Abstract
A structural interpretation of the thermodynamic stability of proteins requires an understanding of the structural properties of the unfolded state. High-pressure small-angle x-ray scattering was used to measure the effects of temperature, pressure, denaturants, and stabilizing osmolytes on the radii of gyration of folded and unfolded state ensembles of staphylococcal nuclease. A set of variants with the internal Val-66 replaced with Ala, Tyr, or Arg was used to examine how changes in the volume and polarity of an internal microcavity affect the dimensions of the native state and the pressure sensitivity of the ensemble. The unfolded state ensembles achieved for these proteins with high pressure were more compact than those achieved at high temperature, and were all very sensitive to the presence of urea and glycerol. Substitutions at the hydrophobic core detectably altered the conformation of the protein, even in the folded state. The introduction of a charged residue, such as Arg, inside the hydrophobic interior of a protein could dramatically alter the structural properties, even those of the unfolded state. The data suggest that a charge at an internal position can interfere with the formation of transient hydrophobic clusters in the unfolded state, and ensure that the pressure-unfolded form of a protein occupies the maximum volume possible. Only at high temperatures does the radius of gyration of the unfolded state ensemble approach the value for a statistical random coil.
Introduction
The structural origins of the thermodynamic stability of proteins continue to be of great interest, owing especially to the role of protein unfolding, misfolding, and aggregation in disease (1–4). Partial unfolding of proteins often leads to fibril formation (1–3), which is suspected to be the source of plaque involved in the pathogenesis of several diseases, including Alzheimer's disease and prion diseases (4). Hence, a deeper understanding of how proteins maintain their conformational stability and how that stability is affected by mutations and environmental factors such as temperature, pressure, and cosolvents is needed. Mutations that affect the hydrophobic core are particularly pernicious and may significantly change the stability and perhaps also the conformation of the protein (5–9). In this study we analyzed the effects of single amino acid substitutions and temperature, pressure, and urea or glycerol on conformation and stability. The mutants we studied are variants of the highly stable Δ+PHS form of staphylococcal nuclease (SNase). In these variants, Val-66, which is buried in the main hydrophobic core of the protein, was substituted with Ala, Tyr, and Arg. Structural, thermodynamic, and kinetic aspects of the unfolding of SNase and many of its variants have been studied previously in great detail (5–14). The study presented here is unique (to our knowledge) in that it provides insight into the average size and shape of folded- and unfolded-state ensembles of these proteins under a variety of environmental conditions, and a detailed analysis of the pressure sensitivity of these ensembles. This level of insight is necessary to improve our understanding of the structural basis of stability.
Heat, acid, and chemical denaturants have been used extensively to unfold proteins. However, protein unfolding by high hydrostatic pressures has not received as much attention (15). The use of pressure to unfold proteins is a unique approach because, in contrast to a temperature increase that simultaneously causes changes in both total energy and volume, unfolding induced by high pressure is determined solely by the volume change of the system and is driven predominantly by the disruption of voids and cavities inside the protein (16). For this reason, pressure is an excellent denaturant with which to investigate the effects of amino acid substitutions in the hydrophobic cores of proteins and examine the nature of compact structures of unfolded proteins. A further advantage is that the formation of protein aggregates, which often plagues temperature-induced unfolding, is suppressed at high pressures (15,17).
Small-angle x-ray scattering (SAXS) is a powerful tool for accurately studying changes in the tertiary structure of proteins and the size and shape of proteins (18–21). Studies employing specialized high-pressure cells with flat diamond windows have also used SAXS to study the pressure sensitivity of structural properties of folded and unfolded proteins (22–24). In some ways, pressure is a gentler variable for shifting populations of equilibrium conformational states. Partly for this reason, it is of interest to examine how different structural ensembles of proteins respond to pressure. The results of this study illustrate the utility of high-pressure SAXS as a tool to examine the fundamental structural properties of proteins.
Material and Methods
SAXS measurements
For monodisperse and highly diluted protein solutions, the SAXS signal I(q) is the spherically averaged squared modulus of the Fourier transform of the sample's electron density, (25):
| (1) |
where n denotes the protein number density, P(q) is the protein's form factor, and is the spherical average. The wave vector transfer depends on the scattering angle 2Θ and the wavelength λ of the incident radiation.
For small q, the scattered intensity can be expressed by the so-called Guinier approximation (26):
| (2) |
The radius of gyration, Rg, which describes the characteristic size of a scattering object, is given by (25):
| (3) |
where Dmax is the maximum extension of the protein, and p(r) is the so-called pair-distance distribution function that characterizes the shape and size of the particle. The Guinier approximation is valid for q-values up to about qmax ≤ 1.3/Rg (26).
Sample preparation and experimental setup
The proteins studied were the V66A, V66Y, and V66R variants of the highly stable form of SNase known as Δ+PHS. Δ+PHS differs from wild-type SNase by a set of substitutions (P117G, H124L, S128A, G50F, and V51N) and one deletion (Δ44-49) (12). All proteins were produced and purified as described previously (27).
Protein solutions were prepared just before the measurements were performed. To avoid pressure-induced changes in the pH value, bis-Tris buffer (50 mM) was used at a pH value of 5.5 (28). All buffers were prepared by adding the appropriate amount of buffer salt to deionized water. The pH value was adjusted by HCl. Cosolvent solutions were prepared by adding the adequate amount of glycerol (2.5 M) and urea (1.5 M, 2.5 M) to the stock solution. The protein concentrations of the solutions were 10.0 mg/mL. At these concentrations, the single scattering approximation is still valid (29).
