Abstract
The activation and differentiation of T cells are dependent upon numerous initiating events that are influenced by the immune environment, nature of the antigen, as well as the activation state of APCs. In the present studies we have investigated the role of a specific notch ligand, delta-like 4 (Dll4). In particular, our data have indicated that Dll4 is inducible by pathogen-associated signals through TLR activation on DC but not early response inflammatory cytokines, IL-1 and IL-18 that also activate cells via MyD88 adapter pathway. Our observations from in vitro cultures with ovalbumin specific TCR transgenic cells (DO11.10) confirmed earlier reports demonstrating that Dll4 inhibits Th2 cytokine production. Furthermore, Dll4 enhances the generation of IL-17 producing T cells in the presence of additional skewing cytokines, IL-6 and TGFβ. In the absence of notch signals IL17 production was significantly reduced even under specific skewing conditions. These studies further demonstrate that Dll4 upregulates RORγt expression in T cells and that both RORγt and IL17 gene promoters are direct transcriptional notch targets that augment the differentiation of Th17 cell populations. Thus, facilitation of efficient T cell differentiation may depend upon the activation of T cells via specific notch ligand stimulation.
Introduction
The induction of acquired immune responses during pathogenic insult depends upon the rapid recognition of pathogenic signals followed by the initiation of the proper acquired immune response. Specific signals have been shown to induce these signals after recognition of pathogen associated molecular patterns (PAMPs) that promote strong activation signals to induce innate cell activation. Toll-like receptors (TLR) have been implicated in the initial activation scheme that quickly promotes activation of the immune responses, including innate cytokine production, MHC molecule expression, and costimulatory molecule activation (1-3). Subsequent antigen presentation and T cell activation is dictated by the APC after these initial events. The TLR-induced events depend predominantly on specific signaling pathways initiated by MyD88 adaptor protein dependent activation that leads to the maturation of DCs and other APC populations (2, 4-6). A more recently identified family of MyD88 dependent molecules that are induced on APCs are Notch ligands that can alter the course of T cell activation (7-10).
Notch is a receptor system that was originally shown to be involved in cell differentiation and survival. Notch signaling is initiated by the ligand engagement of the notch receptor. There are four notch receptors (N1-4) and five notch ligands (Delta like 1, 3 and 4; Jagged 1 and 2) (11, 12). Upon binding by either delta like or jagged ligands, Notch undergoes proteolytic cleavages catalyzed by Adam proteases and gamma secretase complex, leading to the translocation of the notch intracellular domain (N-ICD) into the nucleus. Notch interacts with the transcriptional repressor RBPj-κ (CSL). N-ICD interaction with RBPj-κ displaces transcriptional corepressors from RBPj-κ and also recruits Mastermind (MAML) protein. The new transcriptional complex of N-ICD-RBPj-κ-MAML converts RBPj-κ from a repressor to a transcriptional activator (11-13). Over the past several years this complex signaling and activation system has been shown to determine fate decisions within immune cell populations, including DC, B cells, and T cells (13). It appears that depending upon what ligands on DCs (delta-like or jagged) are used to engage notch receptors on T cells during activation, a specific course of T cell differentiation can be dictated (8, 14). Still other studies using genetic models of defective Notch activation via conditional deletion of RBPj-κ or expression of dominant negative form of MAML on CD4+ T cells have demonstrated that Th2 type, but not Th1 type responses are dependent upon notch signaling pathways and upon GATA3 activation (7, 10, 15). Although no defects were seen by conditional deletion of RBPj-κ or expression of dominant negative form of MAML on Th1 (IFNγ) responses, γ secretase inhibitor (GSI) mediated inhibition of Notch revealed abrogation of IFNγ production (9). More recent studies have demonstrated that delta-like 1 and 4 appear to regulate Th2 cytokine production and therefore regulate the differentiation potential of peripheral effector T cells (16, 17). Overall, however, little defined information as to the role of notch signaling for the regulation of T cell activation events exist. This is further exemplified by a number of studies that have demonstrated that Notch can be involved in Th1, Th2 as well as Treg cell differentiation (7-10, 18, 19), depending on the ligand, immune environment, genetic makeup or disease model in which the responses were derived.
A series of studies examining CD4+ T cell differentiation have further defined a novel class of T cells based upon their ability to produce IL-17, IL-21 and IL-22 (20, 21). This lineage of T cell is known as Th17 and has been shown to be pathogenic in autoimmunity, psoriasis, and in several other chronic disorders. Along with its ability to induce rapid neutrophil accumulation, the production of IL-17 may provide a means for preferential innate immunity during bacterial infections at mucosal sites. Although the regulation of Th17 cells has not been fully defined, IL-6 and other signal transducer and activator of transcription-3 (STAT3) initiating cytokines in concert with TGFβ are necessary for Th17 differentiation. The transcription factor RORγt also appears to be required for Th17 differentiation in addition to the cytokine signals (22). Thus, a large body of literature has emerged on the role and regulation of Th17 during chronic immune responses.
In the present studies we provide evidence of that Dll4 provides additional skewing signals to drive Th17 differentiation through the upregulation of RORγt while at the same time limiting Th2 cytokine production. IL17 production was notch dependent and CSL directly associated with RORγ-t and IL17 promoter regions that contained CSL binding sites. Thus, these studies may define a critical activating scheme for generation of this class of T cell during chronic diseases, such as autoimmunity, potentially via TLR mediated signals that would specifically drive Dll4 upregulation on APC.
