Abstract
Histone Deacetylases (HDACs) are intimately involved in the epigenetic regulation, and thus are one of the key therapeutic targets for cancer, and two HDAC inhibitors, namely suberoylanilide hydroxamic acid (SAHA) and romidepsin have been recently approved for the cancer treatment. Since the screening and detailed characterization of HDAC inhibitors has been time consuming, we synthesized Coumarin-SAHA (c-SAHA) as a fluorescent probe for determining the binding affinities (Kd) and the dissociation off-rates (koff) of the enzyme-inhibitor complexes. The determination of the above parameters relies on the changes in the fluorescence emission intensity (λex = 325 nm, λem = 400 nm) of c-SAHA due to its competitive binding against other HDAC inhibitors, and such determination neither requires employment of polarization accessories nor is dependent on the fluorescence energy transfer from the enzyme’s tryptophan residues to the probe. Our highly sensitive and robust analytical protocol presented herein is applicable to most of the HDAC isozymes, and it can be easily adopted in a high-throughput mode for screening the HDAC inhibitors as well as for quantitatively determining their Kd and koff values.
Keywords: Histone deacetylase, Inhibior, Binding affinity, Dissociation “off rate”, Fluorescent probe
INTRODUCTION
It has been clear that a variety of physiological and pathophysiological processes are under epigenetic control, and they are manifested via dynamic modulation of the chromatin structure [1]. The expression of chromosomal DNA is intimately regulated by its interaction (or lack thereof) with its cognate histone proteins, which undergo different types of covalent modifications (viz., methylation, phosphorylation, acetylation, ubiquitination, etc.) [2]. Of these, the acetylation/deacetylation of selected lysine residues in core histone proteins alters their net positive charges and thus perturbs their binding affinities with negatively charged cognate DNA sequences [3]. Whereas the acetylation of histones is catalyzed by histone acetyltransferases, their deacetylation reactions are accomplished by histone deacetylases (HDACs)1. Since the HDAC catalyzed deacetylation yields positively charged histone proteins, their avidity for the negatively charged cognate DNA sequences increases, which result in the transcriptional repression of certain genes [4, 5]. Circumstantial evidence has led to the suggestion that HDAC inhibitors inhibit cell proliferation and angiogenesis, induce cell differentiation, promote apoptosis etc., and thus such inhibitors have been identified for the treatment of different types of cancers [6]. Due to its wide-spread applicability in human diseases, many pharmaceutical/biotechnology industries are working on the inhibitor design against HDACs. Presently, several HDAC inhibitors are at different stages of clinical trials, and two such inhibitors namely, suberoylanilino hydroxamic acid (SAHA) and romidepsin have been approved for the treatment of cancer [7-9].
Based on the sequence homology, phylogeny, as well as cofactor requirement, human HDACs have been classified into class I (HDAC1,2,3,8), class II (HDAC4,5,7,9,10), class III (Sirt1,2,3,4,5,6,7), and class IV (HDAC11) subfamilies [10]. Of these, whereas class I, II, and IV HDACs are Zn2+ metalloproteins, class III HDAC require NAD+ as the cofactor and these enzymes are known as sirtuins [11, 12]. The cellular localizations and their general/specific physiological functions have started to emerge in recent years [13].
Due to challenging assay system of HDACs, there has been an ongoing effort in developing highly sensitive and simple methods for screening the enzyme inhibitors, particularly in high throughput settings (HTS) [13], as well as for quantitatively determining their binding affinities (Kd) and dissociation off-rate (koff) values to assess their potency [9]. Toward these goals, both enzyme activity assays and competitive ligand binding methods have been developed over the past 7 years [14-19]. The most widely utilized activity based screening of HDAC (irrespective of the isozyme type) inhibitors is the small peptide substrate containing C-terminal acetyl lysine conjugated with 7-amino-4-methylcoumarin (AMC) as a fluorophore, and such substrate is commercially available from BioMol International (now Enzo Life Sciences). This assay relies on the initial deacetylation of lysine by HDAC followed by the cleavage of the lysine-AMC amide linkage by trypsin to produce the fluorescent AMC product [15, 16]. Although the above approach can be standardized to serve as the continuous (direct) assay of the enzyme [20], it suffers from an initial lag phase, long assay time, and the potential of inhibition of the coupling enzyme, trypsin, by the compounds being screened as the HDAC inhibitors, yielding false positive results. Recently, competitive binding assay using fluorescent inhibitors of HDAC or HDAC homolog have been described [17-19]. The existing competitive ligand binding methodologies either relies on the fluorescence resonance energy transfer (FRET) from the intrinsic tryptophan residues of HADCs to the enzyme bound furylacryloyl hydroxamic acid as the weak fluorophore [17], or changes in the polarization/anisotropy of fluorescent (e.g. fluorescein or Atto700) hydroxamic acid derivatives upon binding to the enzyme [18, 19]. Of these, the FRET based assay may suffer from the fact that many inhibitors (including SAHA and TSA) quenches the intrinsic tryptophan fluorescence of HDAC8 (our unpublished results) and possibly other HDAC isoenzymes, and thus would produce unreliable fluorescent signals in the competitive assay system. On the other hand, the polarization approach is expected to suffers from the low signal to noise ratio (due to diminution of light intensity after passing through the polarizers). The latter approach further requires spectrofluorometers which are equipped with the polarization accessories. These limitations become further accentuated in determining dissociation off-rates of inhibitors, relying on the polarization methods via the stopped-flow system.
In view of the above limitations, we purported to develop a fluorescent based assay system for screening the HDAC inhibitors as well as for determining their binding affinities and dissociation off-rates. The latter parameter was deemed to be more important since the effectiveness of the putative drugs is not only dependent on their binding affinities but also on their time dependent dissociability (koff values) from their cognate receptor/enzyme sites [21], and this feature has been argued to be particularly important as regards to the effectiveness of SAHA for the treatment of cutaneous T cell lymphoma [9].