The SAXS experiments were performed at beamline ID02 of the European Synchrotron Radiation Facility (Grenoble, France) (30) using a photon energy of 12.5 keV (λ = 0.995 Å). To cover a large q range, measurements were carried out at two different sample-to-detector distances. At a distance of 905 mm, a q range from 0.3 to 4.5 nm−1 can be recorded that allows one to monitor the form factor P(q) of the proteins. A distance of 3025 mm covers the Guinier regime within the q range of 0.1–1.5 nm−1 with high resolution. The protein solutions and the corresponding buffer solutions were exposed to the x-ray beam for 0.1 s, with a FReLoN CCD camera used for signal detection. To investigate the protein solutions under high pressure, a special sample cell with two flat diamond windows was used (23). For each protein solution, the pressure was increased in 1 kbar steps. After each pressure increase, an equilibration time of 20 min was allowed to ensure that all relaxation processes were completed. All pressure-dependent measurements were performed at T = 25°C. A pressure range of 1 bar to 4 kbar was covered and all pressure-dependent measurements were found to be fully reversible. To reduce radiation damage to the protein, the sample cell was moved perpendicular to the beam path each time before x-ray exposure. It was recently shown that this procedure reduces the radiation-induced aggregation of proteins (31). To determine the temperature effect on the structural properties of the three mutants at ambient pressure, additional measurements were carried out at three different temperatures (25°C, 34°C, and 64°C).
The two-dimensional scattering patterns were azimuthally averaged using the Fit2D software package (32). The resulting SAXS signals were normalized and corrected for solvent and background scattering by subtracting the scattering curve of the corresponding pure buffer solution. To obtain the Rg, the data in the low-q region that showed a linear curve shape in the q2 versus log(I(q)) representation were fitted according to Guinier's approximation (Eq. 2). The pair-distance distribution functions were obtained by fitting the scattering data using the program GNOM (33); p(r) was also used to obtain the Rg (Eq. 3). Ab initio models of the proteins were also calculated. For each structure, 16 independent calculations were started using the program DAMMIF (34). The resulting 16 ab initio structures were aligned by using the program DAMAVER (35) to build a model structure for each protein.
Results and Discussion
Measurements at atmospheric pressure
The scattering signals of Δ+PHS/V66A, Δ+PHS/V66Y, and Δ+PHS/V66R were measured initially at atmospheric pressure and a temperature of 34°C (Fig. 1). Owing to the low scattering signal at higher q-values, the background subtraction results in slightly different accessible q ranges. The inset in Fig. 1 shows the pair-distance distribution function of the three proteins. The maximum dimension Dmax of Δ+PHS/V66Y is slightly larger than that of Δ+PHS/V66A (see also Table 1). The Rg-values of these two proteins obtained by the fit confirm this result. In contrast, Δ+PHS/V66R appears to be denatured at this temperature; this is obvious from the large Rg and Dmax. Of note, V66A and V66Y differ in Rg by ∼1 Å, and whereas the volume of the Tyr buried inside a protein was found to be 197.1 Å3, the volume of Ala is only 90.1 Å3 (36). The side chains of both residues are in a microcavity inside the protein (37). Thus, it appears that because of Tyr's larger volume, insertion of this residue into the cavity leads to a widening of the cavity and consequently to a small but still well detectable increase of the volume of the folded protein.
Figure 1.
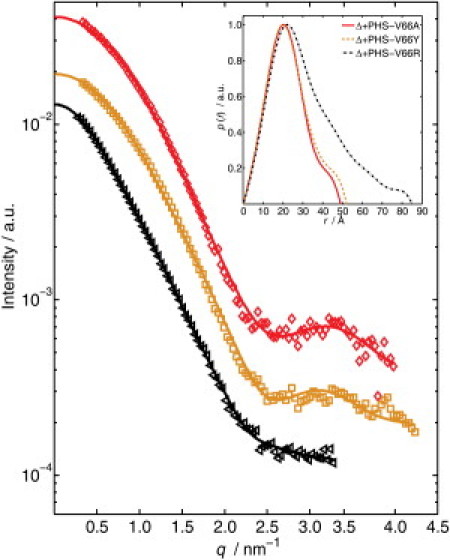
X-ray scattering signal of Δ+PHS/V66A (◊), Δ+PHS/V66Y (□), and Δ+PHS/V66R (▵) at ambient pressure and T = 34°C. Also shown are curves fitted using GNOM (28). Inset: the corresponding pair-distance distribution functions.
Table 1.
Maximum dimension, Dmax, and radii of gyration, Rg, of the three SNase mutants at ambient pressure and T = 34°C
| Variant | Dmax / Å | Rg / Å |
|---|---|---|
| Δ+PHS/V66A | 49 ± 0.5 | 16.8 ± 0.4 |
| Δ+PHS/V66Y | 52 ± 0.5 | 17.7 ± 0.3 |
| Δ+PHS/V66R | 85 ± 1.0 | 24.2 ± 0.5 |
Rg-values were calculated using p(r).
