas of NF-κB involvement are oxidative, immune-inflammatory, and protein stress responses. In addition to these cell-protective responses, NF-κB has been known to have pro-apoptotic functions at times, depending on the severity and genre of stress (Wang et al. 2002).
Many transcription factors, particularly the ones that affect large regions of the genome, have been found to exert epigenetic effects – that is, alteration of the packaging of DNA that subsequently affects transcription of that area. NF-κB has been shown to act in this manner, perhaps contributing to the breadth of genes controlled by it (Vanden Berghe et al. 2006). Epigenetic changes have been shown to contribute to the progression of aging as seen in the retrograde response as well (Pray-Grant et al. 2002; Kim et al. 2004), as dicussed above. Both pathways are further implicated in epigenetic control, involvement in microRNA expression (Taganov et al. 2006, Liang et al. 2009) and in the control of mitochondrial processes through interactions with mtDNA (De Benedictis et al. 2000). Previous work has found that although the epigenetic effects of NF-κB are variegated in their mechanisms, DNA methylation resulting in gene silencing is paramount (Vanden Berghe et al. 2006).
Mitochondrial mutations and damage caused by ROS or otherwise have long been implicated in human aging (Harman 1972; Bandy & Davison 1990; Balaban et al. 2005; Loeb et al. 2005; Navarro & Boveris 2007). It has been suggested that NF-κB, an important regulator of ROS scavengers (Xu et al. 1999), may contribute to ubiquitously observed low ROS levels in aging cells and tissues, but damage such as oxidized proteins can accumulate and constitute another mechanism for NF-κB activation (Kriete et al. 2010). Accordingly, NF-κB has been found constitutively upregulated in various aged tissues (Spencer et al. 1997; Adler et al. 2007) and in resting fibroblasts from older donors (Kriete et al. 2008). Likewise, reduced ATP biosynthesis by damaged mitochondria may introduce an adaptive downregulation of biosynthesis through the TOR pathway, along with a compensatory activation of aerobic glycolysis (Schieke et al. 2006). Under these conditions NF-κB and mTOR would work cooperatively and protectively but trap the cell in a lower-energy supplying state, beginning many of the trends we see in aging. However, without this particular stress response life span is reduced as shown in NF-κB knock-out mice (Lu et al. 2006).
Protein homologies
Rtg1 and Rtg3 are basic helix-loop-helix/leucine zipper (bHLH/zip) transcription factors that heterodimerize to activate transcription from a novel R-box site (Jia et al. 1997b). bHLH/zip proteins comprise a large and important class of transcription factors (Moore et al. 2000), including the Myc/Max/Mad network of proteins that play roles in cell proliferation, differentiation, and death (Grandori et al. 2000). While the structures of the RTG proteins are not available yet, a structural modeling of Rtg1 and Rtg3 by I-TASSER server (Zhang 2008) and the structural alignment using the Vorometric server (Sacan et al. 2008), and protein visualization tools (Krieger et al. 2002; Hanson 2008) confirm the conservation of key bHLH/zip residues between Rtg1 and Myc and between Rtg3 and Max (Figure 1). The residues around the DNA binding sites of Rtg3 and Max are found to be particularly conserved (Figure 2).
Figure 1.
Homology model of yeast proteins Rtg1 (front, red) and Rtg3 (back, red), structurally aligned with the human Myc protein (front, yellow) and Max protein (back, yellow) in complex with DNA (green molecular surface). The Myc/Max/DNA complex is taken from PDB id: 1nkp. The identical residues in the alignment are shown in stick configuration. Structures of Rtg1 and Rtg3 are modeled using I-TASSER server (Zhang, 2008), the structure alignments are obtained using Vorometric (Sacan et al. 2008), and the figure is drawn using Yasara (Krieger et al. 2002).
Figure 2.
A closer look into the structural alignments between Rtg1 (left, red) and Myc (left, yellow), and between Rtg3 (right, red) and Max (right, yellow). Identical residues in the alignments are shown in stick configuration. Myc/Max structures are taken from PDB id: 1nkp. I-TASSER (Zhang, 2008), Vorometric (Sacan et al. 2008), and Jmol (Hanson 2008) are used for modeling, structural alignment, and visualization, respectively.
An amino acid sequence homology analysis shows the microphthalmia-associated transcription factor MITF to be the closest vertebrate homolog of both Rtg1 and Rtg3 (with 32% sequence identity between yeast Rtg1 and mouse MITF and 39% sequence identity between yeast Rtg3 and human MITF). MITF is a member of the MYC family of basic helix-loop-helix leucine zipper transcription factors (Hallsson et al. 2007). It is conserved in both vertebrates and invertebrates, and is important in the development of several different cell types, including melanocytes and retinal pigment epithelial cells (Steingrimsson et al. 2004). The Rtg3 sequence is additionally found to be related to the transcription factor EB (38% identity with human TFEB), Max-like protein isoform X (35% identity with human MLX), transcription factor E3 (33% identity with human TFE3), and upstream stimulatory factor USF (29% identity with human USF2), all of which are bHLH/zip transcription factors. MITF, TFEB, and TFE3, along with TFEC, comprise a transcription factor family (MiT) that regulates key developmental pathways. They have been most widely studied in the context of renal translocation carcinomas (Argani & Ladanyi 2005).
The ubiquitously expressed USF factors are indicated to be key regulators of stress and immune responses, cell cycle and proliferation, and lipid and glucose metabolism. Interestingly, the overexpression of HINT1 tumor repressor gene inhibits both USF and NF-κB activity in human hepatoma cells (Wang et al. 2009), which indicates presence of a common regulation of USF and NF-κB. The structure of USF and its interaction with DNA is highly similar to those of Max (Ferre-D'Amare et al. 1994).
