Abstract
The ARKdb genome databases provide comprehensive public repositories for genome mapping data from farmed species and other animals (http://www.thearkdb.org) providing a resource similar in function to that offered by GDB or MGD for human or mouse genome mapping data, respectively. Because we have attempted to build a generic mapping database, the system has wide utility, particularly for those species for which development of a specific resource would be prohibitive. The ARKdb genome database model has been implemented for 10 species to date. These are pig, chicken, sheep, cattle, horse, deer, tilapia, cat, turkey and salmon. Access to the ARKdb databases is effected via the World Wide Web using the ARKdb browser and Anubis map viewer. The information stored includes details of loci, maps, experimental methods and the source references. Links to other information sources such as PubMed and EMBL/GenBank are provided. Responsibility for data entry and curation is shared amongst scientists active in genome research in the species of interest. Mirror sites in the United States are maintained in addition to the central genome server at Roslin.
INTRODUCTION
Scientists engaged in genome mapping research need access to contemporary summaries of maps and other genome-related data. Genome databases fulfil this requirement and provide mappers with an overview of genome analysis in the species of interest.
The first genome database for a farm animal species was PiGBASE. This was implemented at Roslin Institute by replicating the then current mouse genome database design (GBASE, developed at The Jackson Laboratory) and populating it with pig data. The GBASE model was also cloned and populated with sheep (1) and chicken data to implement genome databases for those species. However, the GBASE model was designed primarily to handle linkage (meta-) data from experiments using inbred lines of mice. We needed richer means of capturing the complexity of experimental data and ways of describing new mapping techniques common within the farmed animal mapping communities but not considered when GBASE was developed. In addition, the original GBASE model was accessed online using a command-line driven forms interface. We wanted to provide a simpler means of disseminating the data to a geographically widespread audience.
In order to address these requirements the Roslin bioinformatics group developed a new genome database model (ARKdb) to handle animal genome mapping data along with tools for data entry and display.
ARKdb DATABASES AND TOOLS
ARKdb uses a relational database management system
The ARKdb genome databases use a commercial relational database management system (RDBMS—INGRES) as the data store. The database schema does not rely on any proprietary extensions and could in theory be implemented in any RDBMS supporting standard SQL.
Web strategy
The users of the ARKdb genome databases are situated around the world and use a variety of operating systems. Thus, from an early stage we adopted a fully Web-operable strategy for all database interface functions: retrieving, submitting and editing data.
Data retrieval
Data are retrieved, either in graphical form through the Anubis genome viewer, or with the WebinTool-based ARKdb browser. WebinTool (2) is a generic tool for building interfaces to relational databases via the Web. Developed entirely in-house, it provides a scripting language to interrogate and/or manipulate a relational database, handling CGI form variables automatically and generating HTML code based on the results of the queries. WebinTool has also been adopted for use in commercial and academic projects not necessarily related to biology and has proved popular amongst the INGRES database community. We have used it to develop our own front end for the textual parts of the database.
The Anubis map viewer was the first genome browser to be fully operable as a fully-fledged GUI (Graphical User Interface) over the WWW. It is currently used as the map viewer for ARKdb genome databases and the INRA BOVMAP database (3–5) (URL http://www.roslin.ac.uk/anubis). The current production release of Anubis (version 2.7) is written in C as a CGI executable generating an image file embedded in HTML. Interactive graphic controls are embedded in the images transferred, and state is stored on the server. Anubis communicates with remote databases using HTTP, connecting with a WebinTool database wrapper that handles Anubis queries. Anubis 2.7 has an (optional) layer of Java to add interactive features impossible under a pure CGI approach. We have recently implemented a prototype replacement version of Anubis (Anubis 4) written entirely in Java.
Data submission and editing
The ARKdb editorial tools also use forms-based CGI. As editing requires a certain degree of cross-referencing of data, a standard dynamic HTML forms approach is quite restrictive. To circumvent this problem, the editorial tools use a combination of WebinTool, JavaScript and frames to provide a flexible multi-window system. Only authorised database editors can access the editorial tools. A parallel submission system is written entirely in Perl, as this is more suitable than WebinTool for the large amount of data pre-processing required. The submission system keeps submitted data in batched form until an editor approves the data—then it is passed into the database. The ARKdb system comes with a number of supporting applications, all developed in-house, for checking data consistency, migrating data from other data sources (databases, spreadsheets, etc.), generating reports, automated bulk loading and mirroring databases.
