Abstract
Numerous studies support the fact that a genetically diverse mouse population may be useful as an animal model to understand and predict toxicity in humans. We hypothesized that cultures of hepatocytes obtained from a large panel of inbred mouse strains can produce data indicative of inter-individual differences in in vivo responses to hepato-toxicants. In order to test this hypothesis and establish whether in vitro studies using cultured hepatocytes from genetically distinct mouse strains are feasible, we aimed to determine whether viable cells may be isolated from different mouse inbred strains, evaluate the reproducibility of cell yield, viability and functionality over subsequent isolations, and assess the utility of the model for toxicity screening. Hepatocytes were isolated from 15 strains of mice (A/J, B6C3F1, BALB/cJ, C3H/HeJ, C57BL/6J, CAST/EiJ, DBA/2J, FVB/NJ, BALB/cByJ, AKR/J, MRL/MpJ, NOD/LtJ, NZW/LacJ, PWD/PhJ and WSB/EiJ, males) and cultured for up to 7 days in traditional 2-dimesional culture. Cells from B6C3F1, C57BL/6J, and NOD/LtJ strains were treated with acetaminophen, WY-14,643 or rifampin and concentration-response effects on viability and function were established. Our data suggest that high yield and viability can be achieved across a panel of strains. Cell function and expression of key liver specific genes of hepatocytes isolated from different strains and cultured under standardized conditions is comparable. Strain-specific responses to toxicant exposure have been observed in cultured hepatocytes and these experiments open new opportunities for further developments of in vitro models of hepatotoxicity in a genetically diverse population.
Keywords: hepatocytes; toxicity testing; mouse genetics; acetaminophen; WY-14,643; rifampin
Introduction
The growing list of chemical substances in commerce and the complexity of the environmental exposures from agents of natural origin represents an enormous challenge with respect to the examination of the adverse health effect potential of exposures (Judson et al., 2009). Current chemical hazard testing procedures can only address a small fraction of agents as single exposures in order to provide sufficient information to meet the extensive data needs under the current regulatory risk assessment guidelines (Judson et al., 2010). Toxicity testing in the 21st century paradigm proposed a gradual transition from the apical end-points to a strategy based on in vitro screening and molecular pathway analysis (National Research Council, 2007). The data generated from the high-throughput assays will provide a substantial proportion of the information needed for environmental decision-making (National Research Council, 2007).
Primary hepatocytes constitute one of the most widely adopted in vitro models for investigative hepatic toxicology and are used to evaluate hepatic toxicity, drug metabolism or genotoxic potential of chemicals (Groneberg et al., 2002). In vitro culture systems offer the potential to greatly reduce the number of animals needed to screen large numbers of compounds for adverse effects (Gebhardt et al., 2003). In spite of the drawbacks of primary hepatocyte cultures, including the absence of organ-specific heterotypic cell-cell interactions and the gradual loss of expression of key liver-specific metabolism genes (Hewitt et al., 2007), they provide useful alternatives to whole animal testing (Acosta et al., 1985; Gebhardt et al., 2003).
Experimental studies that examine adverse effects of toxicants usually are unsuitable to understand genetic factors that affect individual susceptibility to disease, do not take into account the genetic diversity present within populations, and largely ignore gene-environment interactions which affect risk (Harrill and Rusyn, 2008). Several recent studies showed promise for a translational in vivo mouse-to-human research strategy which may aid in identification of the genetic polymorphisms contributing to toxicity (Harrill and Rusyn, 2008). Panels of inbred mouse strains have been used to model a genetically diverse population and discover susceptibility loci and genes (Harrill et al., 2009b), as well as to understand genotype-independent toxicity responses (Harrill et al., 2009a). Liu et al. (Liu et al., 2010), through an integrated genetic, transcriptional and metabolic analysis, demonstrated that multiple genetic loci and an interacting network of metabolic factors affect susceptibility to hepatotoxicity in a panel of 16 inbred mouse strains. Guo et al. (Guo et al., 2006) used a panel of inbred mouse strains to reproduce the known inter-individual variability in the metabolism of warfarin, specifically the generation of 7-hydroxywarfarin, and determined that the phenotypic differences were associated with the polymorphisms in the Cyp2c locus. Furthermore, a study which used liver microsomes isolated from the panel of mouse strains demonstrated that genetic variation in Cyp2b9 and Ugt1a loci played a role in the oxidative metabolism of α-hydroxytestosterone and glucuronidation of irinotecan, respectively (Guo et al., 2007).
While these studies support the notion that a genetically diverse mouse population may be useful as an animal model to understand and predict rare adverse drug events in humans, they are based on the in vivo experiments. In this study, our goal was to establish whether in vitro studies using primary cultured hepatocytes isolated from genetically distinct mouse strains are feasible and can produce data that may be used for studies of in vivo adverse effects of toxicants in a population-based model. We show that standardized isolation and culture conditions can be established for hepatocytes from different mouse inbred strains, and that a comparative in vitro/in vivo analysis of toxicant-induced responses is feasible.
Methods
Animals
Primary hepatocytes were isolated from 4–6 week old male mice (strains A/J, B6C3F1, BALB/cJ, C3H/HeJ, C57BL/6J, CAST/EiJ, DBA/2J, FVB/NJ, BALB/cByJ, AKR/J, MRL/MpJ, NOD/LtJ, NZW/LacJ, PWD/PhJ and WSB/EiJ) from Jackson Laboratories (Bar Harbor, ME). Mice were housed by strain in groups of three in a constant alternating 12-h light and dark cycle and allowed free excess to food and water. All procedures were approved by the Institutional Animal Care and Use Committee.
Isolation and culture of mouse hepatocytes
Primary mouse hepatocytes were isolated using a two-step collagenase perfusion method. In brief, the animals were anesthetized with a cocktail of xylazine (2 mg/ml) and ketamine (20 mg/ml) and the liver was perfused in situ with warmed (37°C) saline solution without magnesium and calcium (4 min) followed by a collagenase-containing buffer (8 min, collagenase type IV, Sigma, St Louis, MO) through a catheter inserted through the atrium of the heart into the superior vena cava. Livers were excised and disassociated in supplemented William’s E culture medium (WEM) containing 10% FBS, 2 µg/ml gentamycin, 15 mM HEPES, 0.1µM dexamethasome, 4 µg/ml insulin (Sigma) and 4 mM glutamax (Invitrogen). Cells were filtered (100 micron nylon mesh) and the yield and viability were determined using a trypan blue exclusion test (Sigma, 0.04%). Hepatocytes were further purified by Percoll gradient centrifugation (Sigma). Viable hepatocytes were washed twice with the same medium, transferred to a T-25 tissue culture flask, placed on an orbital shaker (50 rpms) and allowed to recover for 30 min at 37°C. Hepatocytes were suspended in WEM containing 10% FBS and added to the 24-, or 96-well dishes (precoated with PureCol™ purified collagen or SL collagen type I, Sigma) at a density of 1.5×105 or 2×104 cells/well, respectively. Cells were allowed to attach for 4 hr at 37°C and 5% CO2, medium was changed to WEM without FBS and cells were cultured for another 24 hr before initiating experiments. Medium was replaced on a daily basis.