Materials and Methods
In vitro CD4+ T cell culture
Magnetic bead isolated CD4+CD62L+CD25− splenic naïve T cells were purified utilizing the MACS system (Miltenyi Biotec). Briefly, the CD4+ T cells were isolated using anti-CD4+ magnetic beads by negative selection. The CD62L+ cells were then enriched from the CD4+ T cell population using anti-CD62L+ beads by positive selection. Cells were then plated and cultured in flat-bottomed 96 well plates (luminex analysis) or 6 well plates (western blots and ChIP assays). The cells were activated under different skewing conditions. Th1 conditions: ova-peptide (1 μg/ml), anti IL4 (10 μg/ml) and anti IL12 (10 μg/ml); Th2 conditions: ova-peptide, anti-IFNγ (10 μg/ml); Th17 conditions: plate bound anti-CD3 (1μg/ml) and soluble anti-CD28 (1 μg/ml) for CD4 T cells from Balb/C or C57BL/6 or IL17−/− mice or ova peptide for T cells from DO-11.10 mice together with TGFβ1 (2 ng/ml), IL6 (20ng/ml), anti-IL4 (10 μg/ml) and anti-IFNγ (10 μg/ml). Plate bound recombinant Dll4 (rDll4) was used at a final concentration of 2.5μg/ml.
Generation of BMDCs
BM was harvested from uninfected, naive mice and seeded in T-150 tissue culture flasks at 106 cells/ml in RPMI 1640 – based complete media with GM-CSF/ml (R & D Systems). After 6d, loosely adherent cells were collected and isolated with anti-CD11c by magnetic bead (Miltenyi Biotec) as previously described (23).
Quantification of cytokines and transcription factors
Total RNA was isolated from the CD4 T cells using Trizol (Invitrogen) following the manufacturer’s instructions and was reverse transcribed in a 25 μl volume. mRNA expression was determined in 1 μl of cDNA by TaqMan real-time PCR with a PRISM 7500 sequence detection system using pre developed gene-specific primers for GAPDH, IL17A and RORγ-t (Applied Biosystems). The Sybr primer sets for Dll1, Dll4, Jag1, Jag2 and GATA3 were purchased from Sigma Aldrich and were described previously (8). Results are normalized to GAPDH expression and are presented as the folds increase in mRNA expression over the control group. Protein levels of cytokines were quantified using a Bio-Plex bead-based (luminex) cytokine assay purchased from Bio-Rad Laboratories.
Western Blot
Cytoplasmic or Nuclear extracts were prepared using the NE-PER nuclear and cytoplasmic extraction kit from Pierce (Rockford, IL) following the manufacturer’s protocol. Equal amount of the proteins were separated by SDS-PAGE and transferred on nitrocellulose membrane. The membrane was probed with primary antibodies: rabbit anti cleaved notch 1 (Val 1744) (Cell Signaling); phospho STAT3; total STAT3, (Cell Signaling) anti mouse monoclonal β actin (Sigma-Aldrich). This was followed by incubation with the appropriate peroxidase-conjugated secondary antibody. Membranes were developed with ECL™ Western Blotting Detection Reagent (Pierce). In all the experiments, β- actin was used as the loading control.
Mice
C57Bl/6, Balb/C and DO11.10 mice were obtained from Jackson Laboratories (Bar Harbor, ME). C57Bl/6 IL17−/− mice were the generous gift of Dr. Kate Eaton (University of Michigan, Ann Arbor, MI). All mice were maintained in specific pathogen-free facilities in the Unit for Laboratory Animal Medicine at the University of Michigan. Mice used in experiments were age, strain and sex matched.
ChIP Assay and Primers
The chromatin immunoprecipitation (ChIP) procedure was performed using an assay kit (Upstate Biotechnology) according to the manufacturer’s instructions. Briefly, 1×106 isolated CD4+ T cells were skewed under Th17 conditions in the presence or absence of rDll4 for 20 hours. DNA-protein structure was then cross-linked by 1% formaldehyde for 10 minutes at 37°C. Cells were collected and lysed in 400 μl SDS lysis buffer. The resulting lysates were sonicated to obtain DNA fragments ranging from 200 to 1000 bp (base pairs) using a Branson Sonifier 450 (VWR, West Chester, PA) under the following condition: 4 times for periods of 30 seconds each. After centrifuging, the supernatant containing chromatin was diluted, and an aliquot (4% volume) was saved to indicate the input DNA in each sample. The remaining chromatin fractions were precleared with salmon sperm DNA/protein A agarose beads followed by immunoprecipitation with the following antibodies: anti–RBPj-κ/CSL monoclonal antibody (Cosmo Bio Co., Ltd.) or control anti rat Ig (Jackson Immuno Research) overnight at 4°C with gentle rotation. Cross-linking was reversed for 4 hours at 65°C and was followed by proteinase K digestion. DNA was purified by standard phenol/chloroform and ethanol precipitation and was subjected to Sybr Green real-time PCR. Mouse promoter primers are as follows rorγ-t: forward, 5′ –CCCCTCACCTCTCAATTTGC; reverse, 5′- GCTTCTAGATGCTTCCCATACTTCTG; IL17p1: forward, 5′- TCTGCTTGACTCGATTTTCAGGTA; reverse, 5′- GACGTGTGATGTCATCTCAAAATG; IL17p2: forward, 5′-CAATTGCTCCTCCAAGGACAAG; reverse, 5′- CTGGCTTTGAGAAGAACGGATT; IL17p3: forward, AATTCAAGGAGTTCATGCTTCTCA; reverse, 5′-GCTCACACACACCTCTGATTGC. These sequences were derived based upon the promoter region of IL17 gene as previously described (24, 25).
Recombinant Proteins Antibodies and Chemicals
All the recombinant proteins (Dll4, Jagged-1, TGFβ-1, IL1, IL6, IL12 and IL18) were purchased from R&D systems. Functional antibodies, anti-IL4, anti-IFNγ, anti-IL12, anti-CD3e and anti-CD28 were purchased from eBiosciences. Ovalbumin, LPS, poly I:C and CpG were from Sigma Aldrich. GSI-IX was purchased from Calbiochem.