In pursuit of designing c-SAHA as the fluorescent probe, we noted that the binding affinities of hydroxamate inhibitors for HDACs are drastically enhanced upon incorporation of aromatic “cap” moieties (which sit on the exterior opening of the deep active site pocket of the enzyme) in the ligand structure [22, 23]. For example, in case of SAHA, the anilino group serves as the cap moiety of the inhibitor, and the X-ray crystallographic data reveals that it interacts with the hydrophobic residues at the opening of the enzyme’s active site pocket [22, 23]. In view of the above feature, it appeared logical to replace the anilino group of SAHA by some fluorophore of comparable steric feature so that the binding affinity of the resultant derivative is not compromised. We conceived that that 7-amino-4-methylcoumarin would serve as an ideal fluorescent moiety for replacing the anilino moiety of SAHA to generate coumarin-SAHA as the resultant fluorophore. As will be shown in the subsequent sections, the fluorescence emission spectrum of c-SAHA is quenched upon binding to HDAC8, and such quenching is overcome (resulting in an enhanced fluorescence) upon its competitive displacement by the HDAC inhibitors. Based on the above feature, we have been able to determine the binding affinities as well as dissociation off-rates of selected HDAC inhibitors.
MATERIALS AND METHODS
Materials
SAHA (suberoylanilide hydroxamic acid) was custom synthesized by Biomol Laboratories (Plymouth Meeting, PA). SBHA (Suberoyl bis-hydroxamic acid), M344 (4-Dimethylamino-N-(6-hydroxycarbamoylhexyl)-benzamide) and TSA (Trichostatin) were purchased from Enzo Life Sciences. Nonidet P-40 substitute was from USB Corporation, Cleveland. The plasmid containing the coding sequence of human HDAC8 (pCMV-SPORT6) was obtained from Open Biosystems Huntsville, AL. The ligation independent cloning (LIC) compatible E. coli expression vector pLIC-His [24] was kind gift from Prof. Stephen P. Bottomley (Monash University, Australia). All other chemicals utilized herein were of analytical reagent grade.
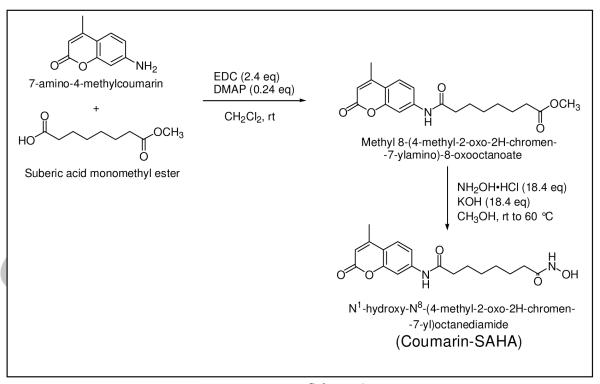
Synthesis of Coumarin-SAHA
Coumarin-SAHA was synthesized via the following two steps, where the second step is similar to the procedure described for the synthesis of SAHA [25]:
Step-I
Suberic acid monomethyl ester (2 mmol), 4-dimethylaminopyridine (DMAP) (0.48 mmol), and 7-amino-4-methylcoumarin (2 mmol) were dissolved in dichloromethane (20 mL) at room temperature. After 30 minutes 1-(3-dimethylamino-propyl)-3-ethylcarbodiimide hydrochloride (EDC) (4.8 mmol) was added, and the mixture was stirred at room temperature for 24 hr. To the reaction mixture was added 50 mL dichloromethane and the solution was washed with water (3 × 50 mL), dried (NaSO4) and evaporated under reduced pressure. The crude product was purified by elution on a short pad of silica gel with ethyl acetate and hexane (10:1) to obtain pure methyl 8-(4-methyl-2-oxo-2H-chromen-7-ylamino)-8-oxooctanoate as a pale yellow solid (366 mg) in 52% yield. HRMS calculated for C19H23NO5 (M+Na)+: 369.1468; Found: 369.0401. The other characteristics of the above intermediate were as follows: Pale yellow solid; mp = 165 °C; IR (film): 3215, 1739, 1716, 1651 cm−1; 1H NMR (CDCl3, 400 MHz): 400 MHz): δ = 1.39-1.34 (m, 4H), 1.61 (tt, J = 7.6, 8.0 Hz, 2H), 1.73 (tt, J = 7.6, 8.0 Hz, 2H) 2.31 (t, J = 7.6 Hz, 2H), 2.40 (s, 3H), 2.43 (t, J = 7.6 Hz, 2H), 3.64 (s, 3H), 6.18 (s, 1H), 7.53 (d, J = 8.4 Hz, 1H), 7.62 (s, 1H), 7.86 (d, J = 8.8 Hz, 1H), 8.25 (s, 1H); 13C NMR (CDCl3 + DMSO-D6, 100 MHz): δ = 16.3, 22.6, 23.0, 26.5, 26.6, 31.5, 34.7, 49.3, 103.7, 110.4, 113.0, 113.3, 123.9, 140.9, 151.1, 152.0, 158.3, 170.2, 171.5.
Step-II
Hydroxylamine hydrochloride (0.76 mmol) in methanol (5 mL) was combined with a solution of KOH (13.8 mmol) in methanol (7 mL) at 40 °C, cooled to 0 °C, and then filtered. Methyl 8-(4-methyl-2-oxo-2H-chromen-7-ylamino)-8-oxooctanoate (0.467 mmol) was then added to the filtrate followed by slow addition (over 30 min) of KOH (0.05 mmol). The mixture was stirred at room temperature for 2 hr and then at 60 °C for 36 h. The reaction mixture was poured into cold water (10 mL) while stirring, and acetic acid was added dropwise until the pH was attained at 7. The precipitate was filtered and the resulting solid product was dried under vacuum. The crude product was purified by silica gel column chromatography eluted with ethyl acetate-dichloromethane (95:5) to furnish pure N1-hydroxy-N8-(4-methyl-2-oxo-2H-chromen-7-yl) octanediamide as a pale brown solid (27 mg) in 17% yield. HRMS calculated for C18H22N2O5 (M+Na)+ was 369.1421, and the observed value was 369.1435. The other characteristics of the final product were as follows: Pale brown solid; mp = 168 - 170 °C; IR (film): 3215, 1718, 1652, 1615 cm−1; 1H NMR (CD3OD, 400 MHz): δ = 1.34-1.40 (m, 4H ), 1.57-1.65 (tt, J = 7.0, 7.6 Hz, 2H,), 1.66-1.72 (q, J = 7.0, 7.6 Hz, 2H), 2.07 (t, J = 7.6 Hz, 2H), 2.39 (t, J = 7.6 Hz, 2H), 2.43 (s, 3H), 6.20 (d, J = 1.2 Hz, 1H), 7.46 (dd, J = 8.8, 2.0 Hz, 1H), 7.67 (d, J = 8.4 Hz, 1H), 7.77 (d, J = 2.0 Hz, 1H); 13C NMR (CDCl3 + DMSO-D6, 100 MHz): δ = 23.5, 30.2, 30.3, 33.7, 33.8, 37.8, 42.0, 111.4, 117.5, 120.1, 120.6, 130.1, 147.9, 157.7, 159.1, 165.9, 175.2, 177.6.