The large Dmax- and Rg-values of Δ+PHS/V66R point to the fact that this variant is already partially unfolded at 34°C. In contrast, at a temperature of 25°C, where the protein is still folded, the Rg of this variant is 18.9 ± 0.5 Å (see below), which is still larger than the Rg of Δ+PHS/V66A and of Δ+PHS/V66Y. This behavior is caused by the Arg at position 66, which has a large and ionizable side chain. In contrast to Lys-66, Asp-66, and Glu-66, which titrated with highly shifted pKa-values (5,6,38,39), the pKa of Arg-66 appears to be quite normal (40). This probably reflects some form of dislocation of the side chain from the hydrophobic core that allows the guanidinium moiety to make contact with bulk water. The increase of Rg by ∼6–7 Å relative to the V66A and V66Y variants may be attributed to the structural reorganization that is expected to occur upon relaxation of the structure in the local neighborhood of Arg-66. Crystallographic studies and computer simulations showing that the first β-strand of the β-barrel is disrupted in the V66R variant are consistent with the large Rg measured by SAXS for this variant (40,41).
The resulting models of the ab initio calculations obtained from SAXS measurements are shown in Fig. 2 together with the ribbon representation of the crystallographic structure of Δ+PHS (42). The structures for the Δ+PHS/V66A and Δ+PHS/V66Y variants have a largely globular body with a smaller cone-shaped feature on top. A comparison of these two models with the crystallographic structure of Δ+PHS suggests that this feature could be due to the disordered C-terminal residues 141–149. The model for Δ+PHS/V66R obtained by the ab initio calculations is much more extended, reflecting that this variant is largely unfolded. Of interest, the main differences between the model of Δ+PHS/V66R and those of the other variants are in the larger part where the mutation should be present.
Figure 2.
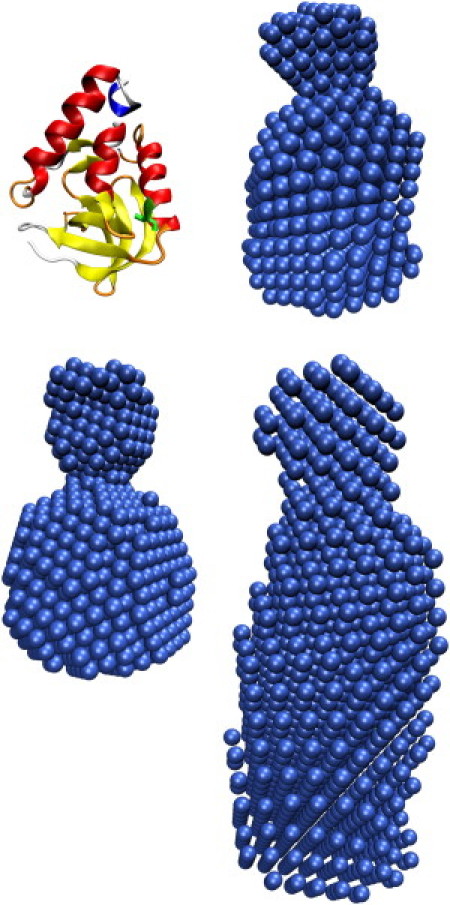
Ribbon representation of the crystal structure of Δ+PHS (3BDC (34)) compared with the models obtained by the ab initio calculations using the SAXS data of the proteins in buffer solution (pH 5.5) at atmospheric pressure and T = 34°C. Top right: Δ+PHS/V66A. Bottom left: Δ+PHS/V66Y. Bottom right: Δ+PHS/V66R.
Measurements at high pressures
The effects of pressure on the scattering intensities of Δ+PHS/V66R at 25°C are shown in the form of a Guinier plot for different pressures in Fig. 3. The decrease in the scattering contrast due to the different compressibilities of the protein and the surrounding water leads to a decrease of the scattering signal with increasing pressure. Thus, the signal/noise ratio at high pressures is lower than the ambient pressure. A significant increase in the slope of the scattering curve with increasing pressure is observed (Fig. 3). This indicates that the protein expands and partially unfolds at high pressures. To obtain the Rg of all three proteins at different pressures, the scattering curves are fitted using the Guinier approximation (see Eq. 2) in the low-q range (solid lines in Fig. 3). The Rg-value measured as a function of pressure for all three SNase variants measured at T = 25°C shows some interesting trends (Fig. 4 a). In the case of Δ+PHS/V66Y, no significant change in Rg is observed upon compression within the experimental error. Hence, this protein is stable up to 4 kbar. For the Δ+PHS/V66A variant, a weak increase in the Rg (by ∼2–3 Å) is observed upon pressurization up to 4 kbar. This effect might be attributed to an expansion, perhaps into a molten globule-like state, or partial unfolding of the protein. In contrast, a drastic increase of Rg to 26.0 ± 0.7 Å is observed for Δ+PHS/V66R at 1 kbar, reflecting a marked unfolding of the SNase mutant. The increase of pressure up to 2 kbar results in a further expansion of the protein, leading to an Rg-value of 31.7 ± 1.2 Å. The transition is 95% complete by 2 kbar (12). Once this value of Rg has been reached, it does not change within the experimental error. Thus, the unfolding process of this mutant induced by high pressure is completed at ∼2 kbar. The pressure-denatured state, however, is not completely unfolded, as the Rg of random-coil-like SNase should be 37.2 ± 1.2 Å (20).
Figure 3.