Evaluation of cell viability and morphology
Phase contrast microscopy of the hepatocytes in the monolayer configuration was performed at days 1, 3, 5 and 7 at a magnification of 200× with an Olympus inverted microscope (Center Valley, PA). In some experiments, hepatocytes were stained with calcein AM and ethidium homodimer-1 (Molecular Probes, Eugene, OR) to qualitatively assess the proportion of live-to-dead cells and imaged using an Olympus microscope equipped with epifluorescence illumination magnification of 200×). Cell culture medium was harvested on a daily basis and centrifuged for 3 min at 14000 rpm; the supernatants were stored at −20°C until assayed. Immediately following medium collection, cells were harvested in 200 µl of TRIzol™ (Invitrogen, Carlsbad, CA) and stored at −80°C. Activity of lactate dehydrogenase (LDH) and production of pyruvate and lactate from cultured cells was assessed by standard enzymatic procedures (Bergmeyer, 1988). Urea synthesis was evaluated using BioAssay Systems (Hayward, CA) QuantiChrom Urea Assay Kit as detailed by the manufacturer. All reported values for pyruvate have been corrected to pyruvate content in the culture media. Cytotoxicity of reference compounds was determined by measuring intracellular adenosine triphosphate (ATP) content, glutathione levels, and caspase 3/7 activity using CellTiter-Glo® Luminescent Cell Viability Assay, GSH-Glo™ Glutathione Assay, or Caspase-Glo® 3/7 Assay (Promega, Madison, WI), respectively, according to the manufacturer’s protocol. Protein quantification was performed using BCA™ Protein Assay Kit (Pierce, Rockford, IL) according to the manufacturer’s protocol.
mRNA Isolation and Gene Expression Analysis by RT-PCR
TRIzol lysates homogenized using a 25 gauge needle and a 1 mL syringe and the RNA was extracted from the samples using the Qiagen RNeasy Mini kit (Qiagen, Valencia, CA). RNA concentrations were measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and quality was verified using the Agilent Bio-Analyzer (Agilent Technologies, Santa Clara, CA). Total RNA (2 µg) was reverse transcribed using random primers and the high capacity cDNA archive kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol. The resulting cDNA was diluted to 1:100 with RNase-free H2O, mixed gently, aliquoted and stored at −20°C until required. The following gene expression assays (Applied Biosystems) were used for quantitative real-time PCR: acyl-coenzyme A oxidase 1 (Acox1, Mm00443579_m1); albumin (Alb, Mm00802090_m1), carbamoyl-phosphate synthetase 1 (Cps1, Mm01256489_m1); cytochrome P450, family 1, subfamily a, polypeptide 2 (Cyp1a2, Mm0048224_m1); cytochrome P450, family 2, subfamily e, polypeptide 1 (Cyp2e1, Mm00491127_m1); cytochrome P450, family 2, subfamily e, polypeptide 1 (Cyp3a11, Mm00731567_m1); cytochrome P450, family 4, subfamily a, polypeptide 10 (Cyp4a10, Mm01188913_g1); glutathione S-transferase, alpha 2 (Gsta2, Mm0083335_mH); hepatic nuclear factor 4, alpha (Hnf4α, Mm00433964_m1); nuclear factor, erythroid derived 2, like 2 (Nrf2, Mm00477784_m1); peroxisome proliferator activated receptor alpha (Pparα, Mm00440939_m1); UDP glucuronosyltransferase 1 family, polypeptide A1 (Ugt1a1, Mm02603337_m1); solute carrier organic anion transporter family, member 1b2 (Slco1b2, Mm00451500_m1); and beta glucuronidase (Gusb, Mm00446753_m1). Reactions were performed in a 96-well assay format. Each plate contained one experimental gene and a housekeeping gene and all samples were plated in duplicate. Reactions were processed using Roche 480 instrument (Roche Applied Science, Indianapolis, IN The cycle threshold (Ct) for each sample was determined from the linear region of the amplification plot. The ΔCt value for all genes relative to the control gene Gusb were determined. The ΔΔCt were calculated using treated group means relative to strain-matched control group means. Fold change data were calculated from the ΔΔCt values.
Data and Statistical Analysis
The software package GraphPad (version 4.0, Prism, La Jolla, CA) was used to plot the time-course and gene expression data and the concentration-response curves, as well as to calculate the EC50 values. Data are represented as mean values plus or minus the standard deviation for the replicate wells for each condition with a minimum of 2 experimental repeats, with each experimental repeat representing cells from a different hepatocyte preparation. One-way ANOVA was used for statistical comparisons. In cases in which more than one variable was compared (i.e., culture day and concentration) a two- way ANOVA with Tukey’s multiple comparison test was utilized. A p value less than 0.05 was selected prior to the study to determine statistical significance between groups.
