Abstract
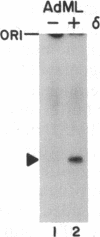
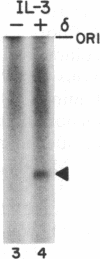
A transcription factor required for synthesis of accurately initiated run-off transcripts by RNA polymerase II has been purified and shown to have an associated DNA-dependent ATPase (dATPase) activity that is strongly stimulated by the TATA region of promoters. This transcription factor, designated delta, was purified more than 3000-fold from extracts of crude rat liver nuclei and has a native molecular mass of approximately 230 kDa. DNA-dependent ATPase (dATPase) and transcription activities copurify when delta is analyzed by hydrophobic interaction and ion-exchange HPLC, arguing that transcription factor delta possesses an ATPase (dATPase) activity. ATPase (dATPase) is specific for adenine nucleotides; ATP and dATP, but not CTP, UTP, or GTP, are hydrolyzed. ATPase (dATPase) is stimulated by both double-stranded and single-stranded DNAs, including pUC18, ssM13, and poly(dT); however, DNA fragments containing the TATA region of either the adenovirus 2 major late or mouse interleukin 3 promoters stimulate ATPase as much as 10-fold more effectively than DNA fragments containing nonpromoter sequences. These data suggest the intriguing possibility that delta plays a critical role in the ATP (dATP)-dependent activation of run-off transcription through a direct interaction with the TATA region of promoters.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bunick D., Zandomeni R., Ackerman S., Weinmann R. Mechanism of RNA polymerase II--specific initiation of transcription in vitro: ATP requirement and uncapped runoff transcripts. Cell. 1982 Jul;29(3):877–886. doi: 10.1016/0092-8674(82)90449-4. [DOI] [PubMed] [Google Scholar]
- Carthew R. W., Samuels M., Sharp P. A. Formation of transcription preinitiation complexes with an amanitin-resistant RNA polymerase II. J Biol Chem. 1988 Nov 15;263(32):17128–17135. [PubMed] [Google Scholar]
- Champoux J. J., McConaughy B. L. Purification and characterization of the DNA untwisting enzyme from rat liver. Biochemistry. 1976 Oct 19;15(21):4638–4642. doi: 10.1021/bi00666a014. [DOI] [PubMed] [Google Scholar]
- Conaway J. W., Bond M. W., Conaway R. C. An RNA polymerase II transcription system from rat liver. Purification of an essential component. J Biol Chem. 1987 Jun 15;262(17):8293–8297. [PubMed] [Google Scholar]
- Conaway J. W., Conaway R. C. A multisubunit transcription factor essential for accurate initiation by RNA polymerase II. J Biol Chem. 1989 Feb 5;264(4):2357–2362. [PubMed] [Google Scholar]
- Conaway R. C., Conaway J. W. ATP activates transcription initiation from promoters by RNA polymerase II in a reversible step prior to RNA synthesis. J Biol Chem. 1988 Feb 25;263(6):2962–2968. [PubMed] [Google Scholar]
- Davison B. L., Egly J. M., Mulvihill E. R., Chambon P. Formation of stable preinitiation complexes between eukaryotic class B transcription factors and promoter sequences. Nature. 1983 Feb 24;301(5902):680–686. doi: 10.1038/301680a0. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983 Mar;32(3):669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- Fire A., Samuels M., Sharp P. A. Interactions between RNA polymerase II, factors, and template leading to accurate transcription. J Biol Chem. 1984 Feb 25;259(4):2509–2516. [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lenardo M. J., Kuang A., Gifford A., Baltimore D. NF-kappa B protein purification from bovine spleen: nucleotide stimulation and binding site specificity. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8825–8829. doi: 10.1073/pnas.85.23.8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue N. F., Kornberg R. D. Accurate initiation at RNA polymerase II promoters in extracts from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8839–8843. doi: 10.1073/pnas.84.24.8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Segall J., Weil P. A., Roeder R. G. Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J Biol Chem. 1980 Dec 25;255(24):11992–11996. [PubMed] [Google Scholar]