Statistics
Results were expressed as means +/− SE. Statistical significance was determined by Students T test or one-way ANOVA with Newman-Keuls post test. Significant differences were regarded as p < 0.05.
Results
Upregulation of Notch ligands Delta-like by TLR but not cytokine responses
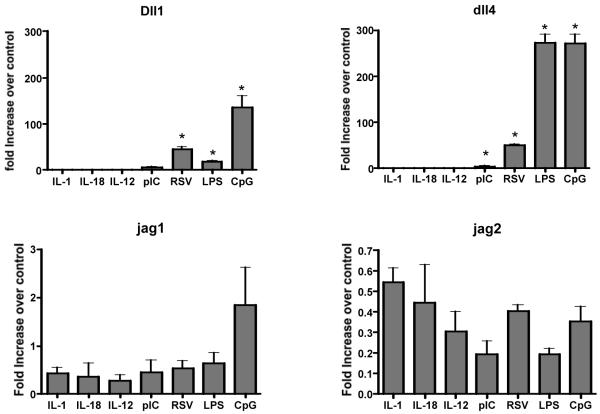
In previous publications the upregulation of notch ligands on DCs was initiated by pathogenic stimuli that demonstrated that delta-like proteins were dependent upon MyD88-mediated signaling (16, 18). To further verify these responses and determine the extent of their regulation, our studies examined a number of stimuli related to viral and/or inflammatory responses. These included TLR-specific signals, Poly I:C (TLR3), LPS (TLR4) and CpG (TLR9), as well as respiratory syncytial virus (RSV), which activates by both RIG-I and TLR-mediated pathways. Our previous studies have outlined the role of Dll4 in regulating Th2 cytokine responses during RSV infection. All of these stimuli significantly upregulated delta-like 4 and to a lesser extent delta-like 1, but did not significantly induce jagged1 and jagged2 expression (Figure 1). Furthermore, we also stimulated the DCs with IL-1β and IL-18, two IL-1R family MyD88-dependent signals. These inflammatory cytokines did not induce the expression of any of the notch ligands. Together these data reflect the idea that Th1 inducing TLR pathogenic signals promote the expression of delta-like but not jagged notch molecules.
Figure 1. Upregulation of notch ligands Delta like by TLR but not inflammatory cytokines.
Bone marrow derived dendritic cells (BMDCs) were stimulated with different Toll like receptor ligands or with individual cytokines for 24 hours. Following stimulation total RNA was harvested, reverse transcribed to cDNA and utilized for Sybr green real time amplification for different notch ligands. The data was normalized to a house keeping gene GAPDH. The data is expressed as the mean ± SE and (*) p<0.05 was considered significant.
TLR-mediated stimulation of IL-17 responses depends upon the stimulus and Dll4
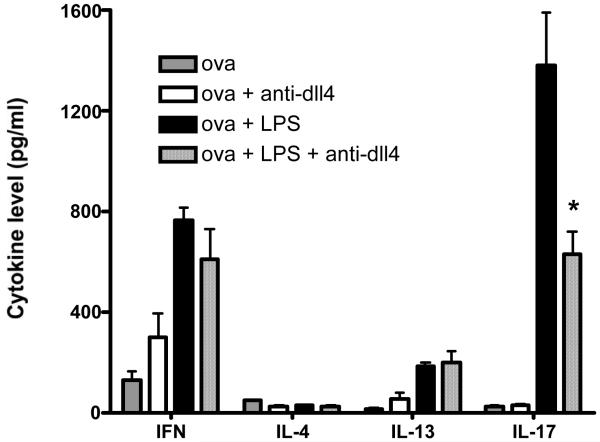
Given the above observations indicating up-regulation of Dll4 following TLR activation we were interested in investigating a TLR stimulation condition that may drive specific cytokine production profiles. We utilized whole splenocytes from TCR transgenic DO-11.10 mice and stimulated them with ova or ova and LPS in the presence or absence of anti Dll4 antibody. The above studies indicated that LPS preferentially induces strong Dll4 expression. The data indicate that stimulating the cells with ova alone is not able to drive IL17 production, correlating to the inability of ova to induce Dll4 in DC (data not shown). However, there was a robust increase in IL17 production in the group receiving both ova and LPS (with control ab), which was significantly inhibited by treating the cells with anti-Dll4 antibody (Figure 2). We also observed a marked increase in IFNγ with ova and LPS (a potent Th1 inducer) however anti-Dll4 had no significant effect in modulating the Th1 response. Taken together the data indicate a novel role of Dll4 in IL17 production that is regulated by TLR activation.
Figure 2. TLR, Dll4 mediated IL17 production.