Results
Assessment of viability and functionality of primary hepatocyte cultures from a panel of strains
We chose 14 inbred mouse strains, representing fixed and reproducible unique genotypes with a broad genetic diversity, comprising the priority group of strains that were resequenced by NIEHS Center for Rodent Genetics (Frazer et al., 2007). In addition, we also used B6C3F1 mice, an F1 cross between C57BL/6J and C3H/HeJ inbred lines, a strain commonly used in toxicological research and by the National Toxicology Program (Allen et al., 2004). Primary hepatocytes were isolated and cultured for up to 7 days to investigate differences in their functionality due to their distinct genetic backgrounds (see Supplemental Figure 1A for the experimental design). Liver parenchymal cell yields (average of 59±13 million cells/mouse) and viability (93±4%) were comparable between strains regardless of the lineage (Supplemental Table 1, Supplemental Figure 2). Strain-to-strain variability in LDH release, pyruvate, lactate and urea production, markers of cell function, was observed over 7-day culture period; however, the overall trends in time-dependent changes were comparable (Supplemental Figure 3). Expression of several liver function-related genes was evaluated in hepatocyte cultures (days 1 and 3) and whole liver samples from 7 randomly selected strains (Supplemental Figure 4). Cells remained functional, as evidenced by preserved levels of expression of Alb and Hnf4α, but became less metabolically active as mRNA for Cyp4a10 and Ugt1a1 decreased markedly by 75–90% within the first 24 hours.
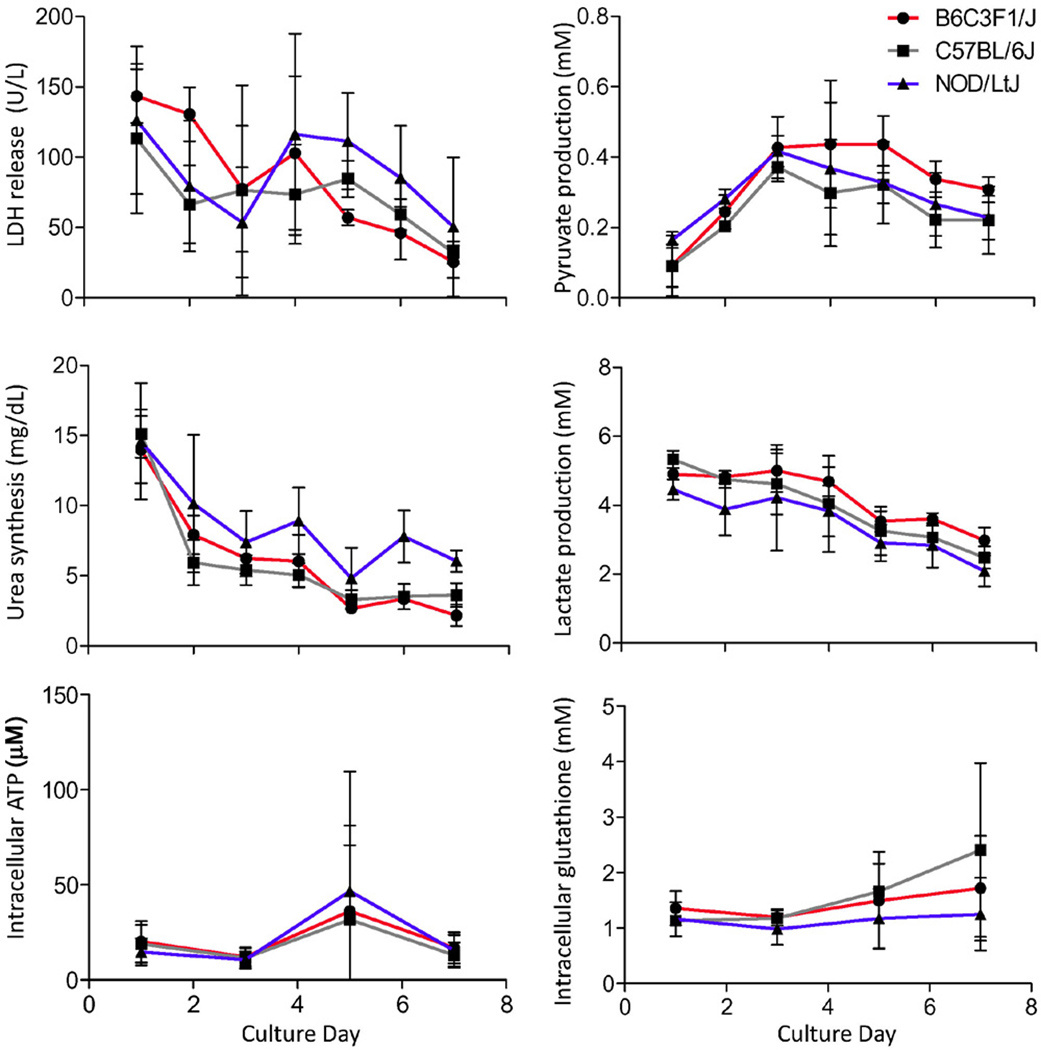
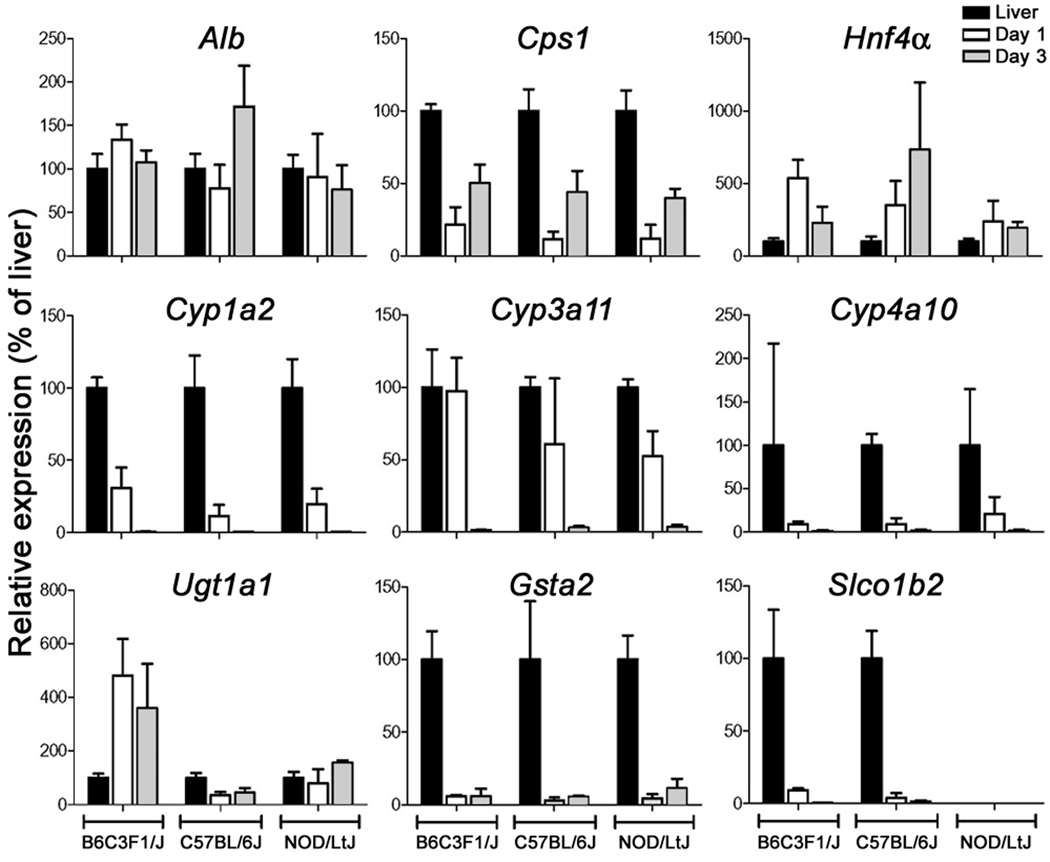
Independent isolations of primary hepatocytes produced consistent results when three separate liver perfusions were performed from B6C3F1/J, C57BL/6J and NOD/LtJ strains (Supplemental Table 1). Considerable consistency between biological replicates and strains was observed in the biochemical endpoints of cell function (Figure 1). Baseline expression of the genes involved in protein and urea metabolism (Alb, Hnf4α and Cps1) remained comparable to that in the whole liver (Figure 2), while liver metabolism genes (Cyp1a2, Cyp3a11, Cyp4a10, Gsta2, and Slco1b2) diminished dramatically with the exception of Ugt1a1 which remained highly expressed. Strain-to-strain variability in expression was observed that was consistent with the known role of genetic polymorphisms on liver gene expression (Gatti et al., 2007).
Figure 1.
Functional characterization of cultured hepatocytes isolated from 3 mouse strains. Hepatocytes were isolated from B6C3F1/J, C57BL/6J and NOD/LtJ (males, n=3) and cultured in 24-well plates in a conventional monolayer up to day 7. Media was harvested daily and activity of lactate dehydrogenase, serum concentrations of urea, pyruvate and lactate, and intracellular concentrations (amount in all cells in each well on days 1, 3 and 7) of ATP and glutathione were quantified. Data shown as mean±SD of independent biological replicates.
Figure 2.
Analysis of reproducibility of gene expression in cultured hepatocytes isolated from 3 mouse strains. mRNA levels of Alb, Cps1, Hnf4α, Cyp1a2, Cyp3a11, Cyp4a10, Ugt1a1, Gsta1, and Slco1b2 was assessed using quantitative RT-PCR in liver tissue (black bar) and hepatocytes cultured for 1 (white bar) or 3 (grey bar) days. Data (mean±SD of biological replicates) was normalized to expression levels in whole liver samples.
In vitro toxicity testing in primary hepatocyte cultures
To investigate strain-specific sensitivity to chemical exposures we used several model toxicants. Hepatocytes were isolated from B6C3F1/J, C57BL/6J and NOD/LtJ mice, cultured and treated (for 24 hrs) with acetaminophen (0.3–30 mM), WY-14,643 (0.1–10 mM), or rifampin (1–100 µM) either 24 or 72 hrs post seeding (for experimental design, see Supplemental Figure 1B). Three independent rounds of cell isolation, plating, and treatment were performed. Cell viability was assessed by ATP production. Intracellular glutathione content and expression of several marker transcripts were also evaluated.
