Abstract
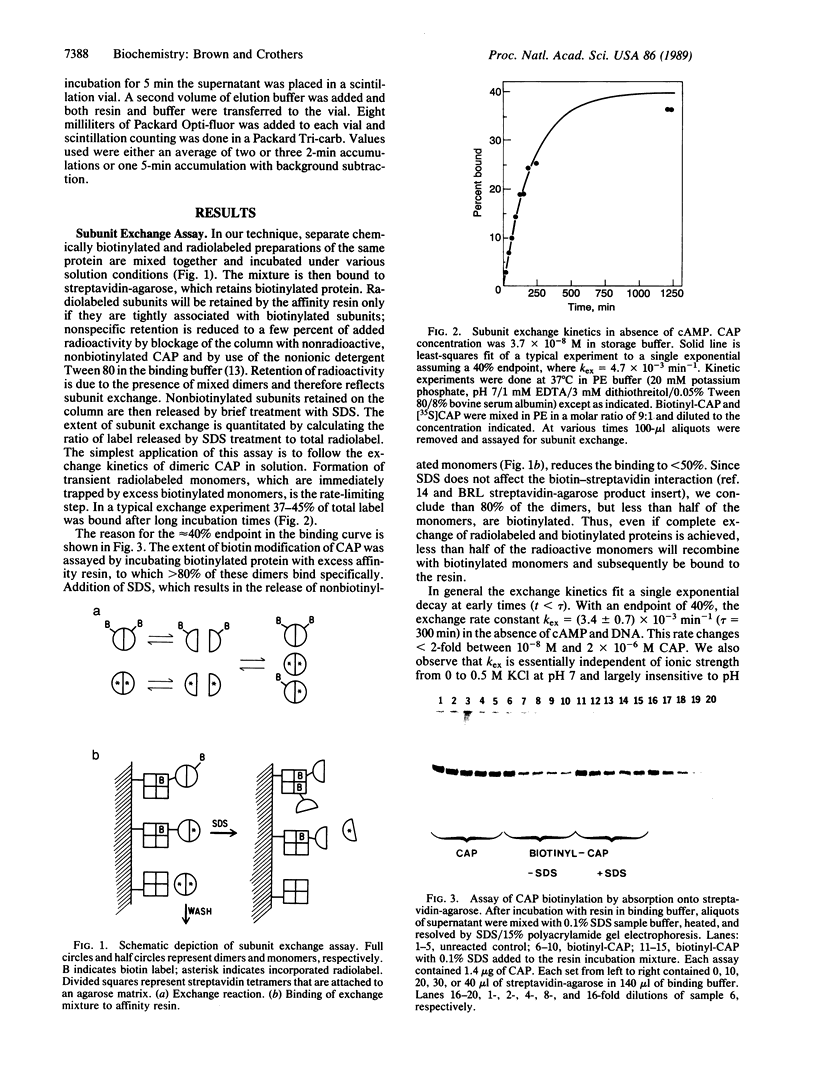
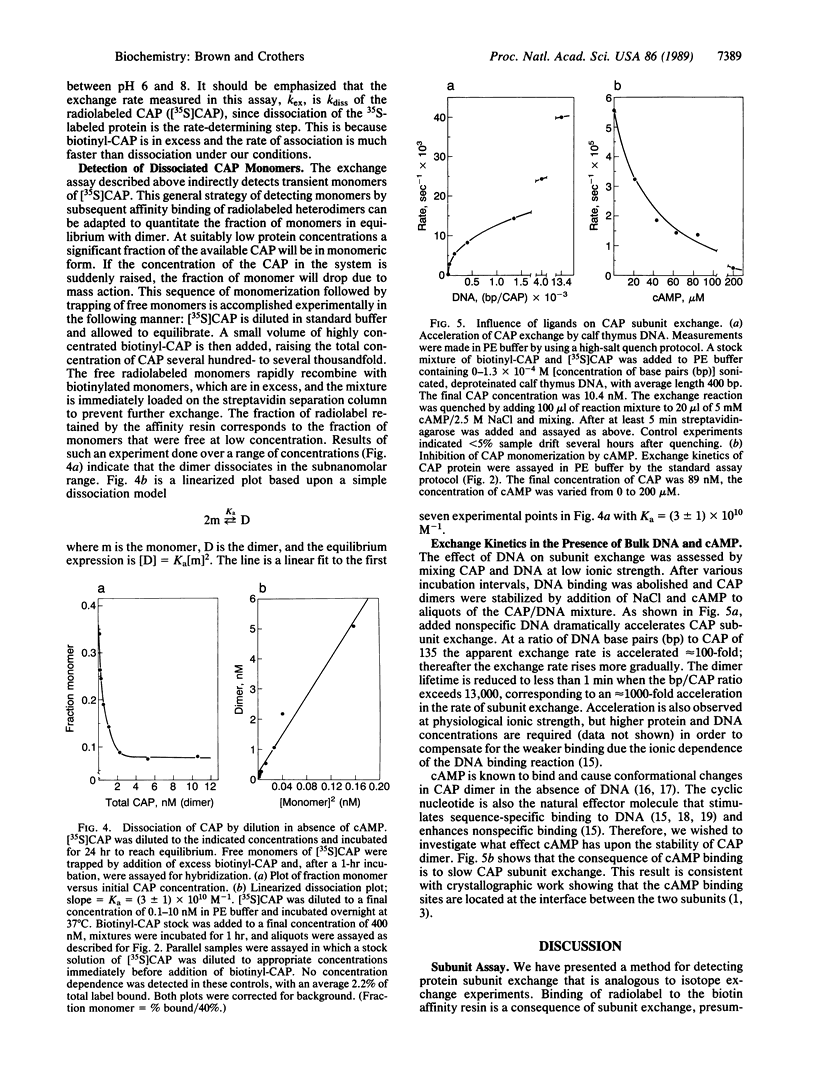
We describe an experimental approach to the measurement of protein subunit exchange in which biotinylated subunits mediate attachment of 35S-labeled subunits to a streptavidin column as a result of the exchange process. Application of the method to Escherichia coli catabolite activator protein (CAP) revealed that in the absence of cAMP, the dimerization equilibrium constant is 3 x 10(10) M-1, with a dimer lifetime of 300 min. Exchange of CAP subunits is accelerated at least 1000-fold by the presence of nonspecific DNA, under low ionic strength conditions. Catalysis of exchange also occurs at physiological ionic conditions. In contrast, physiological concentrations of cAMP stabilize CAP with respect to subunit exchange in either the presence or the absence of DNA. We discuss the functional implications of monomerization of gene-regulatory proteins resulting from kinetic and thermodynamic lability of their dimers.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chodosh L. A., Baldwin A. S., Carthew R. W., Sharp P. A. Human CCAAT-binding proteins have heterologous subunits. Cell. 1988 Apr 8;53(1):11–24. doi: 10.1016/0092-8674(88)90483-7. [DOI] [PubMed] [Google Scholar]
- Chodosh L. A., Olesen J., Hahn S., Baldwin A. S., Guarente L., Sharp P. A. A yeast and a human CCAAT-binding protein have heterologous subunits that are functionally interchangeable. Cell. 1988 Apr 8;53(1):25–35. doi: 10.1016/0092-8674(88)90484-9. [DOI] [PubMed] [Google Scholar]
- Ebright R. H., Le Grice S. F., Miller J. P., Krakow J. S. Analogs of cyclic AMP that elicit the biochemically defined conformational change in catabolite gene activator protein (CAP) but do not stimulate binding to DNA. J Mol Biol. 1985 Mar 5;182(1):91–107. doi: 10.1016/0022-2836(85)90030-0. [DOI] [PubMed] [Google Scholar]
- Eilen E., Krakow J. S. Cyclic AMP-mediated intersubunit disulfide crosslinking of the cyclic AMP receptor protein of Escherichia coli. J Mol Biol. 1977 Jul;114(1):47–60. doi: 10.1016/0022-2836(77)90282-0. [DOI] [PubMed] [Google Scholar]
- Fried M. G., Crothers D. M. Equilibrium studies of the cyclic AMP receptor protein-DNA interaction. J Mol Biol. 1984 Jan 25;172(3):241–262. doi: 10.1016/s0022-2836(84)80025-x. [DOI] [PubMed] [Google Scholar]
- Fried M. G., Crothers D. M. Kinetics and mechanism in the reaction of gene regulatory proteins with DNA. J Mol Biol. 1984 Jan 25;172(3):263–282. doi: 10.1016/s0022-2836(84)80026-1. [DOI] [PubMed] [Google Scholar]