DO-11.10 splenocytes were stimulated with either ova or LPS or both together in the presence or absence of Dll4 antibody for 48 hours. Cytokine levels were measured using a luminex system. The experiment was repeated two times in triplicates and the data represents the mean ± SE. * p ≤ 0.02
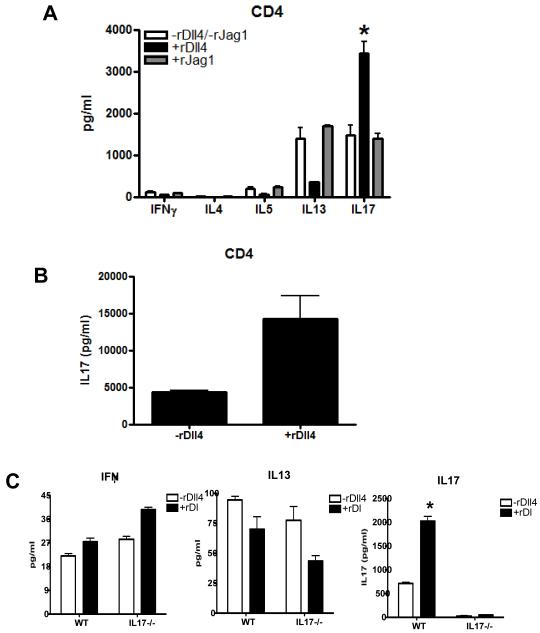
In subsequent experiments set up to assess the role of the specific Dll4 agonist responses in differentiation of immune responses we utilized isolated CD4+ T cells from Balb/c mice activated with anti-CD3 and anti-CD28 under Th17 skewing conditions (IL-6, TGFβ, anti IL4 and anti IFNγ) in the presence or absence of Dll4 or Jagged1 (Jag1) protein, as previous studies have recognized that these two ligands have differential effects on T cell differentiation. In the primary response when Dll4 was provided along with Th17 skewing cytokines, IL-17 production was greatly enhanced and Th2 cytokines IL-5 and IL-13 were markedly inhibited, while IFNγ remained unchanged (Fig. 3A). In contrast, notch ligand Jagged1 had no effect on any of the cytokines, further confirming the specific regulatory role of Dll4 in IL17 production. We also wanted to examine the role of Dll4 on IL-17 production during secondary T cell activation in Th17 skewed cultures. Therefore, we cultured T cells in Th17 skewed conditions in the presence or absence of Dll4 for 5 days followed by a 3 day rest period. Subsequently, T cells were restimulated in unskewed conditions using anti-CD3 and anti-CD28 and demonstrated an increase in IL-17 in the presence of Dll4 (Fig. 3B). Altogether, these results demonstrate a stronger skewing environment and the ability of Dll4 to promote enhanced IL-17 production.
Figure 3. Modulation of T cell responses by Dll4 under Th17 skewed conditions.
A) CD4+T cells were activated under Th17 conditions in the presence or absence of rDll4 (2.5μg/ml) or rJag1 (0.5μg/ml) and the supernatants were analyzed for indicated cytokines. B) CD4+ T cells were differentiated under Th17 conditions in the presence or absence of rDll4 for 5 days, rested for 3 days and then viable cells were re-stimulated with plate bound anti-CD3 and soluble anti-CD28 for 24 hours. Following stimulation the supernatant was analyzed IL17. Each experiment was repeated at least four times with similar results. The data are expressed as the mean± the SEM and (*) p ≤ 0.05 was considered significant. C) MACS purified CD4+ T cells from WT and IL17−/− mice were stimulated under Th17 conditions and after 48 hours the supernatant were analyzed for indicated cytokines using a luminex system. The data represents the mean ± the SEM and * p = 0.004.
Previous studies have demonstrated a role for delta like ligands in the inhibition of the Th2 responses, and we were interested in whether the altered Th2 response could be due to the skewing conditions itself or mediated directly by IL17. To further confirm we examined the potential regulatory role of IL-17 itself. Isolated CD4+ T cell from the spleen of wild type (WT) and IL17 knockout mice (IL17−/−) mice were stimulated with anti-CD3 and anti-CD28 in the presence or absence of Dll4 under Th17 skewing conditions. There was a significant increase in IL17 levels in the wild type and as expected, no increase in the knockout mice (Figure 3C). However, we observed a similar pattern of IL13 production in the WT and IL17−/− mice. Thus, IL17 itself was not having a regulatory effect on the Th2 cytokine response.
Notch activation is necessary for Dll4 induced IL17 production
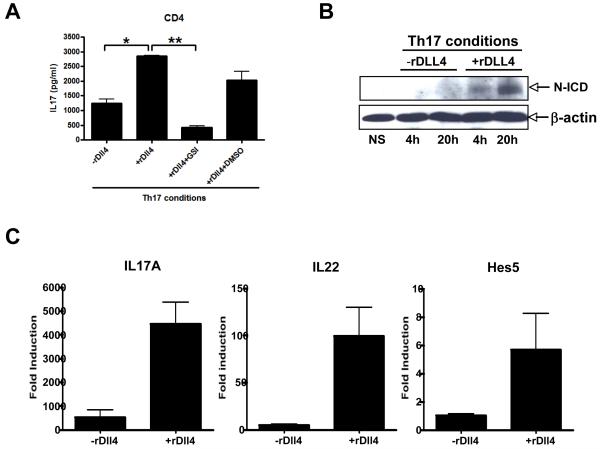
To verify that the addition of Dll4 was inducing the responses through a Notch receptor mediated signaling event our studies utilized GSI in the Th17 skewing cultures. GSI specifically inhibits notch activation by inhibiting the cleavage of notch intracellular domain (N-ICD) from membrane associated notch receptor. Our data indicate that when isolated CD4+ T cells were incubated with GSI in the presence of Dll4 IL-17 production was significantly reduced (Figure 4A). To verify that Dll4 activated the notch-signaling pathway under Th17 conditions lysates were prepared from CD4+ T cells and stimulated in the presence and absence of Dll4. We observed an increase in the nuclear accumulation of the N-ICD with Dll4 stimulation, thus indicating that Dll4 causes the N-ICD to be cleaved under these conditions (Figure 4B). To verify that Dll4 induced other Th17 pathway cytokines and Notch targeted genes we assessed IL-22 and hes5 and found both upregulated along with IL17 in the skewed cultures when Dll4 was present (Figure 4C). Over all our data suggest that Dll4 utilizing notch signaling augments early Th17 differentiation.
Figure 4. Notch activation is necessary for Dll4 enhanced IL17 production in CD4+ T cells.