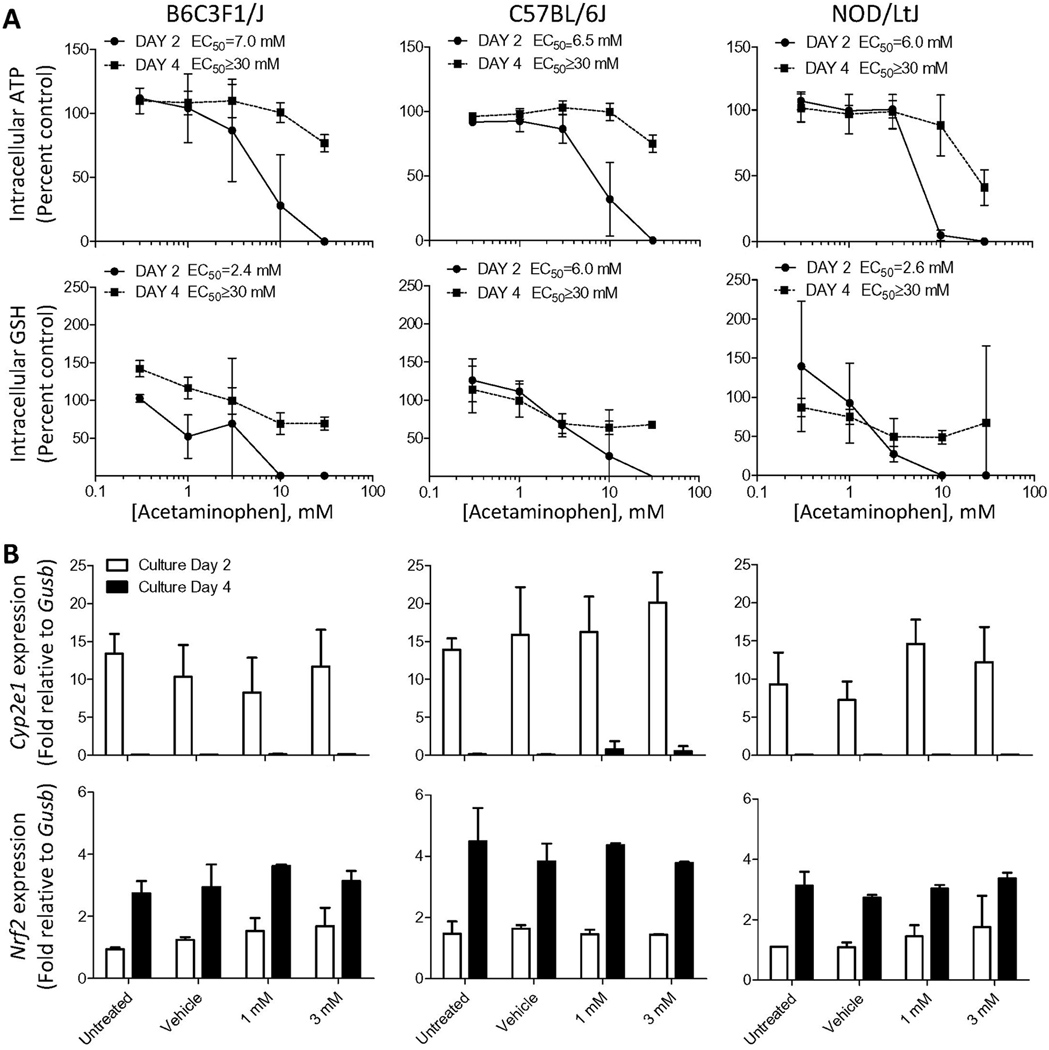
Acetaminophen was selected as a metabolism-dependent hepatotoxicant. Primary hepatocytes originating from 3 different strains exhibited a remarkably similar level of cytotoxicity in 24 hr cultures (average EC50 ranged from 6.0 to 7.0 mM) (Figure 3A). Interestingly, cells from C57BL/6J strain were somewhat more resistant to glutathione depletion (EC50=6.0 mM as compared to 2.4–2.6 mM for other two strains). There were no significant differences in intracellular glutathione content between strains in untreated hepatocytes (Supplemental Figure 5). After 72 hrs in culture, primary cells from all strains lost their sensitivity to acetaminophen in an equally similar manner (EC50≥30 mM for both cell viability and glutathione depletion). Cyp2e1 and Nrf2 are key proteins involved in the mechanism of acetaminophen-induced cytotoxicity in liver with the former necessary for metabolic activation to the reactive thiol, N-acetyl-p-benzo-quinone imine, and the latter playing a role in controlling expression of detoxification genes. While expression of Cyp2e1 was relatively high in 24 hr cultures (and treated with acetaminophen for additional 24 hrs) in all strains, it was barely detectable in cells cultured for the longer duration (Figure 3B), consistent with the observations of cytotoxicity. Expression of Nrf2 increased by about 2-fold with time in culture; however, there were no differences observed between concentrations or strains. No induction of either gene was observed with increasing concentrations of acetaminophen.
Figure 3.
Effects of acetaminophen on hepatocytes derived from 3 mouse strains. Hepatocytes were isolated from B6C3F1/J, C57BL/6J and NOD/LtJ (males, n=3) and cultured in 96-well plates in a conventional monolayer for 24 or 72 hr and exposed to acetaminophen (0.3, 1, 3, 10 and 30 mM) or vehicle (dimethyl sulfoxide, 0.5%) for additional 24 hr. Hepatocytes were harvested on day 2 (filled circles or white bars), or 4 (filled squares or black bars) of culture. (A) Assessment of cytotoxicity using intracellular ATP and glutathione (GSH) levels. Data (mean±SD of biological replicates) was normalized to the values in vehicle-treated cells. (B) mRNA levels of Cyp2e1 and Nrf2 were assessed using quantitative RT-PCR. Data (mean±SD of biological replicates) was normalized to expression level of Gusb.
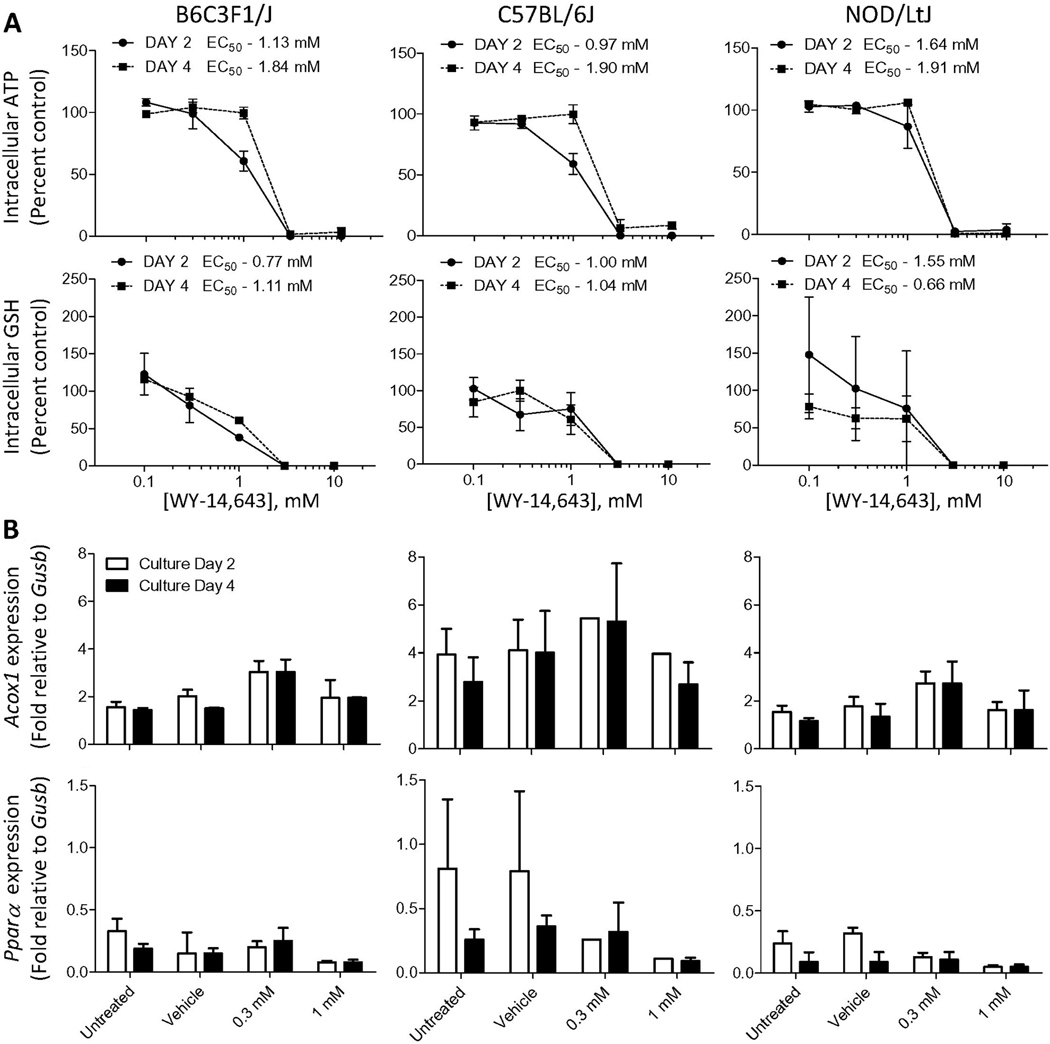
WY-14,643 is a model peroxisome proliferator and inducer of Pparα, and it does not require metabolic activation to exert liver-specific effects. Indeed, the length of time in culture had little effect on cytotoxicity of WY-14,643 (Figure 4A). EC50s for both loss of ATP production and depletion of intracellular glutathione levels were quite consistent between strains and culture duration. No consistent concentration-dependent induction of expression of Pparα was observed (up to 1 mM); however, expression of Acox1 was affected significantly (p<0.05 at 0.3 mM, concentration where to cytotoxicity was observed) by treatment with WY-14,643 in hepatocytes from NOD/LtJ and B6C3F1/J mice (Figure 4B).
Figure 4.