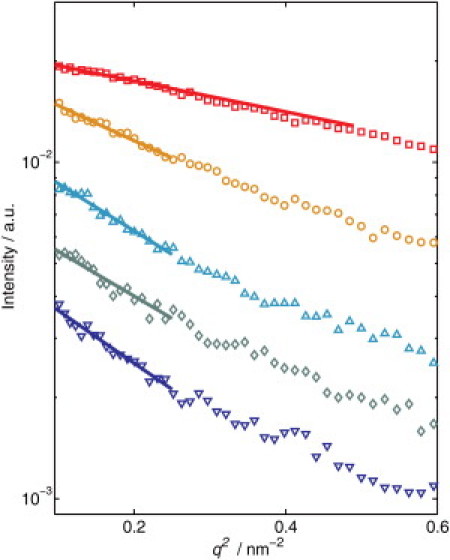
Guinier plots of Δ+PHS/V66R at 1 bar (□), 1 kbar (○), 2 kbar (Δ), 3 kbar (◊), and 4 kbar (∇) at T = 25°C. Also shown are the fits using Eq. 2. For clarity, the curves were shifted by a factor of 1, 1.3, 1, 0.8, and 0.5, respectively.
Figure 4.
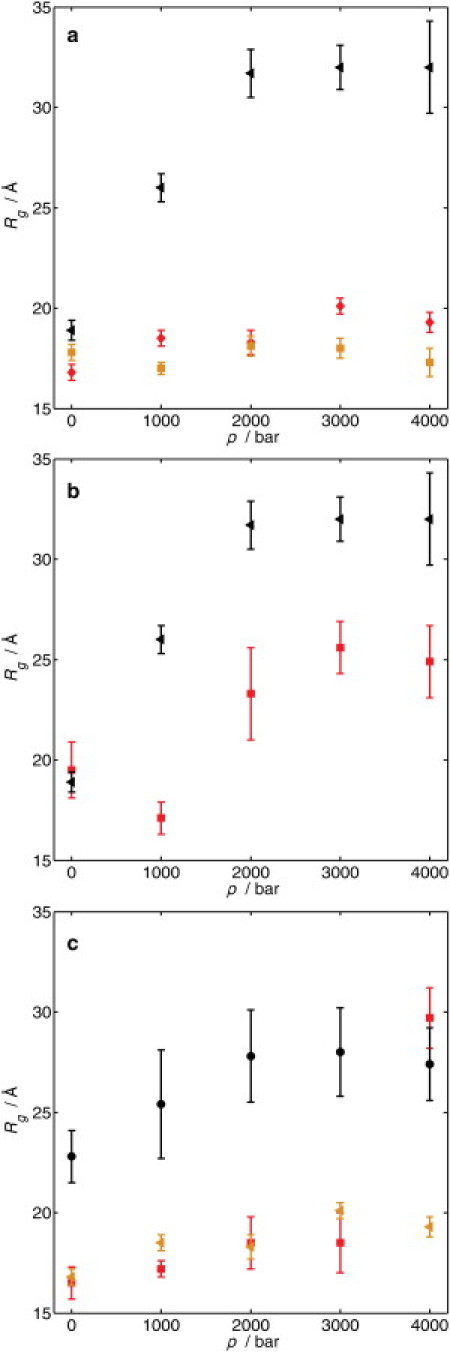
(a) Rg as a function of pressure p at 25°C for the SNase mutants Δ+PHS/V66Y (◃), Δ+PHS/V66A (◊), and Δ+PHS/V66R (◂). (b) Pressure-dependent Rg of Δ+PHS/V66R in pure buffer (◂) and with 2.5 M glycerol (◃). (c) Pressure-dependent Rg of Δ+PHS/V66A in pure buffer (◂), with 1.5 M urea (◃) and with 2.5 M urea (●). Due to the low signal/noise ratio of the scattering signal of the unfolded protein, the error is larger at this urea concentration than for the previous measurements.
The differences in the response to pressure by the three proteins partly reflect the internal packing defects and cavities of the protein. In the case of Δ+PHS/V66Y, the internal cavity in the hydrophobic core is completely filled because Tyr is a large side chain. Thus, high pressure did not lead to significant structural changes. The small increase of Rg in the case of Δ+PHS/V66A suggests that its interior is less densely packed; therefore, increasing pressure led to a minor conformational change. The structural changes of Δ+PHS/V66R in the β-barrel region upon solvent exposure of the charged residue led to a drastic weakening of the protein's conformational stability, which resulted in a further marked increase of Rg with rising pressure. The high-pressure SAXS data are consistent with the presence of a charged residue inside the hydrophobic core drastically reducing the protein's stability (8,9,14). Even more significant is the observation that the Rg-value of the pressure-unfolded Δ+PHS/V66R in water approaches the appropriate value for a statistical random coil. This probably reflects the presence of the charged Arg residue, which must interfere with the formation of transient hydrophobic clusters in the unfolded state, and ensures that the pressure-unfolded form of the protein occupies a larger volume that it would in the absence of the Arg residue.
High-pressure measurements in the presence of cosolvents
It is well known that the stability of proteins is significantly affected by osmolytes (43). SAXS measurements were performed with Δ+PHS/V66A and Δ+PHS/V66R in the presence of glycerol and urea to examine how these additives affected the Rg of folded and unfolded forms of SNase. Glycerol is known to stabilize proteins due to the preferential hydration effect, whereas urea binds preferentially to the backbone of proteins and therefore destabilizes the native state (43). As shown in the previous section, Δ+PHS/V66R is the most unstable of the SNase variants examined in this study. Thus, the influence of glycerol was investigated with Δ+PHS/V66R only (Fig. 4 b). The effects of urea were studied with the Δ+PHS/V66A protein (Fig. 4 c).