Cloning, Expression and Purification of Recombinant Human HDAC8
For expression of recombinant human HDAC8, the coding sequence of human HDAC8 was amplified from the pCMV-SPORT plasmid by PCR using the sense and antisense primers, 5′-CCAGGGAGCAGCCTCGATGGAGGAGGAGCCGGAGGAACCG-3′ and 5′-GCAAAGCACCGGCCTCGTTAGACCACATGCTTCAGATTCCC-3′, respectively. The resulting PCR product was incorporated into the pLIC-His expression plasmid vector using ligation independent cloning carried out as described previously [24]. Incorporation of the coding region of human HDAC8 into the resulting pLIC-His plasmid vector (pLIC-His-HDAC8) was confirmed by sequencing of the plasmid at the University of Chicago, Cancer Research Center. The expression and purification of HDAC8 was performed by following procedure as described earlier [22] with a few modifications. The pLIC-His-HDAC8 plasmid was transformed into E. coli BL21 codon plus DE3 (RIL) cells (Stratagene; La Jolla, CA) following the standard molecular biology protocol [26]. The transformed cells were grown in LB (Luria Bertani) medium, supplemented with 100 μg/mL ampicillin and 30 μg/mL chloramphenicol at 37 °C (shaker speed = 220 rpm) until the optical density of 0.6-0.8 was reached at 600 nm. The expression of HDAC8 was induced by addition of 0.05% (w/v) lactose and the media was supplemented with 100 μM ZnCl2. At this point, the cells were further grown at 18 °C (shaker speed = 220 rpm) overnight and were harvested by centrifugation at 5000 g for 15 min. The pellet was resuspended in a lysis buffer (50 mM Tris-HCl, pH 8, containing 150 mM KCl, 3 mM MgCl2, 1 mM 2-mercaptoethanol, 1 mM PMSF (Phenylmethylsulfonyl fluoride) and 0.25% Nonidet P-40. Following resuspension, the cells were lysed by sonication and the crude extract was centrifuged at 15000 g for 30 min to remove the cellular debris. The resulting supernatant was loaded onto a Ni2+-charged iminodiacetic acid (IDA) resin column, synthesized in our laboratory as described in [27], and pre-equilibrated by the lysis buffer minus PMSF and the detergent. HDAC8 was eluted by a linear gradient of 0-200 mM imidazole in the above buffer, and the fractions containing HDAC8 activity were pooled and dialyzed overnight at 4 °C against the storage buffer (10 mM Tris, pH 7.5, containing 100 mM NaCl, 3 mM MgCl2, 10% glycerol and 1 mM TCEP). The purified enzyme fractions were concentrated by ultrafiltration and the protein concentration was determined by the Bradford method using BSA as the standard protein [28]. The purified enzyme was judged to be homogeneous by the SDS-PAGE analysis (with > 95 % purity), and was stored at −70 °C.
Spectrofluorometric Studies
All the steady-state spectrofluorometric studies were performed on a Perkin-Elmer lambda 50-B spectrofluorometer, equipped with a magnetic stirrer and thermostated water bath using a 4 × 4 mm2 square quartz cuvette. The binding isotherm for the interaction of c-SAHA with HDAC8-complex was determined by titration of a fixed concentration of c-SAHA (0.1 μM) with increasing concentrations of HDAC8 in the storage buffer described above. The fluorescence emission spectrum of c-SAHA was monitored in the range of 400–500 nm (slit = 10) after excitation at 325 nm (slit = 8). The resulting binding isotherm for the HDAC8-c-SAHA complex was analyzed via the complete solution of the quadratic equation as described by Qin and Srivastava [29].
Determination of the Ki values of Competitive Inhibitors
The inhibition constant of HDA8-inhibitor complexes were determined using the trypsin-coupled assay as described by Schultz et al. [20]. The enzyme activity was determined in the assay buffer (50 mM Tris-HCl buffer, pH 7.5, containing 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 1 mg/ml BSA) using 150 μM Biomol’s “Fluor de Lys®” as the fluorogenic substrate, 100 nM Trypsin and appropriately diluted HDAC8. The initial velocity of the enzyme catalyzed reaction was determined by monitoring the time dependent increase in the fluorescence intensity at 500 nm (λex = 365 nm) in the absence and presence of inhibitors, and the Ki values of the inhibitors were determined as described by Morrison [31]. Although Bradner et al. [30] has recently reported that trifluoroacetyllysine containing tripeptide is a better substrate for HDAC8 , we did not experience any problem in determining the Ki values of inhibitors using the classical Fluor de Lys® substrate.
Screening of HDAC Inhibitors and Determining their Dissociation Constants
Since the fluorescence emission intensity of c-SAHA is quenched upon binding to HDAC8 and it is restored upon competitive displacement by the enzyme’s inhibitors (see Results), we utilized the above feature for screening selected HDAC inhibitors (namely, SAHA, TSA, SBHA, and M344). In this endeavor, we measured the fluorescence emission intensity of 0.5 μM SAHA in the absence and presence of 0.5 μM HDAC8 (λex = 325 nm,λem = 400 nm) in 10 mM Tris buffer, pH 7.5, containing 100 mM NaCl, 1 mM TCEP. The mixture containing c-SAHA and HDAC8 was added with 10 μM each of the enzyme inhibitor and the increase in the fluorescence intensity (due to competitive displacement of c-SAHA from the enzyme site) was measured at 400 nm (λex = 325 nm). On consideration that the reasonable HDAC8 inhibitor should have the binding affinity of 1 μM or less, we set 50% (or higher) increase in the fluorescence intensity as the cut-off limiting for selecting putative enzyme inhibitors. As expected (based on their inhibitory potencies), all four test inhibitors, utilized herein, passed the above screening criteria.