Effects of WY-14,643 (0.1, 0.3, 1, 3 and 10 mM) on hepatocytes derived from 3 mouse strains. Cells were isolated, cultured, and data collected as detailed in the legend to Figure 3.
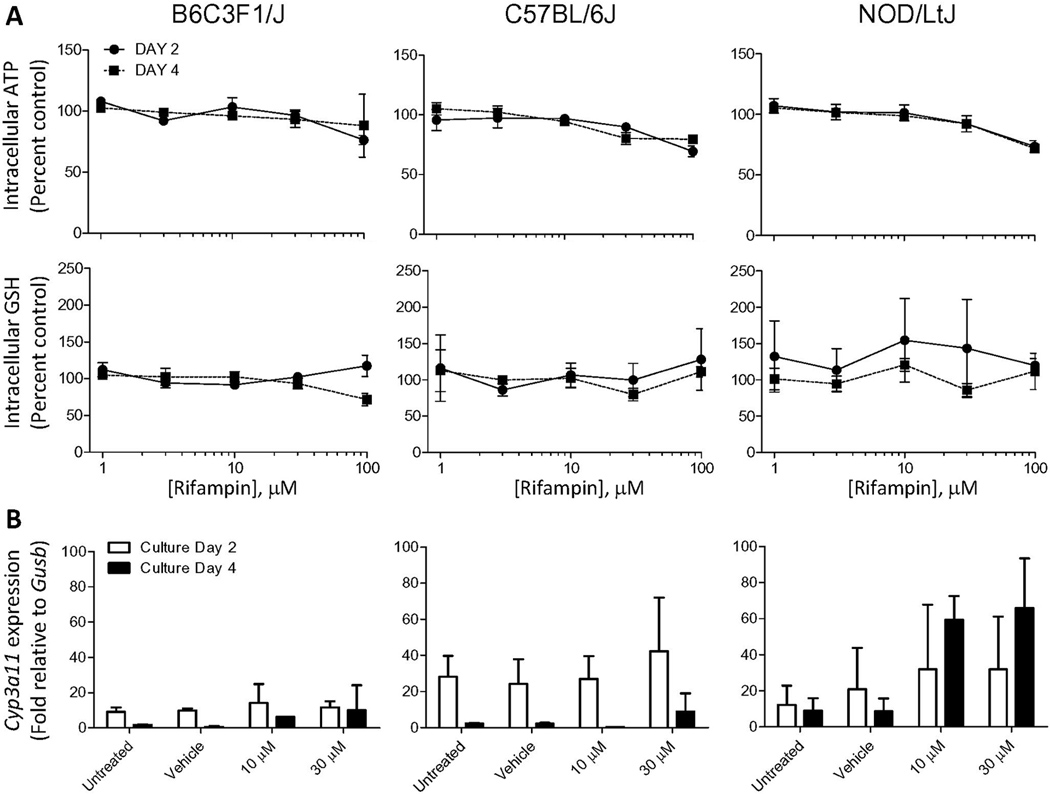
Rifampin is a species-specific hepatotoxicant and its liver toxicity is dependent on activation of pregnane X receptor in human, but not mouse (LeCluyse, 2001b). As expected, little cytotoxicity was observed for rifampin (up to 100 µM) in mouse hepatocytes of either strain with both 24 and 72 hr cultures (Figure 5A). Mouse Cyp3a11 is known to be weakly inducible in rodents by rifampin in vivo (LeCluyse, 2001b) and our data shows that in vitro, induction may be strain-dependent as NOD/LtJ hepatocytes were able to maintain expression of this gene and it was induced in 72 hr cultures (Figure 5B). Concentration of rifampin was found to be a significant (p<0.05) variable for the expression of Cyp3a11 in hepatocytes from NOD/LtJ strain.
Figure 5.
Effects of rifampin (1, 3, 10, 30 and 100 µM) on hepatocytes derived from 3 mouse strains. Cells were isolated, cultured, and data collected as detailed in the legend to Figure 3.
Discussion
Cell-based approaches are well suited for hazard identification, prioritization of environmental chemicals, and drug safety screening (Greer et al., 2010). Furthermore, in vitro toxicity experiments can generate large amounts of information on biological mechanisms of action, including compound’s potential for metabolism-mediated toxicity and inter-individual variability (Mingoia et al., 2007). While inter-individual variability following xenobiotic exposures in primary human hepatocyte cultures is a well-established phenomenon (Goyak et al., 2008), it is frequently viewed as an impediment to in vitro experiments with primary hepatocytes. Our study not only demonstrates the feasibility and utility of a cultured hepatocyte model developed from a mouse diversity panel, but also shows the consistency of viability and function of the cells isolated from the individual animals of the same strain across the panel. Thus, the mouse hepatocytes derived from multiple strains may serve as an infinitely reproducible in vitro model of a genetically diverse population.
While standardized isolation and cell culture conditions are crucial to ensure “phenotypic consistency” of the cultures derived from a panel of genetically distinct mouse strains, there are important limitations to the conventional cultures with regards to maintenance of liver-like function in longer-term cultures (Swift and Brouwer, 2010). In addition, mouse hepatocytes are more difficult to maintain in a functional state than rat or human cells (Swales et al., 1996), likely due to a much higher metabolism rate and oxygen demand. Clearly, much work remains to be done to determine appropriate conditions that would allow maintenance of mouse hepatocytes in culture in a differentiated state past culture day 3.
There are many factors that regulate maintenance of the liver-like phenotype in vitro including cell-cell communications, cell shape, organization and distribution of the cytoskeleton and extracellular matrix interactions (Musat et al., 1993; Stamatoglou and Hughes, 1994; Berthiaume et al., 1996; Oda et al., 2008). Cell-cell contacts help maintain liver-specific function and normal gene expression through adhesion dependent signal transduction pathways mediated by the Rho-family of small GTPases (Etienne-Manneville and Hall, 2002). Indeed, advanced culture systems have been developed to allow for better retention of hepatocyte cytoarchitecture and retention of the in vivo gene expression profiles of liver. These include co-culture with other liver-derived or non-liver cell types, exposing hepatocytes to a gel substratum, culture in collagen gel sandwiches, and culture in spheroids and in 3D bioreactors (LeCluyse, 2001a; Sivaraman et al., 2005; Khetani and Bhatia, 2008; Godoy et al., 2009).
Liver-specific transcription factors, including HNF4α, are involved in the regulation of a number of hepatic genes (Akiyama and Gonzalez, 2003). Cultured hepatocytes in our study were able to maintain the expression of HNF4α out to culture day 3 at levels comparable to whole liver; however, the cells failed to maintain expression of many genes relating to phase I and II metabolism past culture day 3. HNF4α is one of many factors in a complex regulatory network that mediates the expression of many hepatic genes (Chen et al., 1994; Akiyama and Gonzalez, 2003). Monolayer confluency also influences the expression and the inductive capability of several cytochrome P450 enzymes whose expression has been shown to decrease with the loss of confluence (Hewitt et al., 2007), and our observation of loss of confluency due to spindle-like cell transformation may be the key reason for loss of metabolism function.
Despite the challenges in maintaining liver-like phenotype in cell culture, the hepatocytes derived from mice of the same strain exhibited consistent and reproducible features in independent isolations. Furthermore, independent isolations exhibited low variation in cytotoxicity experiments with three model chemicals, acetaminophen, WY-14,643 and rifampin. This outcome points to the potential utility of the in vitro hepatocyte cultures as reproducible proxies for population diversity. Not only can cells be derived repeatedly from the same strain and expected to have a consistent in vitro phenotype, thus eliminating the concerns over the unpredictable nature of batches of human cells which are not renewable, but it is also possible to study the potential haplotype associations since the mouse cells have known genotypes, while the human cells do not.