A) CD4+ T cells were MACS purified from the spleen of Balb/c mice and were skewed under Th17 conditions in the presence or absence of Dll4 or GSI or control (DMSO) for 48 hours. The supernatants were analyzed for IL17 production. The data represents the mean ± the SEM of triplicate determinants and are representative of four independent experiments. * p = 0.004 and ** p = 0.002. B) CD4+ T cells were MACS purified from the spleen Balb/c mice and were skewed under Th17 conditions in the presence or absence of Dll4 for 4 hours or 20 hours. The whole cell lysate was analyzed by Western blot for the notch intracellular domain (N-ICD) and β-actin was used as loading control. The experiment was repeated twice with similar results. C) Using the same conditions as above Dll4 enhances not only IL-17 but also the mRNA expression of IL-22 and the Notch pathway target gene hes5. Data represents the mean ± SE from 3 separate experiments.
Regulation of RORγT transcription factor expression by DLL4
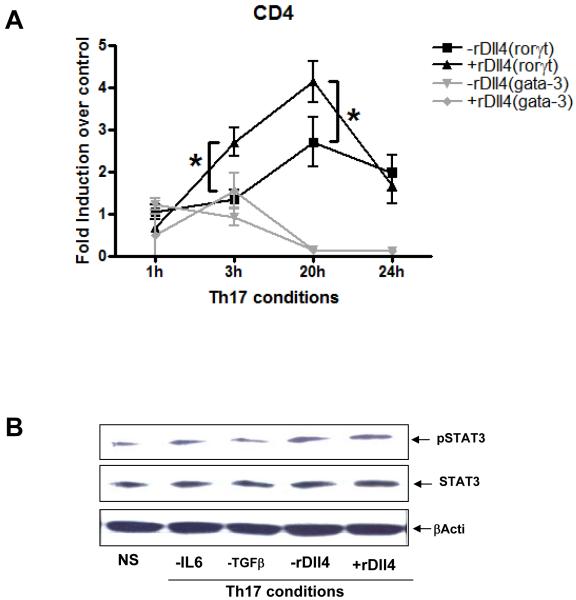
Previous studies have indicated that notch/notch ligand interaction differentially regulates the expression of specific transcription factor expression in T cells. In particular, recent studies using mice genetically unable to express active Notch in CD4+ T cells have suggested that without Notch signaling Th2 cells cannot differentiate, while Th1 responses can proceed (7, 10, 15). The latter studies using genetically modified animals examined this further and concluded that the lack of Th2 development was linked to GATA3 activation. In the present studies we were interested in whether the enhanced expression of IL-17 in the presence of Dll4 was related to RORγT expression, a required transcription factor for Th17 differentiation. When RORγT expression was examined in isolated CD4+ T cells stimulated under Th17 skewing conditions RORγT was significantly induced and further enhanced when cells were incubated with Dll4 in a time dependent fashion (Figure 5A). Moreover there was a coordinated decrease in GATA-3 expression under Th17 skewed conditions but no additional change with Dll4.
Figure 5. Differential regulation of transcription factors rorγ-t and gata3 by Dll4.
A) CD4+ T cells were stimulated under Th17 conditions in the presence or absence of rDll4 for varying time. Total RNA was extracted, reverse transcribed and cDNA was amplified using specific primers for rorγ-t or gata-3. The data is expressed as the mean± the SEM. * p = 0.031. B) CD4+ T cells were stimulated under Th17 conditions in the presence or absence of IL6 or TGFβ or rDll4. The whole cell lysate was the analyzed by western blot for phosphorylation of STAT-3. β-actin was used as a loading control.
A second required factor for differentiation of Th17 is STAT3 protein activation (26, 27). In similar studies as above we examined the activation/phosphorylation of STAT3 during Th17 skewing to determine if Dll4 induced an increase in either total or activated STAT3 in isolated CD4+ T cells. The data in Figure 5B indicates that while STAT3 phosphorylation was enhanced under Th17 skewed conditions; there was no alteration in STAT3 activation in the presence of Dll4 in the Th17 skewed culture conditions. Thus, the increased IL-17 in the presence of Dll4 appears to be associated with increased expression of RORγT, but does not alter STAT3 activation.
Notch interaction with RORγT and IL17 promoter regions that contain CSL binding sites regulates IL17 production
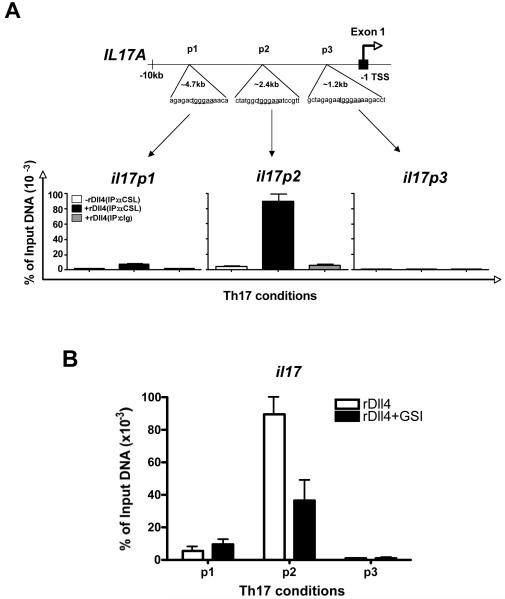
To determine the possibility that notch may regulate RORγT and/or IL17 directly, we searched for CSL (RBPj-κ), transcription factor involved in notch activation) binding sites within the upstream RORγT and IL17 promoter regions. We detected one site on RORγT and three putative sites on IL17A loci. A consensus RBPj-κ/CSL binding site on RORγT was 0.56kb upstream of the translational start site (TSS). Primer sets flanking the RBPj-κ/CSL binding regions on RORγT were designed to amplify the immunoprecipitated (IP) ChIP DNA by quantitative RT-PCR (Figure 6). CD4+ T cells were treated in the presence or absence of Dll4 under Th17 skewed conditions for 20 hours and were immunoprecipitated with anti-RBPj-κ/CSL monoclonal antibody. We chose 20 hours because in our system we detected robust notch activation at that time. Our data indicate that the putative RBPj-κ/CSL binding site on RORγT upstream of the TSS was crucial for RBPj-κ/CSL binding. The abundance of RBPj-κ/CSL binding to RORγT was significantly increased under Th17 skewing conditions in the presence of Dll4 as compared to the Th17 skewing group without Dll4. The specificity of the interaction was ascertained by using a control Ig during IP that had no effect. These data correlated to the ability of Dll4 treatment to increase the expression of rorγt mRNA expression above.