Data curation and updating
Each species database has an editor or editors. Where funding is available curators have also been appointed. The current editors and curators for each of the species served by ARKdb are listed in Table 1. It is the role of the curator (or the editor in the absence of a curator for a particular species) to identify papers relevant to the species of interest as they are published and to enter the relevant details of the publication and the results it contains into the database. The extraction of data from, and the attribution of data to, specific published papers most of which have been subjected to peer review acts as the basic quality control on the data content of the databases. The editors determine policy issues such as nomenclature rules for loci/markers. In general the nomenclature is chosen to conform as closely to human locus symbol nomenclature as possible, although each species is free to adopt its own rules. The editors are recognised experts in genome research relevant to the species of interest. Annotation fields provide the editors with opportunities to comment on specific data.
Table 1. ARKdb genome database editors and curators.
| Database |
Editors |
Curators |
| PiGBASE (ARKdb-pig) | Dr Alan Archibald, Roslin Institute, UK | Mrs Irene Black, Mrs Shirley Leckie, Roslin Institute, UK |
| Dr Max Rothschild, Iowa State University, USA | Yuandan Zhang, Iowa State University, USA | |
| ChickGBASE (ARKdb-chicken) | Dr Dave Burt, Roslin Institute, UK | Mrs Irene Black, Mrs Shirley Leckie, Roslin Institute, UK |
| Dr Hans Cheng, USDA Michigan, USA | ||
| SheepBASE (ARKdb-sheep) | Dr Tom Broad, AgResearch, New Zealand | |
| Dr Noelle Cockett, Utah State University, USA | ||
| Dr Frank Nicholas, University of Sydney, Australia | ||
| BovGBASE (ARKdb-cattle) | Dr Jim Womack, Texas A&M University, USA | Dr Srinivas Kata, TexasA &M University, USA |
| ARKdb-horse | Dr Ernest Bailey, University of Kentucky, USA | Mrs Irene Black, Mrs Shirley Leckie, Roslin Institute, UK |
| Dr Matthew Binns, Animal Health Trust, UK | ||
| ARKdb-tilapia | Dr Thomas Kocher, University of New Hampshire, USA | |
| ARKdb-cat | TBA | Mrs Irene Black, Mrs Shirley Leckie, Roslin Institute, UK |
| ARKdb-turkey | Dr Dave Burt, Roslin Institute, UK | Mrs Irene Black, Mrs Shirley Leckie, Roslin Institute, UK |
| ARKdb-salmon | TBA | Mrs Irene Black, Mrs Shirley Leckie, Roslin Institute, UK |
TBA, to be announced.
New data are initially entered into an editorial copy of the database distinct from the copy viewable through the public web interface. Each week, the editorial copy containing the new data are packaged and collected into a central repository at Roslin. The mirror sites then extract the new copy of the data and install it into the publicly viewable database.
The data
The database consolidates all genome data for a particular species in a central resource. These data include locus/marker data, references/papers, authors, genetic (linkage) map assignments, cytogenetic assignments, experiments, experimental techniques and results, PCR primer information, PCR conditions, hybridisation conditions and enzyme information. We also have the structures for radiation hybrid maps, somatic cell hybrid maps and all supporting data (chromosome content of hybrid probes, locus hybridisations and concordance/correlation summaries). Full cross-referencing of the data allows the user to trace data from the maps, through experiments back to the original papers and primary data sources. Hot links are provided to other data sources such as the EMBL/GenBank databases, SWISS-PROT and MEDLINE (PubMed).
There are four key data types in ARKdb: references, loci, experiments and maps (Table 2).