The increase in the fluorescence signal associated with the displacement of c-SAHA from HDAC8 site (by the enzyme’s competitive inhibitors) was utilized for determining the direct binding affinities of the HDAC8-inhibitor complexes. However, it was realized that both c-SAHA and our selected enzyme inhibitors had fairly high (in the sub-micromolar range) binding affinities, and thus we could not treat “total” concentration of either of the ligands as their “free” concentrations. This feature precluded us from using the quadratic equation developed in our laboratory [32] for determining the binding affinities of weaker inhibitors, which would competitively displace the tight binding fluorescent probe from the enzyme site. With two tight binding (competing) ligands for an enzyme, the algebraic expression required to explain the overall binding isotherm (by combining all partial equilibria and mass balances) has to be “cubic” in nature with possible multiple roots [33], and the use of latter equation is computationally challenging. However, recourse has been made by employing the recursive approach and calculating the overall changes in signal (in the present case contributed by c-SAHA) as a function of the enzyme and ligand concentrations as elaborated by Kuzmic et al. [34] for the binding of cyclosporine A to cyclophilin in competition with a fluorescent analogue. This situation is similar to the competitive binding of c-SAHA to HDAC8 in the presence our test inhibitors, which exhibit comparable tight binding affinity vis a vis the fluorescent ligand (Kdc-SAHA ≈ Kdinhibitor < [E]total). Such recursive-iterative feature for calculating the fluorescence intensity for the competitive binding of the fluorophore and cyclosporine A to cyclophilin has been included as a module (with a script file) in Dynafit software [35]. The details of modifying the script and using the program is elaborated in the Dynafit manual.
The experimental design for determining the binding affinities of test inhibitors using c-SAHA as the fluorescent probe and HDAC8 as the enzyme is similar to that described by Kuzmic et al. [34]. To minimize the interference due to the inner filter effect, we titrated a mixture of an inhibitor and c-SAHA with increasing concentration of HDAC8. Under this situation, at each HDAC8 concentration, the fluorescence intensity was dependent on the fractional occupancy of the competing (fluorescent and non-fluorescent) ligands at the enzyme site, and overall binding isotherm was analyzed via the recursive-iterative approach built in the Dynafit software [35], yielding the binding affinities of all our test inhibitors. It should be mentioned that this approach is also applicable even if an inhibitor falls in the class of the “weak binding” inhibitor (Kdc-SAHA < [E]total < Kdinhibitor), albeit in the latter case, the ratio of c-SAHA to inhibitor has to be adjusted to obtain reliable change in the fluorescence signal.
Transient Kinetic Experiments
To determine the dissociation rate constant of inhibitor from the HDAC8 site, transient kinetic experiments were performed using an Applied Photophysics SX-18MV stopped-flow system. The stopped-flow system (dead time = 1.3 ms) was operated in the fluorescence mode with an emission path length of 2 mm. The time-dependent changes in the c-SAHA fluorescence was monitored by exciting the reaction at 325 nm, and the emission intensity was detected after passing the light through a 395 nm high-pass cut-off filter. All the transient kinetic experiments were performed in triplicate in 10 mM Tris buffer, pH 7.5, containing 100 mM NaCl, 1 mM TCEP and the resultant kinetic traces were analyzed by the data analysis package provided by Applied Photophysics.
RESULTS
Synthesis and Characterization of Coumarin-SAHA as a fluorescent probe for HDAC8
Based on our preliminary modeling results, it appeared that substitution of anilino moiety of SAHA by 7-amino-4-methylcoumarin would neither distort the alkyl chain nor pose any significant steric constraint (particularly in the “cap region” of the enzyme site) in binding of the resultant derivative to the active site pocket of human HDAC8 [22]. The synthesis of the fluorescent probe was accomplished by activation of free carboxyl group of suberic acid monomethyl ester by 1-(3-dimethylamino-propyl)-3-ethylcarbodiimide hydrochloride (EDC) followed by the amidation reaction with 7-amino-4-methylcoumarin in the presence of 4-dimethylaminopyridine (DMAP) and dichloromethane as solvent. The purified form of the resultant product was treated with hydroxylamine under alkaline condition to produce N1-hydroxy-N8-(4-methyl-2-oxo-2H-chromen-7-yl)octanediamide (referred herein as Coumarin-SAHA and abbreviated as c-SAHA) as the final product as summarized in Scheme-1. The crude preparation of c-SAHA was subjected to purification over the silica gel column and its chemical identity was ascertained by a combination of NMR and Mass spectrometry (see Materials and Methods).
Scheme-1.