The pressure-dependent Rg-values of Δ+PHS/V66R in pure buffer and in 2.5 M glycerol solution are compared in Fig. 4 b. Within the experimental error, the Rg-values in both solutions were identical at 1 bar. Owing to the stabilizing effects of glycerol, at a pressure of 1 kbar the protein is still folded; in fact, between 1 bar and 1 kbar the folded protein might undergo a slight compaction and achieve an Rg comparable to that of the other two proteins at 1 bar in the absence of any additives. Unfolding of Δ+PHS/V66R sets in between 1 and 2 kbar. At higher pressures, Rg does not increase anymore. It is noteworthy that the unfolded ensemble of Δ+PHS/V66R is significantly more compact in the presence of glycerol, where the Rg is 24.9 ± 1.8 Å and thus is much smaller than the Rg-value of 31.7 Å found in the absence of glycerol.
From these SAXS data, the volume change of unfolding, ΔV, may also be estimated (22,45). For Δ+PHS/V66R, we find ΔV ≈ −60 ± 7 cm3 mol−1, in good agreement with literature data of ΔV ≈ −67 ± 5 cm3 mol−1 (at pH 6) based on fluorescence spectroscopy data (12). For SNase wild-type in pure buffer solution, a similar value of ΔV = −66 ± 6 cm3 mol−1 has been determined (22,45). Within the experimental error, a value of similar magnitude is obtained for Δ+PHS/V66R in 2.5 M glycerol (ΔV ≈ −51 ± 10 cm3 mol−1). In addition to changes in cavity volume upon pressure-induced partial unfolding, ΔV is also determined by the unfolded-state conformations and the solvation properties of the final state. Hence, both decreasing and increasing ΔV-values may be found for this type of kosmotropic osmolyte in the literature (22,45–47).
The pressure dependence of the Rg of Δ+PHS/V66A in different urea concentrations is shown in Fig. 4 c. In 1.5 M urea between 1 bar and 3 kbar, there was detectable change in the Rg of the protein relative to the Rg in water. Unfolding occurs between 3 and 4 kbar, leading to a state with Rg = 29.7 ± 1.5 Å. An increase of the urea concentration up to 2.5 M leads to partial unfolding and denaturation at 1 bar (Rg = 22.8 ± 1.3 Å). With rising pressure, the unfolding continues until 2 kbar, where a plateau value of Rg is reached. Of interest, the high-pressure denatured state at 4 kbar in 2.5 M urea is not more expanded that that at 1.5 M urea, and is much smaller than that of the unfolded protein in a random-coil like state (Rg ≈ 37 Å). This suggests that in the presence of 2.5 M urea, the high-pressure denatured state is still only partially unfolded. In both urea solutions, the unfolded Δ+PHS/V66A is not as extended as Δ+PHS/V66R in pure buffer solution at 2 kbar, indicating that even under enhanced denaturating conditions Δ+PHS/V66A is still more compact and stable than Δ+PHS/V66R. Rather compact or molten-globule structures have also been observed for various other pressure-induced unfolded proteins (48–51).
Temperature-dependent measurements
To examine the unfolded state structure of the three SNase variants, measurements were also performed up to a temperature of 64°C (accurate SAXS measurements significantly above 70°C are very difficult to perform owing to limitations of the sample cell and increased background noise). The midpoints of temperature unfolding were measured with differential scanning calorimetry (DSC) (Table 2) under solution conditions similar to those reported by Ravindra et al. (44). The unfolding temperature of the Δ+PHS reference protein is ∼72°C and the enthalpy change of unfolding, ΔH, is 380 ± 26 kJ/mol at pH 4.5. For the Δ+PHS/V66A protein, the midpoint of unfolding is 66.7°C and the transition is accompanied by an enthalpy change of 316 ± 30 kJ/mol. The Δ+PHS/V66Y variant is slightly more temperature-stable with Tm = 70°C and ΔH = 367 ± 30 kJ/mol. The protein was drastically destabilized upon introduction of the Arg-66 residue. This is reflected in a low Tm-value of ∼38°C and a lower enthalpy change of unfolding (ΔH = 124 ± 30 kJ/mol).
Table 2.
Midpoint of the unfolding temperature, Tm, of the three SNase mutants determined by DSC measurements at various pH values
| Variant | Tm / °C | pH |
|---|---|---|
| Δ+PHS/V66R | 38.0 ± 0.4 | 5.4 |
| 37.5 ± 0.4 | 4.5 | |
| 38.0 ± 0.4 | 7.0 | |
| Δ+PHS/V66A | 66.7 ± 0.1 | 5.5 |
| Δ+PHS/V66Y | 70.0 ± 0.1 | 5.5 |
| Δ+PHS | 72.7 ± 0.1 | 4.5 |
| 72.3 ± 0.1 | 7.0 |
The Rg of Δ+PHS/V66Y at 64°C increased by ∼3 Å relative to Rg at 25°C (Fig. 5). This reflects small structural changes and partial unfolding before the onset of the unfolding transition with a midpoint at 70°C. At 64°C, Δ+PHS/V66A is in the middle of the unfolding transition with Tm = 66.7°C. This is reflected in the increase in Rg up to 27 Å. In agreement with the DSC data, at 34°C, Δ+PHS/V66R is in the middle of the unfolding transition. At this temperature, Rg has increased up to ∼25 Å. A further increase in temperature results in a continuous growth of Rg, finally reaching 37.7 ± 2.6 Å at 64°C, which is similar to the value obtained for a completely unfolded, random coil-like protein of this size (20).