- Mattoccia E., Attardi D. G., Tocchini-Valentini G. P. DNA-relaxing activity and endonuclease activity in Xenopus laevis oocytes. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4551–4554. doi: 10.1073/pnas.73.12.4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merril C. R., Goldman D., Sedman S. A., Ebert M. H. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science. 1981 Mar 27;211(4489):1437–1438. doi: 10.1126/science.6162199. [DOI] [PubMed] [Google Scholar]
- Nakajima N., Horikoshi M., Roeder R. G. Factors involved in specific transcription by mammalian RNA polymerase II: purification, genetic specificity, and TATA box-promoter interactions of TFIID. Mol Cell Biol. 1988 Oct;8(10):4028–4040. doi: 10.1128/mcb.8.10.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama N., Arai N., Kaziro Y., Arai K. Structural and functional studies of the dnaB protein using limited proteolysis. Characterization of domains for DNA-dependent ATP hydrolysis and for protein association in the primosome. J Biol Chem. 1984 Jan 10;259(1):88–96. [PubMed] [Google Scholar]
- Price D. H., Sluder A. E., Greenleaf A. L. Fractionation of transcription factors for RNA polymerase II from Drosophila Kc cell nuclear extracts. J Biol Chem. 1987 Mar 5;262(7):3244–3255. [PubMed] [Google Scholar]
- Rappaport J., Weinmann R. Purine triphosphate beta-gamma bond hydrolysis requirements for RNA polymerase II transcription initiation and elongation. J Biol Chem. 1987 Dec 25;262(36):17510–17515. [PubMed] [Google Scholar]
- Reinberg D., Horikoshi M., Roeder R. G. Factors involved in specific transcription in mammalian RNA polymerase II. Functional analysis of initiation factors IIA and IID and identification of a new factor operating at sequences downstream of the initiation site. J Biol Chem. 1987 Mar 5;262(7):3322–3330. [PubMed] [Google Scholar]
- Samuels M., Fire A., Sharp P. A. Separation and characterization of factors mediating accurate transcription by RNA polymerase II. J Biol Chem. 1982 Dec 10;257(23):14419–14427. [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Energy requirement for specific transcription initiation by the human RNA polymerase II system. J Biol Chem. 1984 Apr 25;259(8):5321–5326. [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985 Nov;43(1):165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Tolunay H. E., Yang L., Kemper W. M., Safer B., Anderson W. F. Homologous globin cell-free transcription system with comparison of heterologous factors. Mol Cell Biol. 1984 Jan;4(1):17–22. doi: 10.1128/mcb.4.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. Y., Tsai M. J., Kops L. E., Minghetti P. P., O'Malley B. W. Transcription factors from oviduct and HeLa cells are similar. J Biol Chem. 1981 Dec 25;256(24):13055–13059. [PubMed] [Google Scholar]
- Tsuchiya Y., Hattori M., Hayashida K., Ishibashi H., Okubo H., Sakaki Y. Sequence analysis of the putative regulatory region of rat alpha 2-macroglobulin gene. Gene. 1987;57(1):73–80. doi: 10.1016/0378-1119(87)90178-8. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Derbyshire R., Guy A., Molko D., Roget A., Téoule R., Chambon P. Specific in vitro transcription of conalbumin gene is drastically decreased by single-point mutation in T-A-T-A box homology sequence. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7024–7028. doi: 10.1073/pnas.77.12.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. M., Moncollin V., Egly J. M., Chambon P. A general transcription factor forms a stable complex with RNA polymerase B (II). Cell. 1987 Jul 31;50(3):361–368. doi: 10.1016/0092-8674(87)90490-9. [DOI] [PubMed] [Google Scholar]
- Ziegler D. M., Pettit F. H. Microsomal oxidases. I. The isolation and dialkylarylamine oxygenase activity of pork liver microsomes. Biochemistry. 1966 Sep;5(9):2932–2938. doi: 10.1021/bi00873a024. [DOI] [PubMed] [Google Scholar]
- Zurawski S. M., Zurawski G. Identification of three critical regions within mouse interleukin 2 by fine structural deletion analysis. EMBO J. 1988 Apr;7(4):1061–1069. doi: 10.1002/j.1460-2075.1988.tb02914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]