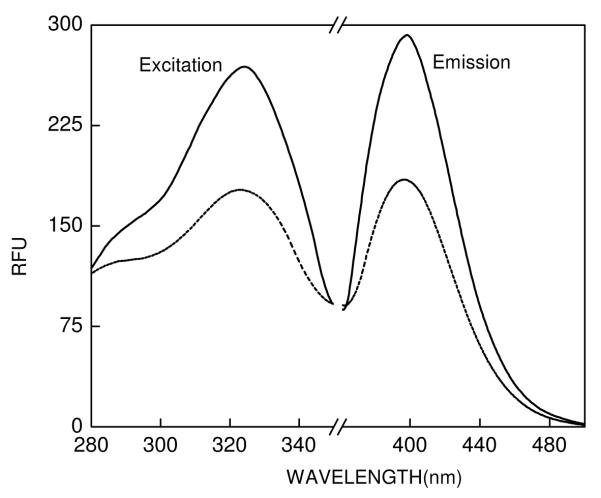
The UV-Vis spectra of c-SAHA showed a prominent absorption band at 325 nm and a minor band at 290 nm (data not Shown). Figure 1 shows the fluorescence excitation and emission spectra of c-SAHA (0.5 μM) in 10 mM Tris buffer, pH 7.5, containing 100 mM NaCl, 1 mM TCEP. Note the fluorescence excitation and emission maxima of free c-SAHA being equal to 325 nm and 400 nm, respectively. We determined the quantum yield of c-SAHA using quinine sulfate as a standard [36] and found its magnitude to be 0.43. Evidently, the quantum yield of free coumarin is not much affected upon conjugated with the aliphatic hydroxamate chain. To probe whether or not the fluorescence profile of c-SAHA is modulated upon binding to HDAC8, we determined the fluorescence emission spectra of the enzyme-fluorophore complex. As shown in Figure 1, although the presence of 2 μM HDAC8 does not change the emission maxima of the fluorophore, it quenches its intensity by about 50% (Figure 1, ----- lines). The steady-state kinetic data revealed that the Ki value of c-SAHA is 160 nM, which is about 5 fold lower than that obtained for its parent compound SAHA.
Figure 1.
Excitation and fluorescence emission spectra of coumarin-SAHA. The solid and the dotted lines represent the spectra of free and enzyme bound forms of c-SAHA, respectively. The spectra were taken in 10 mM Tris buffer pH 7.5 containing 100 mM NaCl, 1 mM TCEP. [c-SAHA] = 0.5 μM, [HDAC] = 2 μM, (λex = 325 nm), and they were corrected by subtracting the spectral contributions of buffer and the enzyme.
Binding Isotherm of HDAC8-c-SAHA
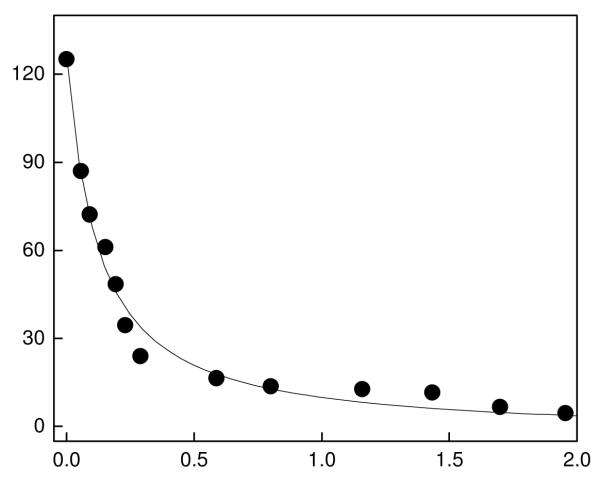
Given that the fluorescence of c-SAHA is quenched upon binding to HDAC8 (see Figure 1), we could utilize such a signal for determining their binding affinity. Figure 2 shows the titration of a fixed concentration of c-SAHA (100 nM) as a function of increasing concentration of HDAC8. Note that at lower concentrations of HDAC8, the fluorescence emission intensity of the c-SAHA decreases almost linearly as a function of the enzyme concentration, suggesting a very strong binding affinity between the interacting species. Hence, the binding affinity of the enzyme-fluorophore complex was determined by analyzing the data by a complete solution of the quadratic equation [29]. The solid smooth line of Figure 2 is the best fit of the data for the Kd value of the HDAC8-c-SAHA complex as being equal to 160± 20 nM.
Figure 2.
Binding isotherm for the interaction of c-SAHA with HDAC8. The decrease in fluorescence emission intensity at 400 nm (λex = 325 nm) for the titration of 100 nM c-SAHA with increasing concentration of HDAC8 (total) is shown. The solid smooth line is the best fit of the experimental data Qin and Srivastava [29] for the Kd value of 0.16 ± 0.02 μM.
Determination of the binding affinities of inhibitors for HDAC8 via the fluorescence displacement method
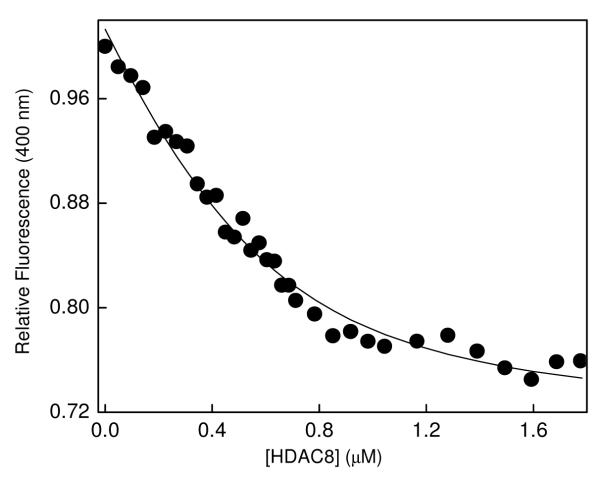
Due to mutually exclusive binding of c-SAHA with respect to non-fluorescent competitive inhibitor of HDAC8, we proceeded to determine the dissociation constants of the enzyme-inhibitor complexes by the fluorescence displacement method. However, in such an approach, we noted that the binding affinities of both c-SAHA and selected HDAC8 inhibitors were fairly high, and they were comparable to the concentration of enzyme (Kdc-SAHA ≈ Kdinhibitor < [E]total) required for obtaining detectable signals. Under this situation, evaluation of “free” versus “bound” concentrations of two ligands (i.e., c-SAHA and putative enzyme inhibitor for which the Kd value was to be determined) would require solutions of a cubic equation, the employment of which is computationally challenging [33]. As described in the Materials and Methods section, recourse was made to perform the competitive binding experiments as elaborated by Kuzmic et al. [34], and the data were analyzed by the aid of the software Dynafit [35]. As a representative example for determining the Kd value of SAHA, we titrated a mixture containing 0.5 μM c-SAHA and 2 μM SAHA with increasing concentration of HDAC8 and monitored the fluorescence signal at 400 nm (λex = 325 nm) as shown in Figure 3. It should be pointed out that at the above excitation and emission wavelengths, there was no interference from the intrinsic tyrosine and tryptophan fluorescence signals of the enzyme. The above titration protocol also minimized the inner filter effect due to employment of low concentration of c-SAHA. The data of Figure 3 shows the decrease in relative fluorescence of c-SAHA in the presence of SAHA as a function of increasing concentration of HDAC8. The solid smooth line is the best fit of the data for the Kd value of HDAC8-SAHA complex as being equal to 0.58 ± 0.14 μM. We performed similar experiment for the binding of other (selected) inhibitors of HDAC, namely TSA (Trichostatin A), M344 (4-Dimethylamino-N-(6-hydroxycarbamoylhexyl)-benzamide), SBHA (Suberoyl bis-hydroxamic acid) via the above protocol and determined their Kd values. The data are summarized in Table 1. To test whether the Kd values of the above inhibitors are similar to their Ki values, we performed the steady-state kinetic experiments using Fluor de Lys® as the fluorogenic substrate and tryspin as the coupling enzyme as described by Schultz et al. [20]. It should be pointed out that the Ki values of all four test inhibitors, determined under our experimental condition, are comparable to the IC50 values reported in the literature [37-39]. It should be further pointed out that the Kd and Ki values of all four test inhibitors (Table 1) are similar, attesting to the authenticity our analytical protocol.