Table 2. ARKdb genome databases: contentsa.
| Database |
Number of references |
Number of
loci |
Number of experiments |
| ARKdb-pig | 906 | 2094 | 3075 |
| ARKdb-chicken | 443 | 2345 | 2264 |
| ARKdb-sheep | 503 | 1464 | 2873 |
| ARKdb-cattle | 437 | 2542 | 5790 |
| ARKdb-horse | 158 | 683 | 882 |
| ARKdb-deer | 161 | 231 | 416 |
| ARKdb-tilapia | 241 | 243 | 135 |
| ARKdb-salmon | 70 | 156 | 226 |
| ARKdb-cat | 56 | 150 | 144 |
| ARKdb-turkey | 16 | 101 | 138 |
aSummary contents on September 1, 2000.
References
As the stored information has been extracted from the published literature, each observation is attributed to a reference or source. Details of the publication (title, authors, journal or other source) and a brief abstract are stored and links are maintained to MEDLINE (PubMed).
Loci
Since the ARKdb genome databases have been developed to support genome mapping projects loci are key objects. Unfortunately, in the current implementation the definition or resolution of loci is limited. Thus, for example, multiple polymorphic nucleotide or restriction enzyme recognition sites within a single transcription unit or gene are regarded as a single locus. Similarly, a locus revealed by in situ hybridisation with a cloned coding sequence is treated as synonymous with a polymorphic site within the same gene. In the original design, no provision was made directly for locus accession numbers. However, each locus has a unique identifier (uid) within the database. Although loci/markers are retrieved solely by symbol, a search with an old locus symbol will retrieve the correct locus complete with its currently accepted symbol and any aliases. Thus data are never ‘lost’ to a researcher when a locus symbol changes. The locus symbol or name defined in the original paper is recorded initially, but this may subsequently be changed to conform to the species nomenclature rules. As the resolution of mapping in the target species improves we will need to refine the locus aspect of the ARKdb model.
Experiments
ARKdb records details of experimental methods and results. For example, for a fluorescence in situ hybridisation (FISH) experiment details of the method: probe, hybridisation conditions and banding treatment of the chromosomes may be stored, depending on the available information. For probes, details of the clone from which it was developed and the DNA library from which the clone itself was developed may be recorded. The details of the results of a FISH experiment may include the chromosomal localisation (chromosome arm, bands, FLPter values). For some experiments there is only a method, e.g. a description of a polymorphic marker. For others, only the results are stored, e.g. for linkage data (two point linkage results and linkage maps).
Maps
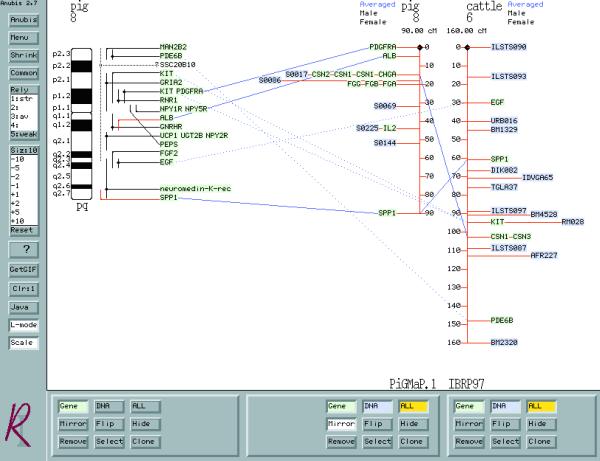
Cytogenetic, linkage and radiation hybrid maps are stored. Multiple maps of the same chromosome or of different chromosomes in different species can be displayed and compared using the Anubis map viewer (Fig. 1). Anubis generates the map displays from the latest data stored in the database(s), a capability most readily revealed with the cytogenetic maps where the map objects are mapped relative to common chromosomal landmarks—chromosome bands. Because the maps are generated ‘on-the-fly’, as new loci are mapped, the data are instantly available for viewing. Where loci are mapped in separate experiments to different positions, the data are handled in one of two ways. Wherever possible, an editorial decision will be made as to which of the locations is the more reliable. If this distinction can be made then the most reliable position is noted as such within the database and this datum will be used when drawing map images. If no distinction can be made, the map displayed will flag the locus symbol with a question mark. Since each locus symbol tag on the map is a ‘hot link’ to the locus details, the disparate mapping data can be readily retrieved. For the linkage and radiation hybrid maps where the loci or markers are mapped in relation to one another a particular published linkage map will not change but rather may be replaced with a new map that includes the additional data. In this case, the differentiation between conflicting map locations is not necessary. The older linkage maps are stored and can be compared with the newer maps using Anubis.