Mouse strain-specific responses to acute acetaminophen toxicity have been demonstrated in in vivo studies (Harrill et al., 2009b; Liu et al., 2010). When liver injury was evaluated 24 hrs after a single large dose (300 mg/kg, i.g.) of acetaminophen, NOD/LtJ mice exhibited mild liver necrosis and inflammation (~10%), while B6C3F1 mice had more than 80% of the parenchyma affected, and C57BL/6J strain exhibited an intermediate phenotype (20–25%) (Harrill et al., 2009b). Polymorphisms in inflammatory-mediator response genes were shown to be associated with liver injury in the population of mice, suggesting that the degree of inflammation, defined by the resident macrophages and infiltrating neutrophils, are key factors in the clinical outcome of acetaminophen toxicity. At the same time, this study also reported that elevations of serum alanine aminotransferase activity were comparable between these strains 4 hrs after dosing. Thus, the fact that we observed no discernable strain differences in hepatocyte viability following the drug challenge in vitro further supports the crucial importance of nonparenchymal cells in the liver damage in vivo (DeLeve et al., 1997; Adams et al., 2010).
Hepatic inflammation is commonly found in a variety of liver diseases and during drug-induced liver toxicity and resident hepatic macrophages, Kupffer cells, have been shown to be key players in determining the outcomes of toxicity through their immune-modulating role (Cover et al., 2006). In addition, Kupffer cells can also be involved in mitogenic responses by affecting proliferation of hepatocytes (Rose et al., 1999), or regulating hepatic transport protein expression during drug-induced liver injury (Campion et al., 2008). Other non-parenchymal cells can also play a significant role in mediating hepatic responses to xenobiotics. For example, hepatic stellate cells, when activated, are the major mediators of matrix formation in response to liver injury (Friedman, 2008) and also play a direct role in the inflammatory process as they secrete chemokines and express cell adhesion molecules which can mediate lymphocyte adhesion (Holt et al., 2009). However, capturing complex heterotypic liver cell interactions in vitro is a challenge. The canonical hepatocyte-enriched primary cell isolations obtained by collagenase perfusion and low-speed centrifugation results in a cell population that contains 5–10% non-parenchymal cells within the majority (90–95%) being liver parenchymal cells even though the latter comprise ~60% of liver cells by number in vivo (Michalopoulos et al., 1999; Peters et al., 2000).
Hepatic intracellular glutathione is also a well known modifier of acetaminophen hepatotoxicity (Mitchell et al., 1973). Evaluation of glutathione content in the cultured hepatocytes from the three strains revealed notable differences in the rate of glutathione depletion in response to increasing concentrations of acetaminophen. However, while B6C3F1 hepatocytes showed a steep concentration-dependent loss of reduced glutathione, commensurate with high sensitivity to liver injury in vivo, NOD/LtJ hepatocytes’ response was quite similar, suggesting that hepatocyte monocultures may not be indicative of the degree of overt toxicity which depends on non-parenchymal cells. Additional studies need to be conducted in more complex culture systems to further investigate the relationship between the genotype and sensitivity to acetaminophen.
Furthermore, our results indicate a significant reduction in the cytotoxicity of acetaminophen when tested against primary hepatocytes cultured out to day 3, which suggest a significant change in the metabolic capacity of the cultured hepatocytes. Acetaminophen toxicity is a metabolism dependent pathway via bioactivation by Cyp2e1 which is involved in the formation of the reactive thiol (Kaplowitz, 2005). Indeed, the observed decrease in sensitivity to acetaminophen correlates with the progressive loss of Cyp2e1 expression. These findings are consistent with previous reports which found metabolic capacity, transport functions and other enzyme activities to diminish dramatically in culture (Richert et al., 2002; Luttringer et al., 2002). This relationship between toxicity to hepatocytes in vitro and drug-induced liver injury in vivo remains poorly defined and highlights an important limitation of over-reliance on the in vitro testing alone without appropriate studies in vivo, or in more complex cell culture systems (Greer et al., 2010).
A model peroxisome proliferator WY-14,643 was used as an agent that requires no metabolic biotransformation to exert either in vivo or in vitro effects. Indeed, the number of days in culture, a significant variable in response to treatment with acetaminophen, appeared to have little effect on cytotoxicity of WY-14.643. Even though cytotoxicity was observed, similar to the high dose effects in vivo (Woods et al., 2007), the phenotype of concern is induction of cell proliferation and hepatomegaly due to peroxisome proliferation. While Acox1 expression was maintained in mouse hepatocytes, it was not inducible by treatment, contrary to the observation in rat hepatocytes (Berthou et al., 1995). Increase in DNA synthesis caused by peroxisome proliferators in vivo is dependent on the activation of Kupffer cells (Rose et al., 2000), and co-cultured systems may be necessary to reproduce this phenotype. Parzefall et al demonstrated that peroxisome proliferators do not influence DNA synthesis in isolated rat hepatocytes, but addition of cytokines can replicate the in vivo effect in primary cultures (Parzefall et al., 2001).
Treatment with increasing concentrations of rifampin had little effect on ATP and glutathione in cells from all strains. Even at the highest concentration (100 µM) tested, only about a 20–25% effect was observed irrespective of the time in culture. There are significant species differences in response to pregnane X receptor ligands between humans and rodents (Jones et al., 2000). Rifampin and other drugs are known to activate the human receptor, but are weak activators of the rodent receptor. Interestingly, expression of Cyp3a11 was weakly inducible in response to treatment with rifampin. Other investigators (Tsutsui et al., 2006) have shown that in 129/sv mice-derived hepatocytes Cyp3a11 expression is diminished over 2.5 days in culture, a trend which was similar to one observed here; however, the inducibility of this gene by a weak ligand shows that the pregnane x receptor is functional, similar to the observations of Shenoy et al (Shenoy et al., 2004).
In summary, our current work supports the utility of an in vitro toxicogenetic model for studying mechanisms of drug-induced hepatotoxicity. While our data supports the concept that cultures of hepatocytes derived from a mouse diversity panel can potentially be utilized as a tool to conduct population-based toxicity studies in vitro, it also highlights the need for further improvements through co-culture (Khetani and Bhatia, 2008) or 3-dimesional flow (Griffith and Swartz, 2006) techniques. On one hand, it is essential to establish an in vitro model which may allow research on the biological mechanisms mediated by underlying genetic determinants and elucidation of the factors contributing to inter-individual responses to toxicity. Indeed, population-based in vitro models, albeit not liver-specific, are being used to provide insights into the genetic factors contributing to inter-individual susceptibility in humans by identifying the unique genotype-specific drug-induced toxicity and gene expression (Welsh et al., 2009). On the other hand, our results suggest that development of a multi-cell liver-derived in vitro model which preserves baseline hepatic function is necessary. While this manuscript demonstrates the importance of optimizing cell isolation and culture conditions for hepatocytes, additional studies which incorporate non-parenchymal cells are needed to construct population-based in vitro models that may accurately predict in vivo responses to a wide range of hepato-toxicants.
Supplementary Material
Acknowledgments
Funding: Supported, in part, by grants from NIH (R01 ES015241) and US EPA (RD833825).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest:
The authors declare no conflicts other than E. LeCluyse declaring that he was employed by and consulted for Invitrogen/LifeTechnologies, a supplier of primary hepatocytes for commercial purposes, when this study was contacted.