The MLX protein mentioned above encodes a MAX-like protein X and belongs to the class of bHLH/zip-transcription factors. The MondoA-MLX heterodimers have been proposed as candidate sensors of glucose concentration (Sans et al. 2006) and may be considered a phylogenetically-related counterpart of Rtg1/3 in higher eukaryotes. Unlike other Myc proteins, MLX and its binding partner MondoA localize in the cytoplasm and are thought to be associated with the outer mitochondrial membrane (Sans et al. 2006). MondoA:MLX complexes accumulate in the nucleus in response to glucose and activate a broad spectrum of metabolic genes (Stoltzman et al. 2008). Note that MondoA:MLX has been proposed as a sensor for glucose levels and a direct correspondence between MondoA:MLX and RTG complex has not been previously suggested. It is not known if MondoA:MLX can also respond to mitochondrial dysfunction.
It is worth noting that there are several difficulties in drawing parallels between related transcription factors across different organisms. Firstly, there is a lack of knowledge of their upstream regulators. Genetic screens have been particularly fruitful in determining some of the important players, such as Mks1p, a negative regulator of RTG-dependent gene expression (Liu et al. 2003). However, the biochemical activity of these additional proteins is yet to be determined. Secondly, the genes identified in one organism may not have identifiable homologs in the other. For example, there is currently no known counterpart in higher eukaryotes for the Rtg2 protein as a sensor of mitochondrial dysfunction and a transducer of mitochondrial signals that activate Rtg1/3-like transcription factors. All that is known is that the sensors exist, as there are downstream transcription factors such as NF-κB that are associated with them. Thirdly, even for known homologs, not all of the protein domains are well characterized. For example, for Rtg1/3 proteins, the homology with Myc-related proteins is limited to the bHLH/zip domain and we have little knowledge regarding the structure and function of the regions flanking that domain. Finally, the downstream targets of signaling modules diverge during evolution, while the modules themselves remain intact.
Conservation of the retrograde response
Is there a retrograde response that plays a role in determining life span in organisms other than yeast? It has been known for quite some time that with age there are changes in metabolism in Caenorhabditis elegans that resemble those found in the retrograde response, and similar changes are induced in daf mutants that display an increased life span (Vanfleteren & De Vreese 1995). More recently, the use of RNAi to knock down gene expression has demonstrated the wide involvement of mitochondrial function in determining the life span of the worm (Dillin et al. 2002; Lee et al. 2003). The decline in mitochondrial function appears to trigger a retrograde response that increases life span (Cristina et al. 2009). There may, however, be more than one pathway associated with this retrograde signaling in the worm (Yang & Hekimi 2010).
In animal cells, the operation of a retrograde response has been discerned for some time (Butow & Avadhani 2004), and is the topic of this article. The difficulty has been to find the common threads that link mitochondrial dysfunction with the induced patterns of gene expression in different cell types. A comparative study has found that there are some commonalities across cell types with many more differences noted (Miceli & Jazwinski 2005b; Miceli & Jazwinski 2005a). The common changes in gene expression involve adaptations to the glycolytic production of energy necessary for cell survival, which bears significant similarities to the retrograde response in yeast. Furthermore, there is evidence that the induction of a retrograde response in human cells in culture results in a delay in replicative senescence and thus perhaps aging as well (Passos et al. 2007).
As we develop evidence for relationships between human and yeast proteins participating in a retrograde response, we also find related pathways in yeast that are conserved in humans. One of the strongest relationships between the RTG genes and the oxidative stress-sensitive pathways lies in the TOR pathway; a protein linking the retrograde response and TOR, Lst8, provides this connection (Butow & Avadhani 2004). Depending on the nature of the nutritional source, the degree of mitochondrial dysfunction, and the amount of Lst8p activity which acts as a switch-protein (Rosner et al. 2009), different target genes downstream of TOR1 and RTG1/2/3 are activated (Giannattasio et al. 2005). Both the kinase TOR and its mammalian counterpart mTOR have two functional complexes TORC1/2 and mTORC1/2 (Frias et al. 2006). These complexes are dormant during stress and active when nutrients abound in the environment (Schmelzle & Hall 2000; Wullschleger et al. 2006). While TOR is usually bound to Lst8 or its mammalian ortholog, GβL (Diaz-Troya et al. 2008), during stress it is separated, which inactivates the complex (TORC1/2) and leads to stress responses that contrast with the default TOR-mediated cellular functions. Notably, another mitochondrial back-signaling mechanism investigated in yeast involves the transcription factor and metabolic regulator SFP1 also interacting with the TORC1 kinase complex (Heeren et al. 2009). In the NF-κB pathway, mTOR activates IKK resulting in activation of NF-κB. GβL is known to suppress TNFα-induced NF-κB signaling by interacting with IKKβ (Kim et al. 2008). Conversely, AKT can stimulate IKK activity directed toward the phosphorylation of IκBα and RelA/p65 in cancer cells, and it in turn is controlled by mTOR (Dan et al. 2008).