Figure 6. RBPj-κ (CSL) directly interacts with rorγ-t promoter.
Diagram for rorγ-t promoter with CSL binding sites (underlined) 5′ upstream of exon 1 is shown and forward and reverse primers were designed to amplify the indicated regions. MACS purified splenic CD4+ T cells were stimulated under Th17 conditions for 20 hours and fixed for ChIP. Antibodies used for IP are anti-CSL (αCSL) and control Ig (cIg). Total input DNA before IP was used for normalization of data. The graphs represent quantitative PCR analysis of the ratio of enriched rorγ-t promoter with CSL binding sites to the input DNA. CSL binding sites were amplified using rorγ-t specific promoter primers. Data represents mean ± SE of a representative of 3 independent experiments each performed in triplicate.
Similar to the identification of a consensus RBPj-κ/CSL binding site on rorγt our studies also examined putative binding sites on the IL-17 promoter region. The RBPj-κ/CSL binding sites on IL17 promoter were identified at approximately 1.2kb, 2.4 kb and 4.7 kb upstream of the translational start site and flanking primer sets were designed to specifically amplify ChIP DNA using quantitative PCR (Figure 7). As above, a twenty-hour time point was utilized to examine anti-RBPj-κ/CSL immunoprecipitated DNA for binding in the presence or absence of Dll4 activation. Examining all three of the consensus RBPj-κ/CSL binding sites, only the binding site 2.4 kb upstream from the transcriptional start site (p2) demonstrated an increase in RBPj-κ/CSL binding after Dll4 treatment (Figure 7A). The 1.2kb (p3) site had no noticeable effect and the 4.8kb site had a very modest increase on the interaction indicating a non-essential RBPj-κ/CSL binding site. To further investigate the specificity of the binding in regards to Notch activation, similar studies were set up with Dll4 treatment groups in the presence or absence of GSI to block the N-ICD cleavage. The data in Figure 7B demonstrate that the increased binding of RBPj-κ/CSL to the IL-17 promoter region at 2.4 kb upstream of the TSS was significantly reduced when GSI was utilized. Taken together our observations indicate two possible novel mechanisms utilized by Notch in regulating IL17 production in CD4+ T cells. Firstly, N-ICD/ RBPj-κ may directly interact and increase the expression of transcription factor RORγT that has been shown to promote Th17 differentiation in CD4+ T cells. Secondly, N-ICD/ RBPj-κ may interact directly with the IL-17 promoter at a putative RBPj-κ/CSL binding site 2.4 kb upstream of exon 1 and upregulate IL17 expression.
Figure 7. RBPj-κ (CSL) directly interacts with IL17 promoter on specific binding sites.
CD4+ T cells were activated under Th17 skewing conditions in the presence of rDll4 or in the presence or absence of GSI for 24 hours. The cells were the fixed for ChIP. IP was carried out using anti CSL antibody. The three CSL binding sites on IL17 promoter (p1, p2 and p3) were amplified and analyzed by quantitative RT-PCR. The data represents the normalized percentage amounts relative to the input DNA before IP. The experiment is one representative of three independent experiments. The data is expressed as the mean ± the SE and is representative of four independent experiments.
Discussion
The regulation of T cell differentiation can be influenced by multiple molecular signals that together fine tune T cell responses. The data generated in these studies demonstrate the enhancing role of Notch ligand Dll4 in Th17 differentiation. We also provide evidence of putative mechanisms by which Dll4 dictates Th17 differentiation. It is clear from numerous studies that Notch family molecules can have a diverse role in determining the fate of T cells during activation. Studies using genetic models have demonstrated that Notch is required for Th2 but not Th1 responses (7, 10, 15), while other results have shown diverse functions of Notch depending upon the ligand and/or the immune environment (8, 14, 16, 17, 28). In the present studies we have confirmed that Dll4 has the ability to alter Th2 cell development in an antigen-specific response. In fact, using a pathogenic stimulus, LPS (TLR4), it appeared the entire phenotype of the skewing response moved away from a primarily Th1 response and toward a mixed cytokine environment, including high levels of IL-17 that were significantly reduced when dll4 was blocked. Interestingly, IL-17 production was only detected when a strong pathogenic (TLR) signal was applied to T cell activation, likely due to the up-regulation of the appropriate skewing signals, as previously demonstrated (18), including IL-6, TGFβ, and now Dll4. To follow these studies we examined the role of Dll4 in naïve CD4+ T cell cultures that were under Th17 skewing conditions and found that rDll4, but not jagged1, could enhance the skewing of T cells toward IL-17 production, as well as IL-22 expression. This appears to manifest itself during development (afferent) and maintained in secondary responses upon subsequent activation. Although it is not likely that notch signaling responses can by themselves influence the direction of immune responses (29), in the present study addition of GSI did inhibit the Dll4 enhanced IL17 production.