Figure 3.
Changes in the fluorescence intensity of c-SAHA upon competitive binding SAHA to HDAC8. The decrease in the relative fluorescence emission intensity of a mixture of c-SAHA and SAHA (λex = 325 nm, λem = 400 nm) as a function of total concentration of HDAC8 has been shown. The concentrations of c-SAHA and SAHA were maintained to be 0.5 μM and 2 μM, respectively. The data were analyzed using the software DynaFit [35], and the solid smooth line represents the best fit of the experimental data for the Kd value of HDAC8-SAHA complex as being equal to 0.58 ± 14 μM
Table-1.
| HADC8 inhibitor | Ki (μM) | Kd (μM) |
|---|---|---|
| TSA | 0.12 ± 0.02 | 0.34 ± 0.12 |
| SAHA | 0.29 ± 0.04 | 0.58 ± 0.14 |
| M344 | 0.18 ± 0.02 | 0.32 ± 0.09 |
| SBHA | 0.71 ± 0.03 | 0.75 ± 0.36 |
Determined via the competitive displacement method using c-SAHA as fluorescent probe
Determined via the steady-state kinetic method
Determination of the Dissociation off-rates of HDC8-inhibitor complexes
An added feature of c-SAHA as a fluorescent probe is its direct applicability in determining the dissociation off rates of HDAC-inhibitor complexes. Since the competitive binding of c-SAHA to HDAC8 (concomitant with displacement of the enzyme bound inhibitors) results in a decrease in the fluorescent signal at 400 nm (λex = 325 nm), the dissociation off rates of the enzyme-inhibitor complexes could be easily determined via the simple stopped-flow method without using polarization accessories. Equation 1 outlines the principle involved in determining the dissociation off-rate of HDAC8-inhibitor complex.
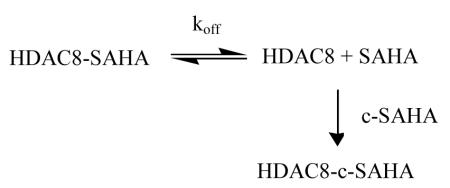 |
(1) |
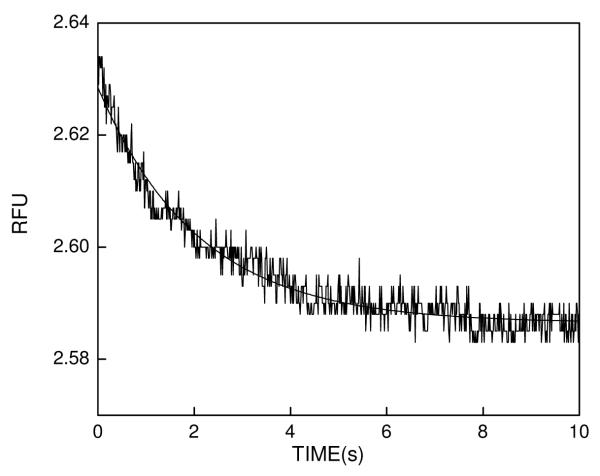
If an enzyme-inhibitor complex is mixed with a high and excessive concentration of c-SAHA, the mass action would drive the overall equilibrium to the right, and under such condition the rate of formation of the enzyme-c-SAHA complex would be given by the dissociation off rate of the inhibitor. We tested this feature for the dissociation off rate of SAHA from HDAC8 by mixing the above species ([HDAC8] = 1 μM and [SAHA] = 10 μM) with 40 μM c-SAHA via the stopped flow syringes. Figure 4 shows the time dependent decrease in fluorescence signal (λex = 325 nm, “cutoff” filter = 395 nm) due to formation of HDAC8-c-SAHA complex. The solid smooth line is the best fit of the data for single exponential rate equation with rate constant of 0.48 ± 0.06 s−1. Since the above rate constant remains unaffected when the concentration of c-SAHA is increased by two fold (data not shown), the above rate constant serves as the measure of the dissociation off rate of SAHA from HDAC8 site. By performing similar transient kinetic experiments, we determined the dissociation rate constants of other test inhibitors, and their magnitudes are summarized in Table 2.
Figure 4.
Representative stopped flow trace for the dissociation of SAHA from the HDAC8 site. The dissociation of the enzyme-bound SAHA was triggered upon mixing of HDAC8-SAHA complex with a high and excessive concentration of c-SAHA. The premixing concentrations of the individual species in the stopped flow syringes were as follows: [HDAC8] = 1.0 μM + [SAHA] = 10 μM (Syringe 1) versus [c-SAHA] = 40 μM (Syringe 2). The solid smooth lines are the best fit of the experimental data according to the single exponential rate equation for a dissociation off rate constant of 0.48 ± 0.06 s−1.
Table 2.