In addition to this connection to the TOR pathway, RTG pathways converge with the Ras pathways as well; these help to control stress resistance as well as life span in S. cerevisiae (Shama et al. 1998). Whereas in humans RAS is best known as a proto-oncogene, it has critical functions in controlling respiration and redox balance in yeast (Heeren et al. 2004). Both RAS genes are GTP binding proteins and activate cAMP-dependent cascades and show strong homology, though evolution has led them to take different roles in each organism (Wigler et al. 1988). As a result of exerting control of cell survival during periods of oxidative stress, as it does in humans, it can extend or limit life span, dependent on transcriptional levels. In fact, much of the pathway itself is similar in humans and yeast in terms of activation, function, and constituents. This is very similar to the effect of NF-κB, which diverts the cell to replicative or quiescent states depending on the amount, duration, and type of stresses faced by the cell (Schoemaker et al. 2002; Wang et al. 2002). Other related pathways have proven homologous or otherwise similar in simpler organisms including sirutins, which have been shown to inhibit NF-κB and inflammation-associated cascades in humans. In S. cerevisiae and C. elegans, they have been shown to be involved in aging (Salminen et al. 2008b). While NF-κB and sirutins have a more complex role in humans, their primordial role in both aging and inflammation has been established in C. elegans. Although no complete homology has been shown between the RTG genes in S. cerevisiae and NF-κB, strong homologies between inhibitors and pathways of both leads one to believe that the retrograde response is a potential predecessor of the now-central stress-regulator, NF-κB.
Intriguingly, the c-MYC protein, which shows homology with Rtg3, has two identified NF-κB binding sites (Duyao et al. 1990a; Duyao et al. 1990b) and is involved in the regulation of glycolysis, suggesting that these mechanisms are indeed related. Furthermore, upregulation of peroxisomal activities in mammalian cells contribute to ROS production, and peroxisome proliferator-activator receptors (PPARs) stimulate NADPH oxidase in macrophages (Teissier et al. 2004), which in unison may represent an evolved defense mechanism under control of NF-κB with ancient roots in the yeast retrograde response, which is known to activate peroxisomal related target genes.
Summary
As described here, the current research has closed in on a relationship between the RTG pathway in yeast and the more sophisticated NF-κB pathway. As an organism becomes more complex, especially in its immune system, it seems that NF-κB takes on a larger role, eventually becoming a central transcription factor in most major stress responses as in humans. Many studies have elaborated on the various patterns of gene expression resulting from different stressors in yeast, while others have looked at progressively less complex organisms and searching for NF-κB or homologous transcription factors. Not only has NF-κB been found in simpler organisms, but its mechanisms of activation and relationship with various stressors have been found to be more primitive as well (Fan et al. 2008). The variants or versions of NF-κB found in simpler organisms like C. elegans have little or no immune involvement, though the human and fly variants are heavily connected with viral responses (Pujol et al. 2001).
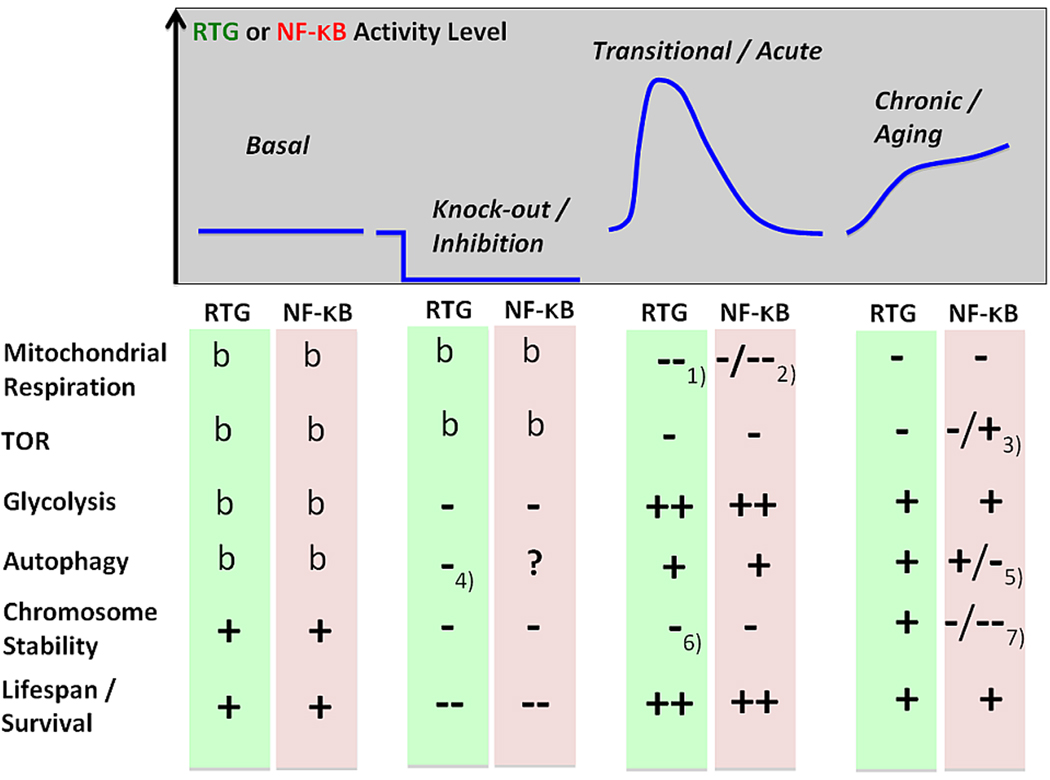
As summarized in Figure 3, a differentiation has to be made under which conditions both stress responses occur. Basal levels of RTG and NF-κB contribute to chromosomal stability. If pharmacologically inhibited or in knock-outs, lifespan in both organisms is compromised. Under nutritional stress and intact mitochondria, RTG facilitates metabolic adaptations and survival. In mammalian cells, periods of inflammation are a comparable transient situation, and without NF-κB the cell would enter apoptosis. In some viral infections this may be accompanied by dysfunctional mitochondria. In aging the respiratory capacity of mitochondria becomes compromised and both pathways become chronically upregulated, which correlates with metabolic adaptations, which may be further modulated by involvement of the TOR pathway. While both pathways provide a pro-survival and adaptive characteristic, an important difference between RTG and NF-κB is their involvement in epigenetic regulation long term. High levels of NF-κB contribute to a tumor promoting chromosomal instability, particularly seen in many age related diseases, whereas in yeast Rtg2 appears to contribute to epigenetic stability if overexpressed.