REFERENCES
- Acosta D, Sorensen EM, Anuforo DC, Mitchell DB, Ramos K, Santone KS, Smith MA. An in vitro approach to the study of target organ toxicity of drugs and chemicals. In Vitro Cell Dev. Biol. 1985;21:495–504. doi: 10.1007/BF02620841. [DOI] [PubMed] [Google Scholar]
- Adams DH, Ju C, Ramaiah SK, Uetrecht J, Jaeschke H. Mechanisms of immune-mediated liver injury. Toxicol Sci. 2010;115:307–321. doi: 10.1093/toxsci/kfq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama TE, Gonzalez FJ. Regulation of P450 genes by liver-enriched transcription factors and nuclear receptors. Biochim. Biophys. Acta. 2003;1619:223–234. doi: 10.1016/s0304-4165(02)00480-4. [DOI] [PubMed] [Google Scholar]
- Allen DG, Pearse G, Haseman JK, Maronpot RR. Prediction of rodent carcinogenesis: an evaluation of prechronic liver lesions as forecasters of liver tumors in NTP carcinogenicity studies. Toxicol Pathol. 2004;32:393–401. doi: 10.1080/01926230490440934. [DOI] [PubMed] [Google Scholar]
- Bergmeyer HU. Methods of enzymatic analysis. New York: Academic Press; 1988. [Google Scholar]
- Berthiaume F, Moghe PV, Toner M, Yarmush ML. Effect of extracellular matrix topology on cell structure, function, and physiological responsiveness: hepatocytes cultured in a sandwich configuration. FASEB J. 1996;10:1471–1484. doi: 10.1096/fasebj.10.13.8940293. [DOI] [PubMed] [Google Scholar]
- Berthou L, Saladin R, Yaqoob P, Branellec D, Calder P, Fruchart JC, Denefle P, Auwerx J, Staels B. Regulation of rat liver apolipoprotein A-I, apolipoprotein A-II and acyl-coenzyme A oxidase gene expression by fibrates and dietary fatty acids. Eur. J Biochem. 1995;232:179–187. doi: 10.1111/j.1432-1033.1995.tb20797.x. [DOI] [PubMed] [Google Scholar]
- Campion SN, Johnson R, Aleksunes LM, Goedken MJ, van Rooijen N, Scheffer GL, Cherrington NJ, Manautou JE. Hepatic Mrp4 induction following acetaminophen exposure is dependent on Kupffer cell function. Am J Physiol Gastrointest Liver Physiol. 2008;295:G294–G304. doi: 10.1152/ajpgi.00541.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Park Y, Kemper B. Differential protein binding and transcriptional activities of HNF-4 elements in three closely related CYP2C genes. DNA Cell Biol. 1994;13:771–779. doi: 10.1089/dna.1994.13.771. [DOI] [PubMed] [Google Scholar]
- Cover C, Liu J, Farhood A, Malle E, Waalkes MP, Bajt ML, Jaeschke H. Pathophysiological role of the acute inflammatory response during acetaminophen hepatotoxicity. Toxicol Appl. Pharmacol. 2006;216:98–107. doi: 10.1016/j.taap.2006.04.010. [DOI] [PubMed] [Google Scholar]
- DeLeve LD, Wang X, Kaplowitz N, Shulman HM, Bart JA, van der HA. Sinusoidal endothelial cells as a target for acetaminophen toxicity. Direct action versus requirement for hepatocyte activation in different mouse strains. Biochem Pharmacol. 1997;53:1339–1345. doi: 10.1016/s0006-2952(97)00048-8. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Eskin E, Kang HM, Bogue MA, Hinds DA, Beilharz EJ, Gupta RV, Montgomery J, Morenzoni MM, Nilsen GB, Pethiyagoda CL, Stuve LL, Johnson FM, Daly MJ, Wade CM, Cox DR. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti D, Maki A, Chesler EJ, Kirova R, Kosyk O, Lu L, Manly KF, Williams RW, Perkins A, Langston MA, Threadgill DW, Rusyn I. Genome-level analysis of genetic regulation of liver gene expression networks. Hepatology. 2007;46:548–557. doi: 10.1002/hep.21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt R, Hengstler JG, Muller D, Glockner R, Buenning P, Laube B, Schmelzer E, Ullrich M, Utesch D, Hewitt N, Ringel M, Hilz BR, Bader A, Langsch A, Koose T, Burger HJ, Maas J, Oesch F. New hepatocyte in vitro systems for drug metabolism: metabolic capacity and recommendations for application in basic research and drug development, standard operation procedures. Drug Metab Rev. 2003;35:145–213. doi: 10.1081/dmr-120023684. [DOI] [PubMed] [Google Scholar]
- Godoy P, Hengstler JG, Ilkavets I, Meyer C, Bachmann A, Muller A, Tuschl G, Mueller SO, Dooley S. Extracellular matrix modulates sensitivity of hepatocytes to fibroblastoid dedifferentiation and transforming growth factor beta-induced apoptosis. Hepatology. 2009;49:2031–2043. doi: 10.1002/hep.22880. [DOI] [PubMed] [Google Scholar]
- Goyak KM, Johnson MC, Strom SC, Omiecinski CJ. Expression profiling of interindividual variability following xenobiotic exposures in primary human hepatocyte cultures. Toxicol Appl Pharmacol. 2008;231:216–224. doi: 10.1016/j.taap.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer ML, Barber J, Eakins J, Kenna JG. Cell based approaches for evaluation of drug-induced liver injury. Toxicology. 2010;268:125–131. doi: 10.1016/j.tox.2009.08.007. [DOI] [PubMed] [Google Scholar]
- Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat. Rev Mol. Cell Biol. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- Groneberg DA, Grosse-Siestrup C, Fischer A. In vitro models to study hepatotoxicity. Toxicol Pathol. 2002;30:394–399. doi: 10.1080/01926230252929972. [DOI] [PubMed] [Google Scholar]
- Guo Y, Lu P, Farrell E, Zhang X, Weller P, Monshouwer M, Wang J, Liao G, Zhang Z, Hu S, Allard J, Shafer S, Usuka J, Peltz G. In silico and in vitro pharmacogenetic analysis in mice. Proc. Natl. Acad Sci U. S. A. 2007;104:17735–17740. doi: 10.1073/pnas.0700724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Weller P, Farrell E, Cheung P, Fitch B, Clark D, Wu SY, Wang J, Liao G, Zhang Z, Allard J, Cheng J, Nguyen A, Jiang S, Shafer S, Usuka J, Masjedizadeh M, Peltz G. In silico pharmacogenetics of warfarin metabolism. Nat Biotechnol. 2006;24:531–536. doi: 10.1038/nbt1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill AH, Ross PK, Gatti DM, Threadgill DW, Rusyn I. Population-based discovery of toxicogenomics biomarkers for hepatotoxicity using a laboratory strain diversity panel. Toxicol Sci. 2009a;110:235–243. doi: 10.1093/toxsci/kfp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill AH, Rusyn I. Systems biology and functional genomics approaches for the identification of cellular responses to drug toxicity. Expert Opin. Drug Metab Toxicol. 2008;4:1379–1389. doi: 10.1517/17425255.4.11.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill AH, Watkins PB, Su S, Ross PK, Harbourt DE, Stylianou IM, Boorman GA, Russo MW, Sackler RS, Harris SC, Smith PC, Tennant R, Bogue M, Paigen K, Harris C, Contractor T, Wiltshire T, Rusyn I, Threadgill DW. Mouse population-guided resequencing reveals that variants in CD44 contribute to acetaminophen-induced liver injury in humans. Genome Res. 2009b;19:1507–1515. doi: 10.1101/gr.090241.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt NJ, LeCluyse EL, Ferguson SS. Induction of hepatic cytochrome P450 enzymes: methods, mechanisms, recommendations, and in vitro-in vivo correlations. Xenobiotica. 2007;37:1196–1224. doi: 10.1080/00498250701534893. [DOI] [PubMed] [Google Scholar]
- Holt AP, Haughton EL, Lalor PF, Filer A, Buckley CD, Adams DH. Liver myofibroblasts regulate infiltration and positioning of lymphocytes in human liver. Gastroenterology. 2009;136:705–714. doi: 10.1053/j.gastro.2008.10.020. [DOI] [PubMed] [Google Scholar]
- Jones SA, Moore LB, Shenk JL, Wisely GB, Hamilton GA, McKee DD, Tomkinson NC, LeCluyse EL, Lambert MH, Willson TM, Kliewer SA, Moore JT. The pregnane X receptor: a promiscuous xenobiotic receptor that has diverged during evolution. Mol. Endocrinol. 2000;14:27–39. doi: 10.1210/mend.14.1.0409. [DOI] [PubMed] [Google Scholar]