Figure 5.
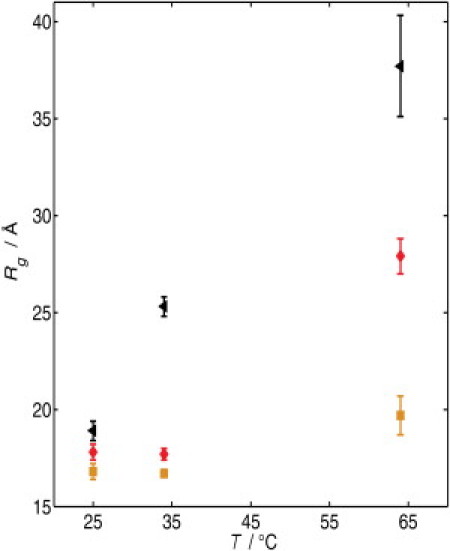
Temperature dependence of the Rg of Δ+PHS/V66Y (□), Δ+PHS/V66A (◊), and Δ+PHS/V66R (◂) in buffer solution (50 mM bis-Tris, pH 5.5) at atmospheric pressure.
Conclusions
The effects of temperature, pressure, denaturants, and protective osmolytes on the Rg of folded and unfolded proteins were examined with the use of V66A, V66Y, and V66R variants of SNase. Glycerol and urea were used to modulate the protein's pressure stability, to effect change in the dimensions of the unfolded ensemble, and to test whether the effect of these osmolytes would also prevail under high-pressure conditions.
When the internal cavity in the hydrophobic core of SNase was occupied with Tyr-66, a small increase in the size of the folded protein relative to the case with Ala-66 was detected at ambient pressure. Presumably, this effect is related to the larger volume of the aromatic side chain of Tyr. An even larger Rg was found with Arg-66, reflecting marked structural changes that were detectable even at ambient temperature and pressure, and probably originated from the reorganization required to accommodate a large and charged side chain in the hydrophobic interior of the protein.
The application of high hydrostatic pressure made it possible to identify differential stabilities for the variants under conditions of constant thermal energy. In the case of the V66Y variant, no changes were detectable in the pressure range studied, whereas the V66A variant showed a slight increase with rising pressure (ΔRg ≈ 3 Å up to 4 kbar). In contrast, the variant with V66R unfolded continuously up to 2 kbar upon pressurization, where Rg-values of 31.7 Å are reached. This protein at high pressure did not adopt a random-coil like state. In contrast, its temperature-unfolded state reached an Rg of ∼37 Å, which is compatible with the random coil. As expected, the addition of glycerol stabilized the V66R variant. The Rg of the unfolded state of this protein in 2.5 M glycerol was smaller by ∼7 Å than its Rg in water.
The effects of urea on the unfolding of the V66A protein in the pressure range covered were clearly detectable. In 2.5 M urea, the protein was partially unfolded at 1 bar and achieved an Rg of 22.8 Å. The unfolding continued with increasing pressure until 2 kbar, where a plateau value of Rg was reached. The high-pressure denatured state above 2 kbar had an Rg of 27 Å, which is considerably smaller than the value of the statistical random coil. This shows that even in the presence of 2.5 M urea, the high-pressure denatured state is still only partially unfolded. At both 1 M and 2.5 M urea, the unfolded state of the V66A variant is not as extended as that of the V66R variant in pure buffer solution at 2 kbar, indicating that even under enhanced denaturing conditions, the V66A variant is still more compact and stable than the variant with V66R. The insertion of the charged Arg-66 in the hydrophobic cluster appears to prevent the interactions responsible for the compactness in the unfolded state of SNase.
Acknowledgments
The authors thank the ID02 beamline group for their help, and the European Synchrotron Radiation Facility machine group for providing the synchrotron radiation.
This work was supported by the Federal Ministry of Education and Research (project 05 KSPE1) and German Research Foundation (project TO169/14-1). R.W. received support from the German Research Foundation and North Rhine-Westphalia. B.G.M.E. received grant MCB-0743422 from the National Science Foundation.