Figure 1.
Comparison of (from L to R) a cytogenetic map of porcine chromosome 8, a linkage map of porcine chromosome 8 and a linkage map of bovine chromosome 6. The ‘gene’ only option has been selected for the cytogenetic map in order to reduce the complexity of the map. The ‘mirror’ option has been selected for the central map in order to aid comparison of the two linkage maps.
IMPLEMENTATION
The ARKdb generic single species database model has been implemented for genome mapping data from 10 species. These are pig, chicken, sheep, cattle, horse, deer, turkey, cat, salmon and tilapia (Table 3). The full cluster of ARKdb databases are mounted on the genome server at the Roslin Institute (http://www.thearkdb.org) with subsets mounted at Texas A&M University and at Iowa State University in the USA.
Table 3. ARKdb genome databases: descriptions and URLsa.
| Database name |
Description |
URL(s) |
| ARKdb | Genome databases—contains interactive map displays, experimental details, references | http://www.roslin.ac.uk/bioinformatics/databases.html http://www.roslin.ac.uk/arkdb/about_ARK.html |
| ARKdb-pig(PiGBASE) | Pig genome database, ARKdb series, contains interactive map displays and experimental details, links to EMBL, SWISS-PROT, PubMed and references | UK (primary) node: http://www.roslin.ac.uk/bioinformatics/databases.html USA node: http://www.genome.iastate.edu/cgi-bin/arkdb/browsers/browser.sh?species=pig |
| ARKdb-sheep(SheepBASE) | Sheep genome database, ARKdb series, contains interactive map displays and experimental details, links to EMBL, SWISS-PROT, PubMed and references | UK (primary) node: http://www.roslin.ac.uk/bioinformatics/databases.html USA node: http://bos.cvm.tamu.edu/cgi-bin/arkdb/browsers/browser.sh?species=sheep |
| ARKdb-chicken(ChickBASE) | Chicken genome database, ARKdb series, contains interactive map displays and experimental details, links to EMBL, SWISS-PROT, PubMed and references | UK (primary) node: http://www.roslin.ac.uk/bioinformatics/databases.html USA node: http://www.genome.iastate.edu/cgi-bin/arkdb/browsers/browser.sh?species=chicken |
| ARKdb-cattle(BovGBASE) | Cattle genome database, ARKdb series, contains interactive map displays and experimental details, links to EMBL, SWISS-PROT, PubMed and references | USA (primary) node: http://bos.cvm.tamu.edu/cgi-bin/arkdb/browsers/browser.sh?species=cattle UK node: http://www.roslin.ac.uk/genome_mapping.html |
| ARKdb-tilapia | Tilapia genome database, ARKdb series, contains interactive map displays and experimental details, | UK: http://www.roslin.ac.uk/genome_mapping.html |
| ARKdb-horse | Horse genome database, ARKdb series, contains interactive map displays and experimental details, links to EMBL, SWISS-PROT, PubMed and references | UK (primary) node: http://www.roslin.ac.uk/genome_mapping.html USA node: http://bos.cvm.tamu.edu/cgi-bin/arkdb/browsers/browser.sh?species=horse |
| ARKdb-turkey | Turkey genome database, ARKdb series, contains experimental details, links to EMBL, SWISS-PROT, PubMed and references; no maps as yet | UK: http://www.roslin.ac.uk/genome_mapping.html |
| ARKdb-deer | Deer genome database, ARKdb series, contains interactive map displays and experimental details, links to EMBL, SWISS-PROT, PubMed and references | UK: http://www.roslin.ac.uk/genome_mapping.html |
| ARKdb-cat | Cat genome database, ARKdb series, contains experimental details, links to EMBL, SWISS-PROT, PubMed and references; no maps as yet | UK: http://www.roslin.ac.uk/genome_mapping.html |
| ARKdb-salmon | Salmonids genome database, ARKdb series, contains experimental details, links to EMBL, SWISS-PROT, PubMed and references; no maps as yet | UK: http://www.roslin.ac.uk/genome_mapping.html |
aThe URLs cited are correct at October 15, 2000. We are currently simplifying the Web addressing for these databases. Under the new scheme the URLs will be http://roslin.thearkdb.org; http://iowa.thearkdb.org and http://texas.thearkdb.org for access to each ARKdb site. For direct access to a specific database the URLs will take the form http://roslin.thearkdb.org/common_name_for_species e.g. http://roslin.thearkdb.org/pig.