Figure 3.
Comparison of the RTG and NF-κB pathways. The four phases showing different levels of activity in the basal state, inhibition or knock-out, a transitional acute phase of nutrional stress in yeast and acute inflammation in mammalian cells, and chronic elevation in aging. Influential of both pathway are mitochondrial respiration and TOR activity, while they modulate glycolysis, autophagy, chromosome stability and life span. Activity is coded by b = basal levels, + enhanced, ++ strongly enhanced, − suppressed and − − strongly suppressed. Explanations: 1) change in nutritional source disables mitochondrial respiration; 2) In infection, mitochondria are working either at highest capacity, which activates the endoplasmic overdose response (EOR) and NF-kB, or the mitochondria are dysfunctional, but without either response cells go into apoptosis; 3) TOR can initially decrease, but later increase activity in senescence and age-related diseases; 4) Inhibition of Rtg3 reduces autophagy; 5) Results from TOR in 3); 6) Rtg2 is unavailable when strongly participating in the retrograde response; 7) Chromosomal instability high in aging and highest in cancer.
Activation of the mitochondrial-to-nucleus pathway both in yeast and mammals, and the bi-genomic regulation of mitochondrial proteins opens the intriguing possibility that these mechanisms are connected, providing a closed feedback loop. Therefore, in times of stress involving the mitochondria, the retrograde response could, in principle, feedback to the mitochondria. In response to deficient mitochondria some studies of cells harboring deleterious mtDNA mutations had indicated an increase in expression of genes encoding mitochondrial proteins to promote mitochondrial biogenesis (Heddi et al. 1999), while in yeast ρ0 petite cells no upregulation of these genes has been found (Epstein et al. 2001). Similarly in mammalian cells, NF-κB has been shown to have a role in mitochondrial biogenesis involving the transcription factor YY1 (Sui 2009), and at the same time, YY1 also activates c-MYC. In aging however, reports on downregulation of mitochondrial genes encoded in the nucleus are ubiquitous throughout the literature, albeit counterintuitive. Lowering mitochondrial respiration under stress conditions reduces oxidative phosphorylation but promotes glycolysis for ATP production and prevents high ROS levels. Indeed, application of mild heat stress in yeast has shown a downregulation of mitochondrial genes encoded in the nucleus (Sakaki et al. 2003) and inhibition of mitochondrial respiration prolongs longevity in nematodes (Cristina et al. 2009). Since mitochondrial respiration is under bi-genomic regulation, all COX genes have been found to be under control of NRF as well as under regulation of the mitochondrial genome. Notably, IkBα/p50 dimers have been found to be directly located in the mitochondria. If activated by external stress factors, such as TNFα, p50 homodimers are formed interacting with the transcriptional machinery of the mitochondria and downregulating cytochromes and cox III encoded by the mitochondrial genome (Cogswell et al. 2003). Furthermore, IKBα has been found to interact with ANT, the mitochondrial ATP/ADP translocator upon induction of apoptosis (Bottero et al. 2001).
Although no complete homology has been shown between the RTG genes in S. cerevisiae and NF-κB, strong homologies between inhibitors and pathways of both leads one to believe that the retrograde response is a potential predecessor of the now-central stress-regulator, NF-κB. We have pointed to the function of LST8p as a conserved connecting element of the TOR pathway and RTG/NF-κB complexes. Furthermore, we have mentioned 14-3-3 proteins, relevant for the nucleocytoplasmic transport of IKBα and p65, and the interaction of Mks1p with the yeast homolog of 14-3-3, Bmh1p. We also have discussed the c-MYC protein, which shows homology with Rtg3 and the role of the retrograde response in peroxisomal processes. Therefore, insight from investigations from either side may help to identify and decipher hitherto unknown mechanisms.
Acknowledgments
The research in SMJ’s laboratory was supported in part by grant AG006168 from the National Institute on Aging (NIH).