References
- 1.Dobson C.M. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Uversky V.N., Fink A.L. Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochim. Biophys. Acta. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Jahn T.R., Radford S.E. The yin and yang of protein folding. FEBS J. 2005;272:5962–5970. doi: 10.1111/j.1742-4658.2005.05021.x. [DOI] [PubMed] [Google Scholar]
- 4.Selkoe D.J. Folding proteins in fatal ways. Nature. 2003;426:900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- 5.García-Moreno B., Dwyer J.J., Stites W.E. Experimental measurement of the effective dielectric in the hydrophobic core of a protein. Biophys. Chem. 1997;64:211–224. doi: 10.1016/s0301-4622(96)02238-7. [DOI] [PubMed] [Google Scholar]
- 6.Karp D.A., Gittis A.G., García-Moreno E B. High apparent dielectric constant inside a protein reflects structural reorganization coupled to the ionization of an internal Asp. Biophys. J. 2007;92:2041–2053. doi: 10.1529/biophysj.106.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karp D.A., Stahley M.R., García-Moreno B. Conformational consequences of ionization of Lys, Asp, and Glu buried at position 66 in staphylococcal nuclease. Biochemistry. 2010;49:4138–4146. doi: 10.1021/bi902114m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isom D.G., Cannon B.R., García-Moreno B. High tolerance for ionizable residues in the hydrophobic interior of proteins. Proc. Natl. Acad. Sci. USA. 2008;105:17784–17788. doi: 10.1073/pnas.0805113105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isom D.G., Castañeda C.A., García-Moreno E.B. Charges in the hydrophobic interior of a protein. Proc. Natl. Acad. Sci. USA. 2010 doi: 10.1073/pnas.1004213107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panick G., Vidugiris G.J., Royer C.A. Exploring the temperature-pressure phase diagram of staphylococcal nuclease. Biochemistry. 1999;38:4157–4164. doi: 10.1021/bi982608e. [DOI] [PubMed] [Google Scholar]
- 11.Ravindra R., Winter R. On the temperature—pressure free-energy landscape of proteins. ChemPhysChem. 2003;4:359–365. doi: 10.1002/cphc.200390062. [DOI] [PubMed] [Google Scholar]
- 12.Brun L., Isom D.G., Royer C.A. Hydration of the folding transition state ensemble of a protein. Biochemistry. 2006;45:3473–3480. doi: 10.1021/bi052638z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitra L., Hata K., Royer C.A. V(i)-value analysis: a pressure-based method for mapping the folding transition state ensemble of proteins. J. Am. Chem. Soc. 2007;129:14108–14109. doi: 10.1021/ja073576y. [DOI] [PubMed] [Google Scholar]
- 14.Mitra L., Rouget J.B., Winter R. Towards a quantitative understanding of protein hydration and volumetric properties. ChemPhysChem. 2008;9:2715–2721. doi: 10.1002/cphc.200800405. [DOI] [PubMed] [Google Scholar]
- 15.Silva J.L., Foguel D., Royer C.A. Pressure provides new insights into protein folding, dynamics and structure. Trends Biochem. Sci. 2001;26:612–618. doi: 10.1016/s0968-0004(01)01949-1. [DOI] [PubMed] [Google Scholar]
- 16.Royer C.A. Revisiting volume changes in pressure-induced protein unfolding. Biochim. Biophys. Acta. 2002;1595:201–209. doi: 10.1016/s0167-4838(01)00344-2. [DOI] [PubMed] [Google Scholar]
- 17.Perrett S., Zhou J.M. Expanding the pressure technique: insights into protein folding from combined use of pressure and chemical denaturants. Biochim. Biophys. Acta. 2002;1595:210–223. doi: 10.1016/s0167-4838(01)00345-4. [DOI] [PubMed] [Google Scholar]
- 18.Kataoka M., Nishii I., Goto Y. Structural characterization of the molten globule and native states of apomyoglobin by solution X-ray scattering. J. Mol. Biol. 1995;249:215–228. doi: 10.1006/jmbi.1995.0290. [DOI] [PubMed] [Google Scholar]
- 19.Svergun D.I., Koch M.H.J. Small-angle scattering studies of biological macromolecules in solution. Rep. Prog. Phys. 2003;66:1735–1782. [Google Scholar]
- 20.Kohn J.E., Millett I.S., Plaxco K.W. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc. Natl. Acad. Sci. USA. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segel D.J., Bachmann A., Kiefhaber T. Characterization of transient intermediates in lysozyme folding with time-resolved small-angle X-ray scattering. J. Mol. Biol. 1999;288:489–499. doi: 10.1006/jmbi.1999.2703. [DOI] [PubMed] [Google Scholar]
- 22.Panick G., Malessa R., Royer C.A. Structural characterization of the pressure-denatured state and unfolding/refolding kinetics of staphylococcal nuclease by synchrotron small-angle X-ray scattering and Fourier-transform infrared spectroscopy. J. Mol. Biol. 1998;275:389–402. doi: 10.1006/jmbi.1997.1454. [DOI] [PubMed] [Google Scholar]
- 23.Krywka C., Sternemann C., Winter R. Effect of osmolytes on pressure-induced unfolding of proteins: a high-pressure SAXS study. ChemPhysChem. 2008;9:2809–2815. doi: 10.1002/cphc.200800522. [DOI] [PubMed] [Google Scholar]
- 24.Ando N., Barstow B., Gruner S.M. Structural and thermodynamic characterization of T4 lysozyme mutants and the contribution of internal cavities to pressure denaturation. Biochemistry. 2008;47:11097–11109. doi: 10.1021/bi801287m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindner P., Zemb T., editors. Neutrons, X-Rays and Light: Scattering Methods Applied to Soft Condensed Matter. Elsevier; North Holland: 2002. [Google Scholar]