- Ghosaini L. R., Brown A. M., Sturtevant J. M. Scanning calorimetric study of the thermal unfolding of catabolite activator protein from Escherichia coli in the absence and presence of cyclic mononucleotides. Biochemistry. 1988 Jul 12;27(14):5257–5261. doi: 10.1021/bi00414a046. [DOI] [PubMed] [Google Scholar]
- King L., Weber G. Conformational drift and cryoinactivation of lactate dehydrogenase. Biochemistry. 1986 Jun 17;25(12):3637–3640. doi: 10.1021/bi00360a024. [DOI] [PubMed] [Google Scholar]
- King L., Weber G. Conformational drift of dissociated lactate dehydrogenases. Biochemistry. 1986 Jun 17;25(12):3632–3637. doi: 10.1021/bi00360a023. [DOI] [PubMed] [Google Scholar]
- Kouzarides T., Ziff E. The role of the leucine zipper in the fos-jun interaction. Nature. 1988 Dec 15;336(6200):646–651. doi: 10.1038/336646a0. [DOI] [PubMed] [Google Scholar]
- Kumar V., Chambon P. The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell. 1988 Oct 7;55(1):145–156. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- Majors J. Specific binding of CAP factor to lac promoter DNA. Nature. 1975 Aug 21;256(5519):672–674. doi: 10.1038/256672a0. [DOI] [PubMed] [Google Scholar]
- McKay D. B., Steitz T. A. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981 Apr 30;290(5809):744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- McKay D. B., Weber I. T., Steitz T. A. Structure of catabolite gene activator protein at 2.9-A resolution. Incorporation of amino acid sequence and interactions with cyclic AMP. J Biol Chem. 1982 Aug 25;257(16):9518–9524. [PubMed] [Google Scholar]
- Miller A. M., MacKay V. L., Nasmyth K. A. Identification and comparison of two sequence elements that confer cell-type specific transcription in yeast. Nature. 1985 Apr 18;314(6012):598–603. doi: 10.1038/314598a0. [DOI] [PubMed] [Google Scholar]
- Ohlendorf D. H., Anderson W. F., Lewis M., Pabo C. O., Matthews B. W. Comparison of the structures of cro and lambda repressor proteins from bacteriophage lambda. J Mol Biol. 1983 Sep 25;169(3):757–769. doi: 10.1016/s0022-2836(83)80169-7. [DOI] [PubMed] [Google Scholar]
- Olesen J., Hahn S., Guarente L. Yeast HAP2 and HAP3 activators both bind to the CYC1 upstream activation site, UAS2, in an interdependent manner. Cell. 1987 Dec 24;51(6):953–961. doi: 10.1016/0092-8674(87)90582-4. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Ruan K., Weber G. Dissociation of yeast hexokinase by hydrostatic pressure. Biochemistry. 1988 May 3;27(9):3295–3301. doi: 10.1021/bi00409a026. [DOI] [PubMed] [Google Scholar]
- Ruan K., Weber G. Hysteresis and conformational drift of pressure-dissociated glyceraldehydephosphate dehydrogenase. Biochemistry. 1989 Mar 7;28(5):2144–2153. doi: 10.1021/bi00431a028. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Pabo C. O., Meyer B. J., Ptashne M., Backman K. C. Regulatory functions of the lambda repressor reside in the amino-terminal domain. Nature. 1979 May 31;279(5712):396–400. doi: 10.1038/279396a0. [DOI] [PubMed] [Google Scholar]