References
- Adler AS, Sinha S, Kawahara TL, Zhang JY, Segal E, Chang HY. Motif module map reveals enforcement of aging by continual NF-kappaB activity. Genes Dev. 2007;21:3244–3257. doi: 10.1101/gad.1588507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera C, Fernandez-Majada V, Ingles-Esteve J, Rodilla V, Bigas A, Espinosa L. Efficient nuclear export of p65-IkappaBalpha complexes requires 14-3-3 proteins. J Cell Sci. 2006;119:3695–3704. doi: 10.1242/jcs.03086. [DOI] [PubMed] [Google Scholar]
- Amuthan G, Biswas G, Ananadatheerthavarada HK, Vijayasarathy C, Shephard HM, Avadhani NG. Mitochondrial stress-induced calcium signaling, phenotypic changes and invasive behavior in human lung carcinoma A549 cells. Oncogene. 2002;21:7839–7849. doi: 10.1038/sj.onc.1205983. [DOI] [PubMed] [Google Scholar]
- Argani P, Ladanyi M. Translocation carcinomas of the kidney. Clin Lab Med. 2005;25:363–378. doi: 10.1016/j.cll.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Bandy B, Davison AJ. Mitochondrial mutations may increase oxidative stress: implications for carcinogenesis and aging? Free Radic Biol Med. 1990;8:523–539. doi: 10.1016/0891-5849(90)90152-9. [DOI] [PubMed] [Google Scholar]
- Barros MH, Bandy B, Tahara EB, Kowaltowski AJ. Higher respiratory activity decreases mitochondrial reactive oxygen release and increases life span in Saccharomyces cerevisiae. J Biol Chem. 2004;279:49883–49888. doi: 10.1074/jbc.M408918200. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Rolfsmeier ML, Dixon MJ, Wagoner K, Lahue RS. Identification of RTG2 as a modifier gene for CTG*CAG repeat instability in Saccharomyces cerevisiae. Genetics. 2002;162:579–589. doi: 10.1093/genetics/162.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G, Adebanjo OA, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, Kotlikoff M, Avadhani NG. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 1999;18:522–533. doi: 10.1093/emboj/18.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G, Tang W, Sondheimer N, Guha M, Bansal S, Avadhani NG. A distinctive physiological role for IkappaBbeta in the propagation of mitochondrial respiratory stress signaling. J Biol Chem. 2008;283:12586–12594. doi: 10.1074/jbc.M710481200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghouts C, Benguria A, Wawryn J, Jazwinski SM. Rtg2 protein links metabolism and genome stability in yeast longevity. Genetics. 2004;166:765–777. doi: 10.1534/genetics.166.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottero V, Rossi F, Samson M, Mari M, Hofman P, Peyron JF. Ikappa b-alpha, the NF-kappa B inhibitory subunit, interacts with ANT, the mitochondrial ATP/ADP translocator. J Biol Chem. 2001;276:21317–21324. doi: 10.1074/jbc.M005850200. [DOI] [PubMed] [Google Scholar]
- Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- Cogswell PC, Kashatus DF, Keifer JA, Guttridge DC, Reuther JY, Bristow C, Roy S, Nicholson DW, Baldwin AS., Jr NF-kappa B and I kappa B alpha are found in the mitochondria. Evidence for regulation of mitochondrial gene expression by NF-kappa B. J Biol Chem. 2003;278:2963–2968. doi: 10.1074/jbc.M209995200. [DOI] [PubMed] [Google Scholar]
- Cristina D, Cary M, Lunceford A, Clarke C, Kenyon C. A regulated response to impaired respiration slows behavioral rates and increases lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000450. doi: 10.1371/journal.pgen.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan HC, Cooper MJ, Cogswell PC, Duncan JA, Ting JP, Baldwin AS. Akt-dependent regulation of NF-{kappa}B is controlled by mTOR and Raptor in association with IKK. Genes Dev. 2008;22:1490–1500. doi: 10.1101/gad.1662308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedictis G, Carrieri G, Garasto S, Rose G, Varcasia O, Bonafe M, Franceschi C, Jazwinski SM. Does a retrograde response in human aging and longevity exist? Exp Gerontol. 2000;35:795–801. doi: 10.1016/s0531-5565(00)00169-8. [DOI] [PubMed] [Google Scholar]
- Diaz-Troya S, Florencio FJ, Crespo JL. Target of rapamycin and LST8 proteins associate with membranes from the endoplasmic reticulum in the unicellular green alga Chlamydomonas reinhardtii. Eukaryot Cell. 2008;7:212–222. doi: 10.1128/EC.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Dilova I, Aronova S, Chen JC, Powers T. Tor signaling and nutrient-based signals converge on Mks1p phosphorylation to regulate expression of Rtg1.Rtg3p-dependent target genes. J Biol Chem. 2004;279:46527–46535. doi: 10.1074/jbc.M409012200. [DOI] [PubMed] [Google Scholar]
- Dougherty MK, Morrison DK. Unlocking the code of 14-3-3. J Cell Sci. 2004;117:1875–1884. doi: 10.1242/jcs.01171. [DOI] [PubMed] [Google Scholar]
- Duyao MP, Buckler AJ, Sonenshein GE. Interaction of an NF-kappa B-like factor with a site upstream of the c-myc promoter. Proc Natl Acad Sci U S A. 1990a;87:4727–4731. doi: 10.1073/pnas.87.12.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyao MP, Kessler DJ, Spicer DB, Sonenshein GE. Binding of NF-KB-like factors to regulatory sequences of the c-myc gene. Curr Top Microbiol Immunol. 1990b;166:211–220. doi: 10.1007/978-3-642-75889-8_27. [DOI] [PubMed] [Google Scholar]