The differentiation of T cell subsets is dependent upon the expression of specific transcription factors. The expression of RORγt that demarcates Th17 cells was significantly upregulated in the presence of Dll4. The simultaneous decrease in GATA3 expression, the signature regulatory transcription factor for Th2 cytokines, displayed no additional alteration in the presence of Dll4 under Th17 skewing conditions. Previous studies have demonstrated that Dll4 downregulates GATA3 under Th2 skewing conditions (16). Interestingly, these latter observations suggest a complicated regulatory pattern for GATA3, as two independent studies have shown that Notch signaling is required for the expression of GATA3 and Th2 cytokines (7, 15). Future investigations will surely need to define how the different Notch ligands and receptors interact and control the divergent responses for the regulation of the T cell responses. One other possibility in the present studies was a direct regulation of the Th2 cytokines by IL17 itself in an autocrine or paracrine fashion. However, our data with CD4+ T cells from the IL17−/− mice demonstrated that even under the skewed conditions IL17 does not play a role in down regulating Th2 cytokines under Th17 skewed conditions. The coordinated temporal downregulation of GATA3 with the increased expression of RORγt under Th17 skewing conditions may suggest that RORγt may provide direct regulation of Th2 responses as previously suggested (30).
In CD4+ T cells RORγt and STAT3 are predominant regulators of the IL17 gene. Importantly, these studies indicate a direct connection between Dll4-induced notch activation and IL17 gene regulation with RBPj-κ/CSL. Notch activation and interaction of N-ICD with the transcription factor RBPj-κ/CSL converts RBPj-κ/CSL from a transcriptional repressor to a transcriptional activator leading to the expression of notch dependent genes. We found a consensus RBPj-κ/CSL binding site on the IL17 promoter 2.4 kb from the TSS that may regulate IL17 expression after Dll4 co-activation. Moreover the interaction was completely lost when notch signaling was inhibited by GSI. Previously it has been shown that RBPj-κ/CSL can directly regulate GATA3 and IL4 genes by in an inductive model of Notch-mediated Th2 signaling (16). Our studies also revealed a similar direct connection between Dll4-induced RBPj-κ/CSL and RORγ-t through a consensus RBPj-κ/CSL binding site, suggesting that Dll4 augments IL17 production by specific multiple interactive mechanisms. In contrast, notch activation via Dll4 did not alter STAT3 phosphorylation.
These novel findings demonstrating enhanced Th17 skewing with Dll4-induced N-ICD release are likely influenced by the immune environment created by the TLR stimuli that would include the generation of high levels of IL-6. It appears that Dll4 enhances the skewing effect in the presence of other Th17 relevant signals as indicated using specific skewing conditions for Th17 cell generation. In this regard, Dll4-induced Notch signals may only function to modify the immune responses dependent upon the contextual nature that the signal is provided. Thus, while Notch activation in T cells is an important signal it may serve primarily as a guide for activating/regulating the T cell for a particular immune environment. In the case of Dll4, which is dependent upon pathogenic signals and PAMPs through TLR activation (16, 18), the immune environment would skew away from Th2 type responses facilitating pathogen clearance. While beneficial in most cases if an individual had an underlying immune dysfunction, such as autoimmunity, a detrimental bystander effect could result in exacerbation of disease. This would potentially promote disease-altering effects in a system that is biased toward development of anti-pathogen defenses. Under these conditions exacerbation of disease by pathogenic challenges could lead to an exacerbated scenario of severe tissue/organ damage. Evidence of this is reflected in recent studies that have observed differential changes in development of autoimmune responses upon manipulation of different notch ligands or in the overall blockade of notch activation (31, 32). In fact, recent studies have drawn direct correlations between specific Notch receptors and IL-17 production in autoimmunity (31).
Overall, these studies extend our understanding of how specific notch ligand signals can direct the immune responses by alteration of specific gene regulation. The coordination of these responses, while complex, depends upon the innate immune system and recognition of specific pathogenic cues.
Acknowledgements
The Authors wish to thank Dr. Beau Carson for providing primers for rorγt promoter sequences used in this manuscript and Dr. Ivan Maillard for helpful discussions. This work was funded in part by NIH Grant AI073876.
Footnotes
The authors have no conflicting financial interests.