The International Society for Animal Genetics (ISAG), which acts as the scientific body ‘governing’ genome mapping in farm animals, currently recognises the ARKdb databases as the primary genome databases for pigs, chickens and sheep. This recognition is facilitated by the sharing of the editorial responsibilities on an international basis and the commitment to maintaining multiple sites (see Tables 1 and 3).
The ARKdb genome databases attempt to provide broadly the same functionality as the Genome Database (GDB, human data) (6) and the Mouse Genome Database (MGD) (7) for their respective animal species. We have also developed resource databases and tools to handle data from QTL and linkage mapping experiment (resSpecies http://www.roslin.ac.uk/bioinformatics/databases.html) and for radiation hybrid mapping using the Cambridge–Roslin pig and cattle radiation hybrid mapping panels (http://www.resgen.com/products/RHMAP.php3#relatedproducts and http://www. roslin.ac.uk/radhyb/). Whilst the ARKdb genome databases are freely accessible to all, access to these complementary resources is restricted. However, it is our intention to integrate these restricted access databases with the public ARKdb genome databases thus reducing the lag between map construction and publication.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to our colleagues and collaborators who contribute to the development and maintenance of bioinformatics resources for farm animals. We acknowledge the financial support from the European Commission (1992–1995), the UK Medical Research Council (1994–1995) and currently from the Biotechnology and Biological Sciences Research Council (1996–2003 PAGA and GAIT Bioinformatics grants).
References
- 1.Sise J.A., Hillyard,A.L. and Montgomery,G.W. (1996) The sheep gene map database (SheepBase) is now available on the World Wide Web. Mamm. Genome, 7, 1. [DOI] [PubMed] [Google Scholar]
- 2.Hu J., Nicholson,D., Mungall,C., Hillyard,A.L. and Archibald,A.L. (1996) WebinTool: A generic Web to database interface building tool. Proceedings of the 7th International Conference and Workshop on Database and Expert Systems (DEXA 96), Zurich, September 9–13, 1996. pp. 285–290.
- 3.Mungall,C. and Hu,J. (1997) The Anubis genome viewer: towards a component architecture. Objects in Bioinformatics 97, June 1997. European Bioinformatics Institute.
- 4.Mungall C. (1996) Visualisation tools for genome mapping—the Anubis map manager. XXVth International Conference on Animal Genetics, 21–25 July 1996, Tours, France. Animal Genet., 27, Suppl. 2, 56. [Google Scholar]
- 5.Gas S., Eggen,A., Samson,F., Christophe,C., Mungall,C., Bessieres,P. and Leveziel,H. (1996) The BOVMAP database. XXVth International Conference on Animal Genetics, 21–25 July 1996, Tours, France. Animal Genet., 27, Suppl. 2, 59. [Google Scholar]
- 6.Letovsky S.I., Cottingham,R.W., Porter,C.J. and Li,P.W.D. (1998) GDB: the human genome database. Nucleic Acids Res., 26, 94–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blake J.A., Eppig,J.T., Richardson,J.E., Davisson,M.T. and the Mouse Genome Database Group. (2000) The mouse genome database (MGD): expanding genetic and genomic resources for the laboratory mouse. Nucleic Acids Res., 28, 108–111. Updated article in this issue: Nucleic Acids Res. (2001), 29, 91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]