- 26.Guinier A., Fournet G. Wiley; New York: 1955. Small Angle Scattering of X-Rays. [Google Scholar]
- 27.Shortle D., Meeker A.K. Mutant forms of staphylococcal nuclease with altered patterns of guanidine hydrochloride and urea denaturation. Proteins. 1986;1:81–89. doi: 10.1002/prot.340010113. [DOI] [PubMed] [Google Scholar]
- 28.Neumann R.C., Jr., Kauzmann, and W., Zipp A. Pressure-dependence of weak acid ionization in aqueous buffers. J. Phys. Chem. 1973;77:2687–2691. [Google Scholar]
- 29.Javid N., Vogtt K., Winter R. Protein-protein interactions in complex cosolvent solutions. ChemPhysChem. 2007;8:679–689. doi: 10.1002/cphc.200600631. [DOI] [PubMed] [Google Scholar]
- 30.Narayanan T., Diat O., Bösecke P. SAXS and USAXS on the high brilliance beamline at the ESRF. Nucl. Instrum. Methods A. 2001;467:1005–1009. [Google Scholar]
- 31.Hong X., Hao Q. Measurements of accurate x-ray scattering data of protein solutions using small stationary sample cells. Rev. Sci. Instrum. 2009;80:014303. doi: 10.1063/1.3069285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammersley A.P., Svensson S.O., Häusermann D. Two-dimensional detector software: From real detector to idealised image or two-θ scan. High Press. Res. 1996;14:235–248. [Google Scholar]
- 33.Svergun D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- 34.Franke D., Svergun D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volkov V.V., Svergun D.I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harpaz Y., Gerstein M., Chothia C. Volume changes on protein folding. Structure. 1994;2:641–649. doi: 10.1016/s0969-2126(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 37.Schlessman J.L., Abe C., García-Moreno E B. Crystallographic study of hydration of an internal cavity in engineered proteins with buried polar or ionizable groups. Biophys. J. 2008;94:3208–3216. doi: 10.1529/biophysj.107.122473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fitch C.A., Karp D.A., García-Moreno E B. Experimental pK(a) values of buried residues: analysis with continuum methods and role of water penetration. Biophys. J. 2002;82:3289–3304. doi: 10.1016/s0006-3495(02)75670-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dwyer J.J., Gittis A.G., García-Moreno E B. High apparent dielectric constants in the interior of a protein reflect water penetration. Biophys. J. 2000;79:1610–1620. doi: 10.1016/S0006-3495(00)76411-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karp, D. A. 2007. Structural and energetic consequences of the ionization of internal groups in staphylococcal nuclease. Ph.D. dissertation. Johns Hopkins University, Baltimore, MD.
- 41.Damjanović A., Wu X., Brooks B.R. Backbone relaxation coupled to the ionization of internal groups in proteins: a self-guided Langevin dynamics study. Biophys. J. 2008;95:4091–4101. doi: 10.1529/biophysj.108.130906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castañeda C.A., Fitch C.A., García-Moreno B.E. Molecular determinants of the pKa values of Asp and Glu residues in staphylococcal nuclease. Proteins. 2009;77:570–588. doi: 10.1002/prot.22470. [DOI] [PubMed] [Google Scholar]
- 43.Auton M., Bolen D.W. Predicting the energetics of osmolyte-induced protein folding/unfolding. Proc. Natl. Acad. Sci. USA. 2005;102:15065–15068. doi: 10.1073/pnas.0507053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ravindra R., Royer C., Winter R. Pressure perturbation calorimetic studies of the solvation properties and the thermal unfolding of staphylococcal nuclease. Phys. Chem. Chem. Phys. 2004;6:1952–1961. doi: 10.1039/b516608j. [DOI] [PubMed] [Google Scholar]
- 45.Herberhold H., Royer C.A., Winter R. Effects of chaotropic and kosmotropic cosolvents on the pressure-induced unfolding and denaturation of proteins: an FT-IR study on staphylococcal nuclease. Biochemistry. 2004;43:3336–3345. doi: 10.1021/bi036106z. [DOI] [PubMed] [Google Scholar]
- 46.Oliveira A.C., Gaspar L.P., Silva J.L. Arc repressor will not denature under pressure in the absence of water. J. Mol. Biol. 1994;240:184–187. doi: 10.1006/jmbi.1994.1433. [DOI] [PubMed] [Google Scholar]
- 47.Tan C.-Y., Xu C.-H., Ruan K.C. Pressure equilibrium and jump study on unfolding of 23-kDa protein from spinach photosystem II. Biophys. J. 2005;88:1264–1275. doi: 10.1529/biophysj.104.050435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng X., Jonas J., Silva J.L. Molten-globule conformation of Arc repressor monomers determined by high-pressure 1H NMR spectroscopy. Proc. Natl. Acad. Sci. USA. 1993;90:1776–1780. doi: 10.1073/pnas.90.5.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silva J.L., Silveira C.F., Pontes L. Dissociation of a native dimer to a molten globule monomer. Effects of pressure and dilution on the association equilibrium of arc repressor. J. Mol. Biol. 1992;223:545–555. doi: 10.1016/0022-2836(92)90669-b. [DOI] [PubMed] [Google Scholar]
- 50.Kobashigawa Y., Sakurai M., Nitta K. Effect of hydrostatic pressure on unfolding of α-lactalbumin: volumetric equivalence of the molten globule and unfolded state. Protein Sci. 1999;8:2765–2772. doi: 10.1110/ps.8.12.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruan K., Lange R., Balny C. A stable partly denatured state of trypsin induced by high hydrostatic pressure. Biochem. Biophys. Res. Commun. 1997;239:150–154. doi: 10.1006/bbrc.1997.7313. [DOI] [PubMed] [Google Scholar]