- Epstein CB, Waddle JA, Hale Wt, Dave V, Thornton J, Macatee TL, Garner HR, Butow RA. Genome-wide responses to mitochondrial dysfunction. Mol Biol Cell. 2001;12:297–308. doi: 10.1091/mbc.12.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan ZH, Wang XW, Lu J, Ho B, Ding JL. Elucidating the function of an ancient NF-kappaB p100 homologue, CrRelish, in antibacterial defense. Infect Immun. 2008;76:664–670. doi: 10.1128/IAI.00948-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre-D'Amare AR, Pognonec P, Roeder RG, Burley SK. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994;13:180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira Junior JR, Spirek M, Liu Z, Butow RA. Interaction between Rtg2p and Mks1p in the regulation of the RTG pathway of Saccharomyces cerevisiae. Gene. 2005;354:2–8. doi: 10.1016/j.gene.2005.03.048. [DOI] [PubMed] [Google Scholar]
- Frias MA, Thoreen CC, Jaffe JD, Schroder W, Sculley T, Carr SA, Sabatini DM. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol. 2006;16:1865–1870. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Gerschman R, Gilbert DL, Nye SW, Dwyer P, Fenn WO. Oxygen poisoning and x-irradiation: a mechanism in common. Science. 1954;119:623–626. doi: 10.1126/science.119.3097.623. [DOI] [PubMed] [Google Scholar]
- Giannattasio S, Liu Z, Thornton J, Butow RA. Retrograde response to mitochondrial dysfunction is separable from TOR1/2 regulation of retrograde gene expression. J Biol Chem. 2005;280:42528–42535. doi: 10.1074/jbc.M509187200. [DOI] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Hallsson JH, Haflidadottir BS, Schepsky A, Arnheiter H, Steingrimsson E. Evolutionary sequence comparison of the Mitf gene reveals novel conserved domains. Pigment Cell Res. 2007;20:185–200. doi: 10.1111/j.1600-0749.2007.00373.x. [DOI] [PubMed] [Google Scholar]
- Hanson R. Jmol: an open-source java viewer for chemical structures in 3d, version 11.8. 2008 http://www.jmol.org/
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Harman D. The biologic clock: the mitochondria? J Am Geriatr Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Heddi A, Stepien G, Benke PJ, Wallace DC. Coordinate induction of energy gene expression in tissues of mitochondrial disease patients. J Biol Chem. 1999;274:22968–22976. doi: 10.1074/jbc.274.33.22968. [DOI] [PubMed] [Google Scholar]
- Heeren G, Jarolim S, Laun P, Rinnerthaler M, Stolze K, Perrone GG, Kohlwein SD, Nohl H, Dawes IW, Breitenbach M. The role of respiration, reactive oxygen species and oxidative stress in mother cell-specific ageing of yeast strains defective in the RAS signalling pathway. FEMS Yeast Res. 2004;5:157–167. doi: 10.1016/j.femsyr.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Heeren G, Rinnerthaler M, Laun P, von Seyerl P, Kossler S, Klinger H, Jarolim S, Simon-Nobbe B, Hager M, Schuller C, Carmona-Gutierrez D, Breitenbach-Koller L, Muck C, Jansen-Durr P, Criollo A, Kroemer G, Madeo F, Breitenbach M. The mitochondrial ribosomal protein of the large subunit, Afo1p, determines cellular longevity through mitochondrial back-signaling via TOR1. Aging (Albany NY) 2009;1:622–636. doi: 10.18632/aging.100065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Janssens S, Tschopp J. Signals from within: the DNA-damage-induced NF-kappaB response. Cell Death Differ. 2006;13:773–784. doi: 10.1038/sj.cdd.4401843. [DOI] [PubMed] [Google Scholar]
- Jazwinski SM. Metabolic control and gene dysregulation in yeast aging. Ann N Y Acad Sci. 2000;908:21–30. doi: 10.1111/j.1749-6632.2000.tb06632.x. [DOI] [PubMed] [Google Scholar]
- Jazwinski SM. The retrograde response links metabolism with stress responses, chromatin-dependent gene activation, and genome stability in yeast aging. Gene. 2005;354:22–27. doi: 10.1016/j.gene.2005.03.040. [DOI] [PubMed] [Google Scholar]
- Jia Y, Rothermel B, Thornton J, Butow RA. A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol. 1997a;17:1110–1117. doi: 10.1128/mcb.17.3.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Rothermel B, Thornton J, Butow RA. A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol. 1997b;17:1110–1117. doi: 10.1128/mcb.17.3.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journo D, Mor A, Abeliovich H. Aup1-mediated regulation of Rtg3 during mitophagy. J Biol Chem. 2009;284:35885–35895. doi: 10.1074/jbc.M109.048140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kim S, Benguria A, Lai CY, Jazwinski SM. Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:3125–3136. doi: 10.1091/mbc.10.10.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Ohkuni K, Couplan E, Jazwinski SM. The histone acetyltransferase GCN5 modulates the retrograde response and genome stability determining yeast longevity. Biogerontology. 2004;5:305–316. doi: 10.1007/s10522-004-2568-x. [DOI] [PubMed] [Google Scholar]
- Kim YL, Kim JE, Shin KJ, Lee S, Ahn C, Chung J, Kim DH, Seong JY, Hwang JI. GbetaL regulates TNFalpha-induced NF-kappaB signaling by directly inhibiting the activation of IkappaB kinase. Cell Signal. 2008;20:2127–2133. doi: 10.1016/j.cellsig.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Kirchman PA, Kim S, Lai CY, Jazwinski SM. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics. 1999;152:179–190. doi: 10.1093/genetics/152.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H, Oda K. Robustness trade-offs and host-microbial symbiosis in the immune system. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100039. 2006 0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA--a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- Kriete A, Bosl WJ, Booker G. Rule-based cell systems model of aging using feedback loop motifs mediated by stress responses. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000820. e1000820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriete A, Mayo KL. Atypical pathways of NF-kappaB activation and aging. Exp Gerontol. 2009;44:250–255. doi: 10.1016/j.exger.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Kriete A, Mayo KL, Yalamanchili N, Beggs W, Bender P, Kari C, Rodeck U. Cell autonomous expression of inflammatory genes in biologically aged fibroblasts associated with elevated NF-kappaB activity. Immun Ageing. 2008;5:5. doi: 10.1186/1742-4933-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CY, Jaruga E, Borghouts C, Jazwinski SM. A mutation in the ATP2 gene abrogates the age asymmetry between mother and daughter cells of the yeast Saccharomyces cerevisiae. Genetics. 2002;162:73–87. doi: 10.1093/genetics/162.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Li N, Karin M. Is NF-kappaB the sensor of oxidative stress? FASEB J. 1999;13:1137–1143. [PubMed] [Google Scholar]
- Liang R, Bates DJ, Wang E. Epigenetic Control of MicroRNA Expression and Aging. Curr Genomics. 2009;10:184–193. doi: 10.2174/138920209788185225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X, Butow RA. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- Liu Z, Sekito T, Epstein CB, Butow RA. RTG-dependent mitochondria to nucleus signaling is negatively regulated by the seven WD-repeat protein Lst8p. Embo J. 2001;20:7209–7219. doi: 10.1093/emboj/20.24.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Sekito T, Spirek M, Thornton J, Butow RA. Retrograde signaling is regulated by the dynamic interaction between Rtg2p and Mks1p. Mol Cell. 2003;12:401–411. doi: 10.1016/s1097-2765(03)00285-5. [DOI] [PubMed] [Google Scholar]
- Liu Z, Spirek M, Thornton J, Butow RA. A novel degron-mediated degradation of the RTG pathway regulator, Mks1p, by SCFGrr1. Mol Biol Cell. 2005;16:4893–4904. doi: 10.1091/mbc.E05-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA, Wallace DC, Martin GM. The mitochondrial theory of aging and its relationship to reactive oxygen species damage and somatic mtDNA mutations. Proc Natl Acad Sci U S A. 2005;102:18769–18770. doi: 10.1073/pnas.0509776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZY, Yu SP, Wei JF, Wei L. Age-related neural degeneration in nuclear-factor kappaB p50 knockout mice. Neuroscience. 2006;139:965–978. doi: 10.1016/j.neuroscience.2005.12.062. [DOI] [PubMed] [Google Scholar]
- Matsuura A, Anraku Y. Characterization of the MKS1 gene, a new negative regulator of the Ras-cyclic AMP pathway in Saccharomyces cerevisiae. Mol Gen Genet. 1993;238:6–16. doi: 10.1007/BF00279524. [DOI] [PubMed] [Google Scholar]
- Miceli MV, Jazwinski SM. Common and cell type-specific responses of human cells to mitochondrial dysfunction. Exp Cell Res. 2005a;302:270–280. doi: 10.1016/j.yexcr.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Miceli MV, Jazwinski SM. Nuclear gene expression changes due to mitochondrial dysfunction in ARPE-19 cells: implications for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2005b;46:1765–1773. doi: 10.1167/iovs.04-1327. [DOI] [PubMed] [Google Scholar]
- Moore AW, Barbel S, Jan LY, Jan YN. A genomewide survey of basic helix-loop-helix factors in Drosophila. Proc Natl Acad Sci U S A. 2000;97:10436–10441. doi: 10.1073/pnas.170301897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller I. Experiments on ageing in single cells of Saccharomyces cerevisiae. Arch Mikrobiol. 1971;77:20–25. doi: 10.1007/BF00407985. [DOI] [PubMed] [Google Scholar]
- Navarro A, Boveris A. The mitochondrial energy transduction system and the aging process. Am J Physiol Cell Physiol. 2007;292:C670–C686. doi: 10.1152/ajpcell.00213.2006. [DOI] [PubMed] [Google Scholar]
- Pahl HL. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- Pahl HL, Baeuerle PA. A novel signal transduction pathway from the endoplasmic reticulum to the nucleus is mediated by transcription factor NF-kappa B. EMBO J. 1995;14:2580–2588. doi: 10.1002/j.1460-2075.1995.tb07256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh VS, Morgan MM, Scott R, Clements LS, Butow RA. The mitochondrial genotype can influence nuclear gene expression in yeast. Science. 1987;235:576–580. doi: 10.1126/science.3027892. [DOI] [PubMed] [Google Scholar]
- Passos JF, Saretzki G, Ahmed S, Nelson G, Richter T, Peters H, Wappler I, Birket MJ, Harold G, Schaeuble K, Birch-Machin MA, Kirkwood TB, von Zglinicki T. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol. 2007;5:e110. doi: 10.1371/journal.pbio.0050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, Cook RG, Workman JL, Yates JR, 3rd, Grant PA. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol Cell Biol. 2002;22:8774–8786. doi: 10.1128/MCB.22.24.8774-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol N, Bonnerot C, Ewbank JJ, Kohara Y, Thierry-Mieg D. The Caenorhabditis elegans unc-32 gene encodes alternative forms of a vacuolar ATPase a subunit. J Biol Chem. 2001;276:11913–11921. doi: 10.1074/jbc.M009451200. [DOI] [PubMed] [Google Scholar]
- Rosner M, Fuchs C, Siegel N, Valli A, Hengstschlager M. Functional interaction of mTOR complexes in regulating mammalian cell size and cell cycle. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacan A, Toroslu IH, Ferhatosmanoglu H. Integrated search and alignment of protein structures. Bioinformatics. 2008;24:2872–2879. doi: 10.1093/bioinformatics/btn545. [DOI] [PubMed] [Google Scholar]