- Schmitz A. Cyclic AMP receptor proteins interacts with lactose operator DNA. Nucleic Acids Res. 1981 Jan 24;9(2):277–292. doi: 10.1093/nar/9.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert T., Bartholmes P., Jaenicke R. Influence of cofactor pyridoxal 5'-phosphate on reversible high-pressure denaturation of isolated beta 2 dimer of tryptophan synthase bienzyme complex from Escherichia coli. Biochemistry. 1985 Jan 15;24(2):339–345. doi: 10.1021/bi00323a016. [DOI] [PubMed] [Google Scholar]
- Shepherd J. C., McGinnis W., Carrasco A. E., De Robertis E. M., Gehring W. J. Fly and frog homoeo domains show homologies with yeast mating type regulatory proteins. Nature. 1984 Jul 5;310(5972):70–71. doi: 10.1038/310070a0. [DOI] [PubMed] [Google Scholar]
- Takahashi M., Blazy B., Baudras A., Hillen W. On the origin of selectivity in recognition by cyclic adenosine 3',5'-monophosphate receptor protein of its specific binding site of the lactose promoter region. J Mol Biol. 1983 Jul 15;167(4):895–899. doi: 10.1016/s0022-2836(83)80118-1. [DOI] [PubMed] [Google Scholar]
- Tenenbaum-Bayer H., Levitzki A. The refolding of lactate dehydrogenase subunits and their assembly to the functional tetramer. Biochim Biophys Acta. 1976 Sep 14;445(2):261–279. doi: 10.1016/0005-2744(76)90081-4. [DOI] [PubMed] [Google Scholar]
- Weber G., Drickamer H. G. The effect of high pressure upon proteins and other biomolecules. Q Rev Biophys. 1983 Feb;16(1):89–112. doi: 10.1017/s0033583500004935. [DOI] [PubMed] [Google Scholar]
- Wolberger C., Dong Y. C., Ptashne M., Harrison S. C. Structure of a phage 434 Cro/DNA complex. Nature. 1988 Oct 27;335(6193):789–795. doi: 10.1038/335789a0. [DOI] [PubMed] [Google Scholar]
- Wray W., Boulikas T., Wray V. P., Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981 Nov 15;118(1):197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- Wu F. Y., Nath K., Wu C. W. Conformational transitions of cyclic adenosine monophosphate receptor protein of Escherichia coli. A fluorescent probe study. Biochemistry. 1974 Jun 4;13(12):2567–2572. doi: 10.1021/bi00709a015. [DOI] [PubMed] [Google Scholar]
- Zhang R. G., Joachimiak A., Lawson C. L., Schevitz R. W., Otwinowski Z., Sigler P. B. The crystal structure of trp aporepressor at 1.8 A shows how binding tryptophan enhances DNA affinity. Nature. 1987 Jun 18;327(6123):591–597. doi: 10.1038/327591a0. [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]