Dissociation off-rates of selected HDAC8 inhibitors1
| HADC8 inhibitor | koff (s−1) |
|---|---|
| TSA | 0.12 ± 0.04 |
| SAHA | 0.48 ± 0.06 |
| M344 | 1.05 ± 0.12 |
| SBHA | 1.01 ± 0.03 |
Determined by the stopped-flow transient kinetic method using c-SAHA as a competitive ligand
DISCUSSION
We introduce, for the first time, coumarin-SAHA (c-SAHA) as an ideal fluorescent probe for determining the binding affinity and dissociation off-rates of selected HDAC inhibitors. Aside from its ease in synthesis, it offers several advantages over other protocols developed by other groups toward determining the above parameters [16-19,40]: (1) The fluorescent intensity of c-SAHA is quenched upon binding to HDAC8, and it is enhanced upon competitive displacement by the enzyme inhibitors. (2) Since the excitation maximum of c-SAHA is far removed from those of the enzyme’s tryptophan and tyrosine residues, the above probe can be exclusively excited without any interference from the latter. (3) Since our overall analytical protocol (for determining Kd and koff values of HDAC-inhibitor complexes) via changes in the fluorescence intensity of c-SAHA, there is no need for employment of fluorescence equipment (e.g., steady-state spectrofluorometer and stopped flow system) which are equipped with polarization accessories. (4) Since SAHA is a considered to be a general HDAC inhibitor, with a few exception [30], c-SAHA can be used for screening inhibitors for most of the HDAC isozymes as well as determining their Kd and koff values
We were initially surprised to note that the binding affinity of c-SAHA for HDAC8 was somewhat higher than that of SAHA, suggesting that the coumarin moiety is better accommodated in the “cap region” of the enzyme than the anilino moiety of SAHA. Our transient kinetic studies (to be published subsequently) have revealed that such feature is encoded in the isomerization of the initially formed enzyme-ligand collision complex.
In summary, our newly synthesized c-SAHA serves as an ideal fluorescent probe for determining the dissociation constants as well as the dissociation off-rates of different HDAC8 inhibitors. Since SAHA inhibits most of the HDAC isozymes, with a few exception [30], it is expected that c-SAHA would interact with all those isozymes as well, and thus our analytical protocol would be find application with different HDAC isozymes and their cognate competitive inhibitors as long as the latter do not interfere with the fluorescent signal of c-SAHA. Our approach neither relies on the resonance energy transfer from the enzyme’s tryptophan residues to the probe nor requires polarization accessories for determining the Kd and koff values of the HDAC-inhibitor complexes.
ACKNOWLEDGEMENTS
This research was supported by the NIH grants CA113746 and CA132034 and the National Science Foundation grant DMR-0705767 to DKS, and the NIH COBRE grant NCRR-P20-RR15566 to GC
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
- HDAC
- histone deacetylase
- SAHA
- suberoylanilide hydroxamic acid
- c-SAHA
- coumarin-SAHA
- Kd
- equilibrium dissociation constant
- Ki
- inhibition constant
- koff
- dissociation rate constant
- HTS
- high throughput setting
- AMC
- 7-amino-4-methylcoumarin
- FRET
- fluorescence resonance energy transfer
- TSA
- trichostatin
- M344
- 4-Dimethylamino-N-(6-hydroxycarbamoylhexyl)-benzamide
- SBHA
- suberoyl bis-hydroxamic acid
- LIC
- ligation independent cloning
- DAMP
- 4-dimethylaminopyridine
- PCR
- polymerase chain reaction
- LB
- luria bertani
- PMSF
- phenylmethanesulfonyl fluoride
- IDA
- iminodiacetic acid
- TCEP
- tris(2-carboxyethyl)- phosphine hydrochloride
- BSA
- bovine serum albumin
- EDC
- 1-(3-dimethylamino-propyl)-3-ethylcarbodiimide hydrochloride
- NMR
- nuclear magnetic resonance
- RFU
- relative fluorescence unit
- Uv-vis
- ultraviolet visible
- DTNB
- 5,5′-dithiobis(2-nitrobenzoic acid)
REFERENCES
- [1].Strahl D, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- [2].Marmorstein R. Protein modules that manipulate histone tails for chromatin regulation. Nat. Rev. Mol. Cell Biol. 2001;2:422–432. doi: 10.1038/35073047. [DOI] [PubMed] [Google Scholar]
- [3].Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- [4].Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- [5].Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the Regulation of RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- [7].Ma X, Ezzeldin HH, Diasio RB. Histone deacetylase inhibitors: current status and overview of recent clinical trials. Drugs. 2009;69:1911–1934. doi: 10.2165/11315680-000000000-00000. [DOI] [PubMed] [Google Scholar]
- [8].Grant C, Rahman F, Piekarz R, Peer C, Frye R, Robey RW, et al. Romidepsin: a new therapy for cutaneous T-cell lymphoma and a potential therapy for solid tumors. Expert Rev Anticancer Ther. 2010;10:997–1008. doi: 10.1586/era.10.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- [10].Gregoretti IV, Lee Y, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- [11].Holbert MA, Marmorstein R. Structure and activity of enzymes that remove histone modifications. Curr. Opin. Struct. Biol. 2005;15:673–680. doi: 10.1016/j.sbi.2005.10.006. [DOI] [PubMed] [Google Scholar]
- [12].Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- [13].de Ruijter AJM, van Gennip AH, Caron HN, Kemp S, van Kuilenburg ABP. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wegener D, Hildmann C, Schwienhorst A. Recent progress in the development of assays suited for histone deacetylase inhibitor screening. Mol. Genet. Metab. 2003;80:138–147. doi: 10.1016/j.ymgme.2003.08.008. [DOI] [PubMed] [Google Scholar]