References
- 1.Beutler B, Jiang Z, Georgel P, Crozat K, Croker B, Rutschmann S, Du X, Hoebe K. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu Rev Immunol. 2006;24:353–389. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- 2.O’Neill LA. Signal transduction pathways activated by the IL-1 receptor/toll-like receptor superfamily. Curr Top Microbiol Immunol. 2002;270:47–61. [PubMed] [Google Scholar]
- 3.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 4.Beutler B, Hoebe K, Du X, Janssen E, Georgel P, Tabeta K. Lps2 and signal transduction in sepsis: at the intersection of host responses to bacteria and viruses. Scand J Infect Dis. 2003;35:563–567. doi: 10.1080/00365540310016295. [DOI] [PubMed] [Google Scholar]
- 5.Colonna M. TLR pathways and IFN-regulatory factors: to each its own. Eur J Immunol. 2007;37:306–309. doi: 10.1002/eji.200637009. [DOI] [PubMed] [Google Scholar]
- 6.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, Busslinger M, McCright B, Gridley T, Flavell RA. Direct regulation of gata3 expression determines the T helper differentiation potential of notch. Immunity. 2007;27:89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–526. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- 9.Minter LM, Turley DM, Das P, Shin HM, Joshi I, Lawlor RG, Cho OH, Palaga T, Gottipati S, Telfer JC, Kostura L, Fauq AH, Simpson K, Such KA, Miele L, Golde TE, Miller SD, Osborne BA. Inhibitors of gamma-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat Immunol. 2005;6:680–688. [PubMed] [Google Scholar]
- 10.Tu L, Fang TC, Artis D, Shestova O, Pross SE, Maillard I, Pear WS. Notch signaling is an important regulator of type 2 immunity. J Exp Med. 2005;202:1037–1042. doi: 10.1084/jem.20050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dallman MJ, Champion B, Lamb JR. Notch signalling in the peripheral immune system. Novartis Found Symp. 2003;252:268–276. discussion 276-268. [PubMed] [Google Scholar]
- 12.Maillard I, Adler SH, Pear WS. Notch and the immune system. Immunity. 2003;19:781–791. doi: 10.1016/s1074-7613(03)00325-x. [DOI] [PubMed] [Google Scholar]
- 13.Ehebauer M, Hayward P, Martinez-Arias A. Notch signaling pathway. Sci STKE. 2006;2006:cm7. doi: 10.1126/stke.3642006cm7. [DOI] [PubMed] [Google Scholar]
- 14.Rutz S, Mordmuller B, Sakano S, Scheffold A. Notch ligands Delta-like1, Delta-like4 and Jagged1 differentially regulate activation of peripheral T helper cells. Eur J Immunol. 2005;35:2443–2451. doi: 10.1002/eji.200526294. [DOI] [PubMed] [Google Scholar]
- 15.Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. Notch Directly Regulates Gata3 Expression during T Helper 2 Cell Differentiation. Immunity. 2007;27:100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun J, Krawczyk CJ, Pearce EJ. Suppression of Th2 cell development by Notch ligands Delta1 and Delta4. J Immunol. 2008;180:1655–1661. doi: 10.4049/jimmunol.180.3.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaller MA, Neupane R, Rudd BD, Kunkel SL, Kallal LE, Lincoln P, Lowe JB, Man Y, Lukacs NW. Notch ligand Delta-like 4 regulates disease pathogenesis during respiratory viral infections by modulating Th2 cytokines. J Exp Med. 2007;204:2925–2934. doi: 10.1084/jem.20070661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skokos D, Nussenzweig MC. CD8-DCs induce IL-12-independent Th1 differentiation through Delta 4 Notch-like ligand in response to bacterial LPS. J Exp Med. 2007;204:1525–1531. doi: 10.1084/jem.20062305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ostroukhova M, Qi Z, Oriss TB, Dixon-McCarthy B, Ray P, Ray A. Treg-mediated immunosuppression involves activation of the Notch-HES1 axis by membrane-bound TGF-beta. J Clin Invest. 2006;116:996–1004. doi: 10.1172/JCI26490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677–688. doi: 10.1016/j.immuni.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat Rev Immunol. 2008;8:337–348. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- 22.Ivanov B, II, McKenzie S, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 23.Rudd BD, Luker GD, Luker KE, Peebles RS, Lukacs NW. Type I interferon regulates respiratory virus infected dendritic cell maturation and cytokine production. Viral Immunol. 2007;20:531–540. doi: 10.1089/vim.2007.0057. [DOI] [PubMed] [Google Scholar]
- 24.Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–5972. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- 25.Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;9:1297–1306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishihara M, Ogura H, Ueda N, Tsuruoka M, Kitabayashi C, Tsuji F, Aono H, Ishihara K, Huseby E, Betz UA, Murakami M, Hirano T. IL-6-gp130-STAT3 in T cells directs the development of IL-17+ Th with a minimum effect on that of Treg in the steady state. Int Immunol. 2007;19:695–702. doi: 10.1093/intimm/dxm045. [DOI] [PubMed] [Google Scholar]
- 27.Kimura A, Naka T, Kishimoto T. IL-6-dependent and -independent pathways in the development of interleukin 17-producing T helper cells. Proc Natl Acad Sci U S A. 2007;104:12099–12104. doi: 10.1073/pnas.0705268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tacchini-Cottier F, Allenbach C, Otten LA, Radtke F. Notch1 expression on T cells is not required for CD4+ T helper differentiation. Eur J Immunol. 2004;34:1588–1596. doi: 10.1002/eji.200324337. [DOI] [PubMed] [Google Scholar]
- 29.Ong CT, Sedy JR, Murphy KM, Kopan R. Notch and presenilin regulate cellular expansion and cytokine secretion but cannot instruct Th1/Th2 fate acquisition. PLoS ONE. 2008;3:e2823. doi: 10.1371/journal.pone.0002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tilley SL, Jaradat M, Stapleton C, Dixon D, Hua X, Erikson CJ, McCaskill JG, Chason KD, Liao G, Jania L, Koller BH, Jetten AM. Retinoid-related orphan receptor gamma controls immunoglobulin production and Th1/Th2 cytokine balance in the adaptive immune response to allergen. J Immunol. 2007;178:3208–3218. doi: 10.4049/jimmunol.178.5.3208. [DOI] [PubMed] [Google Scholar]
- 31.Jurynczyk M, Jurewicz A, Raine CS, Selmaj K. Notch3 inhibition in myelin-reactive T cells down-regulates protein kinase C theta and attenuates experimental autoimmune encephalomyelitis. J Immunol. 2008;180:2634–2640. doi: 10.4049/jimmunol.180.4.2634. [DOI] [PubMed] [Google Scholar]
- 32.Elyaman W, Bradshaw EM, Wang Y, Oukka M, Kivisakk P, Chiba S, Yagita H, Khoury SJ. JAGGED1 and delta1 differentially regulate the outcome of experimental autoimmune encephalomyelitis. J Immunol. 2007;179:5990–5998. doi: 10.4049/jimmunol.179.9.5990. [DOI] [PubMed] [Google Scholar]