- Sakaki K, Tashiro K, Kuhara S, Mihara K. Response of genes associated with mitochondrial function to mild heat stress in yeast Saccharomyces cerevisiae. J Biochem. 2003;134:373–384. doi: 10.1093/jb/mvg155. [DOI] [PubMed] [Google Scholar]
- Salminen A, Huuskonen J, Ojala J, Kauppinen A, Kaarniranta K, Suuronen T. Activation of innate immunity system during aging: NF-kB signaling is the molecular culprit of inflamm-aging. Ageing Res Rev. 2008a;7:83–105. doi: 10.1016/j.arr.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Salminen A, Ojala J, Huuskonen J, Kauppinen A, Suuronen T, Kaarniranta K. Interaction of aging-associated signaling cascades: inhibition of NF-kappaB signaling by longevity factors FoxOs and SIRT1. Cell Mol Life Sci. 2008b;65:1049–1058. doi: 10.1007/s00018-008-7461-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans CL, Satterwhite DJ, Stoltzman CA, Breen KT, Ayer DE. MondoA-Mlx heterodimers are candidate sensors of cellular energy status: mitochondrial localization and direct regulation of glycolysis. Mol Cell Biol. 2006;26:4863–4871. doi: 10.1128/MCB.00657-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieke SM, Phillips D, McCoy JP, Jr, Aponte AM, Shen RF, Balaban RS, Finkel T. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem. 2006;281:27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- Schoemaker MH, Ros JE, Homan M, Trautwein C, Liston P, Poelstra K, van Goor H, Jansen PL, Moshage H. Cytokine regulation of pro- and anti-apoptotic genes in rat hepatocytes: NF-kappaB-regulated inhibitor of apoptosis protein 2 (cIAP2) prevents apoptosis. J Hepatol. 2002;36:742–750. doi: 10.1016/s0168-8278(02)00063-6. [DOI] [PubMed] [Google Scholar]
- Sekito T, Liu Z, Thornton J, Butow RA. RTG-dependent mitochondria-to-nucleus signaling is regulated by MKS1 and is linked to formation of yeast prion [URE3] Mol Biol Cell. 2002;13:795–804. doi: 10.1091/mbc.01-09-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekito T, Thornton J, Butow RA. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol Biol Cell. 2000;11:2103–2115. doi: 10.1091/mbc.11.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shama S, Kirchman PA, Jiang JC, Jazwinski SM. Role of RAS2 in recovery from chronic stress: effect on yeast life span. Exp Cell Res. 1998;245:368–378. doi: 10.1006/excr.1998.4276. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Spencer NF, Poynter ME, Im SY, Daynes RA. Constitutive activation of NF-kappa B in an animal model of aging. Int Immunol. 1997;9:1581–1588. doi: 10.1093/intimm/9.10.1581. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- Stoltzman CA, Peterson CW, Breen KT, Muoio DM, Billin AN, Ayer DE. Glucose sensing by MondoA:Mlx complexes: a role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proc Natl Acad Sci U S A. 2008;105:6912–6917. doi: 10.1073/pnas.0712199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui G. Regulation of YY1 in Tumorigenesis and its Targeting Potential in Cancer Therapy. Molecular and Cellular Pharmacology. 2009;1:157–176. [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teissier E, Nohara A, Chinetti G, Paumelle R, Cariou B, Fruchart JC, Brandes RP, Shah A, Staels B. Peroxisome proliferator-activated receptor alpha induces NADPH oxidase activity in macrophages, leading to the generation of LDL with PPAR-alpha activation properties. Circ Res. 2004;95:1174–1182. doi: 10.1161/01.RES.0000150594.95988.45. [DOI] [PubMed] [Google Scholar]
- Vanden Berghe W, Ndlovu MN, Hoya-Arias R, Dijsselbloem N, Gerlo S, Haegeman G. Keeping up NF-kappaB appearances: epigenetic control of immunity or inflammation-triggered epigenetics. Biochem Pharmacol. 2006;72:1114–1131. doi: 10.1016/j.bcp.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Vanfleteren JR, De Vreese A. The gerontogenes age-1 and daf-2 determine metabolic rate potential in aging Caenorhabditis elegans. Faseb J. 1995;9:1355–1361. doi: 10.1096/fasebj.9.13.7557026. [DOI] [PubMed] [Google Scholar]
- Wang J, Jiang JC, Jazwinski SM. Gene regulatory changes in yeast during life extension by nutrient limitation. Exp Gerontol. 2010;45:621–631. doi: 10.1016/j.exger.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li H, Zhang Y, Santella RM, Weinstein IB. HINT1 inhibits beta-catenin/TCF4, USF2 and NFkappaB activity in human hepatoma cells. Int J Cancer. 2009;124:1526–1534. doi: 10.1002/ijc.24072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Kotamraju S, Konorev E, Kalivendi S, Joseph J, Kalyanaraman B. Activation of nuclear factor-kappaB during doxorubicin-induced apoptosis in endothelial cells and myocytes is pro-apoptotic: the role of hydrogen peroxide. Biochem J. 2002;367:729–740. doi: 10.1042/BJ20020752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M, Field J, Powers S, Broek D, Toda T, Cameron S, Nikawa J, Michaeli T, Colicelli J, Ferguson K. Studies of RAS function in the yeast Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 2):649–655. doi: 10.1101/sqb.1988.053.01.074. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Xu Y, Kiningham KK, Devalaraja MN, Yeh CC, Majima H, Kasarskis EJ, St Clair DK. An intronic NF-kappaB element is essential for induction of the human manganese superoxide dismutase gene by tumor necrosis factor-alpha and interleukin-1beta. DNA Cell Biol. 1999;18:709–722. doi: 10.1089/104454999314999. [DOI] [PubMed] [Google Scholar]
- Yang W, Hekimi S. Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans. Aging Cell. 2010 doi: 10.1111/j.1474-9726.2010.00571.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]