- [15].Wegener D, Wirsching F, Riester D, Schwienhorst A. A fluorogenic histone deacetylase assay well suited for high-throughput activity screening. Chem. Biol. 2003;10:61–68. doi: 10.1016/s1074-5521(02)00305-8. [DOI] [PubMed] [Google Scholar]
- [16].Wegener D, Hildmann C, Riester D, Schwienhorst A. Improved fluorogenic histone deacetylase assay for high-throughput-screening applications. Anal. Biochem. 2003;321:202–208. doi: 10.1016/s0003-2697(03)00426-3. [DOI] [PubMed] [Google Scholar]
- [17].Riester D, Hildmann C, Schwienhorst A, Meyer-Almes F. Histone deacetylase inhibitor assay based on fluorescence resonance energy transfer. Anal. Biochem. 2007;362:136–141. doi: 10.1016/j.ab.2006.12.019. [DOI] [PubMed] [Google Scholar]
- [18].Riester D, Hildmann C, Haus P, Galetovic A, Schober A, Schwienhorst A, Meyer-Almes F. Non-isotopic dual parameter competition assay suitable for high-throughput screening of histone deacetylases. Bioorg. Med. Chem. Lett. 2009;19:3651–3656. doi: 10.1016/j.bmcl.2009.04.102. [DOI] [PubMed] [Google Scholar]
- [19].Mazitschek R, Patel V, Wirth DF, Clardy J. Development of a fluorescence polarization based assay for histone deacetylase ligand discovery. Bioorg. Med. Chem. Lett. 2008;18:2809–2812. doi: 10.1016/j.bmcl.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Schultz BE, Misialek S, Wu J, Tang J, Conn MT, Tahilramani R, et al. Kinetics and comparative reactivity of human class I and class IIb histone deacetylases. Biochemistry. 2004;43:11083–11091. doi: 10.1021/bi0494471. [DOI] [PubMed] [Google Scholar]
- [21].Krogsgaard M, Prado N, Adams EJ, He X, Chow D, Wilson DB, Garcia KC, Davis MM. Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T cell activation. Mol. Cell. 2003;12:1367–1378. doi: 10.1016/s1097-2765(03)00474-x. [DOI] [PubMed] [Google Scholar]
- [22].Vannini A, Volpari C, Filocamo G, Casavola EC, Brunetti M, Renzoni D, Chakravarty P, Paolini C, De Francesco R, Gallinari P, Steinkühler C, Di Marco S. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15064–15069. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Somoza JR, Skene RJ, Katz BA, Mol C, Ho JD, Jennings AJ, Luong C, Arvai A, Buggy JJ, Chi E, Tang J, Sang B, Verner E, Wynands R, Leahy EM, Dougan DR, Snell G, Navre M, Knuth MW, Swanson RV, McRee DE, Tari LW. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure. 2004;12:1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- [24].Cabrita LD, Dai W, Bottomley SP. A family of E. coli expression vectors for laboratory scale and high throughput soluble protein production. BMC Biotechnol. 2006;6:1–8. doi: 10.1186/1472-6750-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gediya LK, Chopra P, Purushottamachar P, Maheshwari N, Njar VCO. A new simple and high-yield synthesis of suberoylanilide hydroxamic acid and its inhibitory effect alone or in combination with retinoids on proliferation of human prostate cancer cells. J. Med. Chem. 2005;48:5047–5051. doi: 10.1021/jm058214k. [DOI] [PubMed] [Google Scholar]
- [26].Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning A Laboratory Manual. New York; Cold Spring Harbor Laboratory Press: 2001. [Google Scholar]
- [27].Hemanson GT, Mallia AK, Smith PK. Immobilized Affinity Ligand Techniques. Academic Press; San Diego: 1992. [Google Scholar]
- [28].Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- [29].Qin L, Srivastava DK. Energetics of two-step binding of a chromophoric reaction product, trans-3-indoleacryloyl-CoA, to medium-chain acyl-coenzyme-A dehydrogenase. Biochemistry. 1998;37:3499–3508. doi: 10.1021/bi9720585. [DOI] [PubMed] [Google Scholar]
- [30].Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, et al. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Morrison JF. Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors. Biochim. Biophys. Acta. 1969;185:269–286. doi: 10.1016/0005-2744(69)90420-3. [DOI] [PubMed] [Google Scholar]
- [32].Banerjee AL, Swanson M, Roy BC, Jia X, Haldar MK, Mallik S, Srivastava DK. Protein surface-assisted enhancement in the binding affinity of an inhibitor for recombinant human carbonic anhydrase II. J. Am. Chem. Soc. 2004;126:10875–10883. doi: 10.1021/ja047557p. [DOI] [PubMed] [Google Scholar]
- [33].Wang ZX. An exact mathematical expression for describing competitive binding of two different ligands to a protein molecule. FEBS Lett. 1995;360:111–114. doi: 10.1016/0014-5793(95)00062-e. [DOI] [PubMed] [Google Scholar]
- [34].Kuzmic P, Moss ML, Kofron JL, Rich DH. Fluorescence displacement method for the determination of receptor-ligand binding constants. Anal. Biochem. 1992;205:65–69. doi: 10.1016/0003-2697(92)90579-v. [DOI] [PubMed] [Google Scholar]
- [35].Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- [36].Lackowicz JR. Principles of Fluorescence Spectroscopy. Springer; 2006. [Google Scholar]
- [37 ].Hildmann C, Wegener D, Riester D, Hempel R, Schober A, Merana J, et al. Substrate and inhibitor specificity of class 1 and class 2 histone deacetylases. J. Biotechnol. 2006;124:258–270. doi: 10.1016/j.jbiotec.2006.01.030. [DOI] [PubMed] [Google Scholar]
- [38 ].Hahnen E, Eyüpoglu IY, Brichta L, Haastert K, Tränkle C, Siebzehnrübl FA, et al. In vitro and ex vivo evaluation of second-generation histone deacetylase inhibitors for the treatment of spinal muscular atrophy. J. Neurochem. 2006;98:193–202. doi: 10.1111/j.1471-4159.2006.03868.x. [DOI] [PubMed] [Google Scholar]
- [39 ].Hahnen E, Hauke J, Tränkle C, Eyüpoglu IY, Wirth B, Blümcke I. Histone deacetylase inhibitors:possible implications for neurodegenerative disorders. Expert Opin. Investig. Drugs. 2008;17(2):1–16. doi: 10.1517/13543784.17.2.169. [DOI] [PubMed] [Google Scholar]
- [40].Fatkins DG, Zheng W. A spectrophotometric assay for histone deacetylase 8. Anal. Biochem. 2008;372:82–88. doi: 10.1016/j.ab.2007.08.031. [DOI] [PubMed] [Google Scholar]