- Judson R, Richard A, Dix DJ, Houck K, Martin M, Kavlock R, Dellarco V, Henry T, Holderman T, Sayre P. The toxicity data landscape for environmental chemicals. Environ Health Perspect. 2009;117:685–695. doi: 10.1289/ehp.0800168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RS, Houck KA, Kavlock RJ, Knudsen TB, Martin MT, Mortensen HM, Reif DM, Rotroff DM, Shah I, Richard AM, Dix DJ. In Vitro Screening of Environmental Chemicals for Targeted Testing Prioritization: The ToxCast Project. Environ Health Perspect. 2010;118:485–492. doi: 10.1289/ehp.0901392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplowitz N. Idiosyncratic drug hepatotoxicity. Nat. Rev. Drug Discov. 2005;4:489–499. doi: 10.1038/nrd1750. [DOI] [PubMed] [Google Scholar]
- Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- LeCluyse EL. Human hepatocyte culture systems for the in vitro evaluation of cytochrome P450 expression and regulation. Eur. J Pharm Sci. 2001a;13:343–368. doi: 10.1016/s0928-0987(01)00135-x. [DOI] [PubMed] [Google Scholar]
- LeCluyse EL. Pregnane X receptor: molecular basis for species differences in CYP3A induction by xenobiotics. Chem Biol Interact. 2001b;134:283–289. doi: 10.1016/s0009-2797(01)00163-6. [DOI] [PubMed] [Google Scholar]
- Liu HH, Lu P, Guo Y, Farrell E, Zhang X, Zheng M, Bosano B, Zhang Z, Allard J, Liao G, Fu S, Chen J, Dolim K, Kuroda A, Usuka J, Cheng J, Tao W, Welch K, Liu Y, Pease J, de Keczer SA, Masjedizadeh M, Hu JS, Weller P, Garrow T, Peltz G. An integrative genomic analysis identifies Bhmt2 as a diet-dependent genetic factor protecting against acetaminophen-induced liver toxicity. Genome Res. 2010;20:28–35. doi: 10.1101/gr.097212.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttringer O, Theil FP, Lave T, Wernli-Kuratli K, Guentert TW, de Saizieu A. Influence of isolation procedure, extracellular matrix and dexamethasone on the regulation of membrane transporters gene expression in rat hepatocytes. Biochem Pharmacol. 2002;64:1637–1650. doi: 10.1016/s0006-2952(02)01382-5. [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK, Bowen WC, Zajac VF, Beer-Stolz D, Watkins S, Kostrubsky V, Strom SC. Morphogenetic events in mixed cultures of rat hepatocytes and nonparenchymal cells maintained in biological matrices in the presence of hepatocyte growth factor and epidermal growth factor. Hepatology. 1999;29:90–100. doi: 10.1002/hep.510290149. [DOI] [PubMed] [Google Scholar]
- Mingoia RT, Nabb DL, Yang CH, Han X. Primary culture of rat hepatocytes in 96-well plates: effects of extracellular matrix configuration on cytochrome P450 enzyme activity and inducibility, and its application in in vitro cytotoxicity screening. Toxicol In Vitro. 2007;21:165–173. doi: 10.1016/j.tiv.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Jollow DJ, Potter WZ, Gillette JR, Brodie BB. Acetaminophen-induced hepatic necrosis. IV. Protective role of glutathione. J Pharmacol Exp Ther. 1973;187:211–217. [PubMed] [Google Scholar]
- Musat AI, Sattler CA, Sattler GL, Pitot HC. Reestablishment of cell polarity of rat hepatocytes in primary culture. Hepatology. 1993;18:198–205. [PubMed] [Google Scholar]
- National Research Council. Toxicity Testing in the 21st Century: A Vision and a Strategy. Washington, DC: National Academies Press; 2007. [DOI] [PubMed] [Google Scholar]
- Oda H, Yoshida Y, Kawamura A, Kakinuma A. Cell shape, cell-cell contact, cell-extracellular matrix contact and cell polarity are all required for the maximum induction of CYP2B1 and CYP2B2 gene expression by phenobarbital in adult rat cultured hepatocytes. Biochem Pharmacol. 2008;75:1209–1217. doi: 10.1016/j.bcp.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Parzefall W, Berger W, Kainzbauer E, Teufelhofer O, Schulte-Hermann R, Thurman RG. Peroxisome proliferators do not increase DNA synthesis in purified rat hepatocytes. Carcinogenesis. 2001;22:519–523. doi: 10.1093/carcin/22.3.519. [DOI] [PubMed] [Google Scholar]
- Peters JM, Rusyn I, Rose ML, Gonzalez FJ, Thurman RG. Peroxisome proliferator-activated receptor alpha is restricted to hepatic parenchymal cells, not Kupffer cells: implications for the mechanism of action of peroxisome proliferators in hepatocarcinogenesis. Carcinogenesis. 2000;21:823–826. doi: 10.1093/carcin/21.4.823. [DOI] [PubMed] [Google Scholar]
- Richert L, Binda D, Hamilton G, Viollon-Abadie C, Alexandre E, Bigot-Lasserre D, Bars R, Coassolo P, LeCluyse E. Evaluation of the effect of culture configuration on morphology, survival time, antioxidant status and metabolic capacities of cultured rat hepatocytes. Toxicol In Vitro. 2002;16:89–99. doi: 10.1016/s0887-2333(01)00099-6. [DOI] [PubMed] [Google Scholar]
- Rose ML, Rusyn I, Bojes HK, Belyea J, Cattley RC, Thurman RG. Role of Kupffer cells and oxidants in signaling peroxisome proliferator- induced hepatocyte proliferation. Mutat. Res. 2000;448:179–192. doi: 10.1016/s0027-5107(99)00235-3. [DOI] [PubMed] [Google Scholar]
- Rose ML, Rusyn I, Bojes HK, Germolec DR, Luster M, Thurman RG. Role of Kupffer cells in peroxisome proliferator-induced hepatocyte proliferation. Drug Metab Rev. 1999;31:87–116. doi: 10.1081/dmr-100101909. [DOI] [PubMed] [Google Scholar]
- Shenoy SD, Spencer TA, Mercer-Haines NA, Alipour M, Gargano MD, Runge-Morris M, Kocarek TA. CYP3A induction by liver x receptor ligands in primary cultured rat and mouse hepatocytes is mediated by the pregnane X receptor. Drug Metab Dispos. 2004;32:66–71. doi: 10.1124/dmd.32.1.66. [DOI] [PubMed] [Google Scholar]
- Sivaraman A, Leach JK, Townsend S, Iida T, Hogan BJ, Stolz DB, Fry R, Samson LD, Tannenbaum SR, Griffith LG. A microscale in vitro physiological model of the liver: predictive screens for drug metabolism and enzyme induction. Curr. Drug Metab. 2005;6:569–591. doi: 10.2174/138920005774832632. [DOI] [PubMed] [Google Scholar]
- Stamatoglou SC, Hughes RC. Cell adhesion molecules in liver function and pattern formation. FASEB J. 1994;8:420–427. doi: 10.1096/fasebj.8.6.8168692. [DOI] [PubMed] [Google Scholar]
- Swales NJ, Johnson T, Caldwell J. Cryopreservation of rat and mouse hepatocytes. II. Assessment of metabolic capacity using testosterone metabolism. Drug Metab Dispos. 1996;24:1224–1230. [PubMed] [Google Scholar]
- Swift B, Brouwer KL. Influence of seeding density and extracellular matrix on bile Acid transport and mrp4 expression in sandwich-cultured mouse hepatocytes. Mol Pharm. 2010;7:491–500. doi: 10.1021/mp900227a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui M, Ogawa S, Inada Y, Tomioka E, Kamiyoshi A, Tanaka S, Kishida T, Nishiyama M, Murakami M, Kuroda J, Hashikura Y, Miyagawa S, Satoh F, Shibata N, Tagawa Y. Characterization of cytochrome P450 expression in murine embryonic stem cell-derived hepatic tissue system. Drug Metab Dispos. 2006;34:696–701. doi: 10.1124/dmd.105.007674. [DOI] [PubMed] [Google Scholar]
- Welsh M, Mangravite L, Medina MW, Tantisira K, Zhang W, Huang RS, McLeod H, Dolan ME. Pharmacogenomic discovery using cell-based models. Pharmacol Rev. 2009;61:413–429. doi: 10.1124/pr.109.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CG, Burns AM, Bradford BU, Ross PK, Kosyk O, Swenberg JA, Cunningham ML, Rusyn I. WY-14,643-induced cell proliferation and oxidative stress in mouse liver are independent of NADPH oxidase. Toxicol. Sci. 2007;98:366–374. doi: 10.1093/toxsci/kfm104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.